Abstract
Microtubules are dynamic protein polymers that continuously switch between elongation and rapid shrinkage. They have an exceptional bending stiffness that contributes significantly to the mechanical properties of eukaryotic cells. Measurements of the persistence length of microtubules have been published since 10 years but the reported values vary over an order of magnitude without an available explanation. To precisely measure the rigidity of microtubules in their native growing state, we adapted a previously developed bending mode analysis of thermally driven shape fluctuations to the case of an elongating filament that is clamped at one end. Microtubule shapes were quantified using automated image processing, allowing for the characterization of up to five bending modes. When taken together with three other less precise measurements, our rigidity data suggest that fast-growing microtubules are less stiff than slow-growing microtubules. This would imply that care should be taken in interpreting rigidity measurements on stabilized microtubules whose growth history is not known. In addition, time analysis of bending modes showed that higher order modes relax more slowly than expected from simple hydrodynamics, possibly by the effects of internal friction within the microtubule.
INTRODUCTION
Microtubules are long cylindrically shaped protein filaments that are able to give mechanical strength to a living cell (Alberts et al., 2002; Howard, 2001). They are key structural components of cellular structures like cilia and flagella, and can transmit as well as generate forces in, for example, the mitotic spindle. The cylindrical wall of a microtubule has an outer diameter of 25 nm and contains on average 13 protofilaments, i.e., linear arrays of tubulin dimers (Desai and Mitchison, 1997). Assembly takes place at the microtubule tips from tubulin dimers that have guanosine triphosphate (GTP) bound to them. After assembly this GTP is hydrolyzed to guanosine di-phosphate (GDP) and as a result microtubules stochastically undergo catastrophes, i.e., a switch to a state of rapid shrinkage. The cylindrical construction of a microtubule ensures a large resistance against externally applied forces and bending moments. In vitro measurements have shown that the flexural rigidity (κ) of microtubules, which is a measure for this bending resistance (Landau and Lifshitz, 1986), is in the order of 10 pN μm2. Because microtubules contribute so substantially to the mechanical and elastic properties of eukaryotic cells, it is important to be able to measure this quantity precisely. Such quantitative measurements can then be used to study, for example, the effects of the binding of microtubule associated proteins and drugs such as taxol on the mechanical properties of microtubules (Felgner et al., 1997, 1996; Kurz and Williams, 1995; Mickey and Howard, 1995; Venier et al., 1994).
A precise estimate of the rigidity of microtubules is also essential to calibrate forces in experiments were, because of their high rigidity, microtubules are used as a force probe in the piconewton range. For example, elastic deformations of microtubules were analyzed to measure the force that the microtubule-based motor protein kinesin can exert (Gittes et al., 1996), and to measure the force that microtubules can generate themselves by polymerization (Dogterom and Yurke, 1997; Janson and Dogterom, 2004). A microtubule-based force meter was furthermore used to study the strength of biological receptor/ligand pairs (Hess et al., 2002).
Rigidity measurements on microtubules grown in vitro from purified GTP-tubulin range between 4 and 34 pN μm2, corresponding to a persistence length in the range of 1–8 mm (Cassimeris et al., 2001; Dogterom and Yurke, 1997; Felgner et al., 1997, 1996; Fygenson et al., 1997; Kurz and Williams, 1995; Mickey and Howard, 1995; Venier et al., 1994; Janson and Dogterom, 2004). Apart from systematic errors related to the different measurement methods used (see below), there is at present no good explanation for why these numbers vary so wildly. One possibility is that differences in microtubule growth conditions are a cause for the discrepancy in reported values. It has been shown, for example, that lowering the temperature makes microtubules less rigid (Mickey and Howard, 1995). To investigate possible changes due to growth conditions, we developed a technique that allowed us to measure the rigidity of microtubules in their native growing state. Our data suggest that the rigidity of slow-growing microtubules is higher than the rigidity of fast-growing microtubules. Although we cannot provide a clear microscopic explanation for this at the moment, this finding may help to explain the differences in reported values so far. Below, we first review earlier rigidity measurements before describing, in the main text, our measurement technique and the results we obtained.
Measurements of microtubule rigidity
In the past, different experimental methods have been employed to measure the flexural rigidity of microtubules. Distinction can be made between active and passive techniques. In active techniques, the rigidity is derived from an observed shape response to an experimentally controlled force. A homogeneous flow field was used (Kurz and Williams, 1995; Venier et al., 1994) to introduce a drag force that bends the microtubule. The drag coefficient, which determines the force applied, depends on experimental details such as the distance between microtubule and coverslip (Gittes et al., 1993; Hunt et al., 1994; Mickey and Howard, 1995). These details are hard to estimate and thus introduce an unknown systematic error. Other active measurements are based on optical tweezers by which forces can be applied on objects that differ in refractive index from their surrounding liquid (Ashkin, 1997). Tweezers were used directly on the microtubule to bend the filament (Felgner et al., 1997, 1996). The speed of relaxation to the equilibrium shape was measured after the tweezers were taken away. Relaxation is determined by both the flexural rigidity and the drag coefficient and an unknown drag coefficient thus introduces similar systematic errors. Calibrated optical tweezers were further used to exert a known compressive force on a microtubule via a silica bead coupled to the microtubule (Kurachi et al., 1995; Takasone et al., 2002). Considerable bending moments were also exerted on the microtubule in this geometry hindering data interpretation. Another approach to active rigidity measurements makes use of microtubule buckling inside lipid vesicles (Fygenson et al., 1997). Here, forces are applied on the ends of microtubules by controlling the membrane tension of the vesicle. However, controlling the number of microtubules inside the vesicle is hard in this experimental setup. Recently, atomic force microscope has been applied to deform microtubules on a submicron length scale (de Pablo et al., 2003; Kis et al., 2002). These studies provide important information on local molecular interactions between tubulin dimers, but are less suited to measure the flexural rigidity of the complete microtubule, which can be considered to be a “bulk” property.
In contrast to active techniques, passive techniques do not use an externally applied force. As a consequence of thermal motion, the local curvature of a microtubule changes continuously. These thermal forces are well characterized by the laws of statistical physics (Reif, 1965), and shape fluctuations can be used for rigidity measurements (Cassimeris et al., 2001; Gittes et al., 1993; Kurz and Williams, 1995; Mickey and Howard, 1995; Venier et al., 1994). No calibrated forces need to be applied, nor is an exact knowledge of the drag coefficient in liquid required. Gittes et al. (1993) observed shape fluctuations of stabilized (nongrowing) microtubules in a sample-cell that was made so thin that their motion was effectively confined to two dimensions. Observed bending shapes were projected onto Fourier cosine modes. Each mode gave a statistically independent estimate of the flexural rigidity. Estimates from several modes can be averaged to decrease the experimental error. In practice, depending on microtubule length, only one or two modes could be analyzed because amplitudes rapidly decrease for higher modes (Gittes et al., 1993; Kurz and Williams, 1995).
Most of the measurements reported so far were performed on nongrowing, stabilized, microtubules. Often, however, one is interested in the properties of microtubules in their native, nonstabilized state. Rigidity measurements on growing, nonstabilized microtubules have been performed on microtubules grown from surface-connected nucleation sites (Cassimeris et al., 2001; Kurz and Williams, 1995; Venier et al., 1994). The freely growing end exhibits thermally induced shape fluctuations and the system can be analyzed as a cantilevered beam. To quantify rigidity, thermal fluctuations of a single fixed point on the elongating microtubule were analyzed (Cassimeris et al., 2001; Venier et al., 1994). These studies probe the first bending mode and yield only a single persistence length estimate per microtubule. A mode analysis similar as used for nonsurface-connected microtubules (Gittes et al., 1993) was also applied to the cantilevered beam setup (Kurz and Williams, 1995). In the later study, microtubules were analyzed as if they had a fixed length whereas in reality they were growing. Furthermore, the fluctuations in shape were projected on functions that approximate the motion of a nongrowing microtubule with both ends free (Gittes et al., 1993), ignoring potential errors that are introduced when one end is clamped while the other end is growing.
We will show in this article that a valid mode analysis is possible for microtubules that are fixed at one side while growing substantially at the other side during the observation time. Instead of cosine modes, which cannot be reconciled with the boundary conditions imposed by a clamped end, we used solutions of the equation of motion for the cantilevered beam problem (Wiggins et al., 1998). These solutions show that the dynamics of motion is dramatically slowed down if one end of a microtubule is fixed instead of free. As a consequence the first bending mode is, in our case, unsuitable for rigidity measurements. We applied image analysis methods to automatically quantify the shape of fluctuating microtubules in an unbiased and precise manner, thereby decreasing experimental noise. This allowed us to characterize the amplitude and dynamics of the first five bending modes.
MATERIALS AND METHODS
Synthesis of biotin-labeled nucleation seeds
All reagents were purchased from Sigma-Aldrich (St. Louis, MO), unless otherwise stated. Small, stabilized microtubules with lengths <5 μm served as surface-bound nucleation sites for dynamic GTP microtubules. For this, a 4:1 mixture of bovine brain tubulin and biotin-labeled tubulin (both from Cytoskeleton, Denver, CO; 10 mg/ml) was made. Remnants of glycerol were first removed from this mixture by dilution in MRB80 (80 mM Pipes, 1 mM EGTA, 4mM MgCl2, pH 6.8 with KOH) and subsequent concentration to ∼20 mg/ml total tubulin concentration using a microconcentrator. This solution was then polymerized for 30 min at 35°C in the presence of 0.5 mM GMPCPP (a kind gift of T. J. Mitchison, Harvard Medical School, Boston, MA), generating microtubules that are several microns long. These so-called seeds were diluted 25-fold in MRB80 plus 0.4 mM GMPCPP and 0.62 μM nonlabeled tubulin to extend them at both ends with a stabilized micron-sized biotin-free region (10 min at 35°C). These terminating biotin-free parts of the seeds increased their microtubule-nucleating efficiency when bound to a streptavidin-coated surface. Seeds were stable over a timescale of hours and nucleated microtubules at both sides. Only microtubule plus-ends were analyzed.
Sample preparation
A flow cell was constructed by spacing two acid-cleaned coverslips, sized 24 mm × 24 mm and 24 mm × 60 mm, respectively, 25 μm apart using two lines of vacuum grease and two removable 25 μm thick metal wires. Coverslips displayed little visible roughness, when viewed with the differential interference contrast (DIC) microscope, and gave a good contrast between microtubules and surroundings. Tissue paper was used to blot one solution out of the cell at one side while refilling it at the other side with a new solution. First, the cell was filled with 1 μm diameter latex beads in MRB80, which were allowed to stick nonspecifically to the surface for 5 min. The amount of beads that was added yielded ∼500 beads per mm2 of surface. Next, the cell was filled with 25 μl biotin-labeled bovine serum albumin (biotin-BSA, 2.5 mg/ml in acetate buffer pH 5.2, 5 min incubation), flushed with 50 μl MRB80, refilled with 25 μl streptavidin (Molecular Probes, Leiden, The Netherlands; 1 mg/ml in MRB80, 5 min incubation), flushed again with 50 μl MRB80 and filled with 25 μl biotin-labeled nucleation seeds diluted in MRB80 (5 min incubation). The concentration of seeds was tuned such that <1 microtubule was visible per field of view of the microscope. The intermediate layer of biotin-BSA increased the binding of seeds to streptavidin (Gittes et al., 1996; Janson et al., 2003; Janson and Dogterom, 2004). Finally, 25 μl of a solution containing tubulin (Cytoskeleton, Denver, CO), 1 mM GTP, and 10 mg/ml nonlabeled BSA in MRB80 was flown into the flow cell. BSA was used to compete for tubulin for possible nonspecific binding to surfaces. Measurements were done at two different growth conditions: three samples were analyzed to which we added 28 μM tubulin and an oxygen-scavenging system (OXS system, 4 mM dithiotreitol, 0.2 mg/ml catalase, 0.4 mg/ml glucose-oxydase and 50 mM glucose (Dogterom and Yurke, 1997)) and to one sample we added 26 μM tubulin without OXS system. However, based on the average growth velocities measured in the different samples (see text) and previously measured growth velocities (Janson and Dogterom, 2004; Janson, 2002), we conclude that the tubulin concentration in the last sample was in fact lower. We probably lost some control over the tubulin concentration in the final step of sample preparation when the sample thickness was decreased to only 1 μm. Errors may have been introduced if some parts of the flow cell were not flushed with tubulin-rich solution correctly. Pressure was applied on the top coverslip while blotting excess fluid. This reduced the sample thickness to the 1 μm diameter of the latex beads. Hot candle wax was used to seal the flow cell along the edges, thereby preventing evaporation of fluid.
Video microscopy and image acquisition
Samples were visualized by video-enhanced DIC light microscopy with a Leica DM IRB/E inverted microscope equipped with a 100× oil-immersion objective (numerical aperture 1.3). The sample temperature was maintained at 23°C. An Argus 20 image processor (Hamamatsu, Almere, The Netherlands) was used to perform background subtraction and contrast enhancement of charge-coupled device camera images (CF8/1, Kappa, Gleichen, Germany). The resulting image stream was recorded on Super-VHS videocassettes and digitized at a rate of 1 frame every 6 s (SGI visual workstation with built-in frame grabber; SGI, Mountain View, CA). The field of view of this system was 35 μm × 26 μm.
Microtubule shape digitization
Contrast in DIC microscopy is direction sensitive, exhibiting a maximum along the axis of shear and a minimum in the orthogonal direction (e.g., Fig. 1 a). To achieve maximum contrast we selected microtubules that grew approximately in a direction orthogonal to the axis of shear, which in our setup is horizontally. These microtubules possess a shadow-cast appearance, i.e., one side is dark and the other side is bright. The local vertical position of the microtubule corresponds with the middle of this interference pattern. A computer algorithm (programmed in Interactive Data Language, Research Systems Inc., Boulder, CO) was designed to find this position in every vertical pixel column of the image, thereby quantifying the microtubule-shape in a precise and nonprejudiced manner. The algorithm traced the microtubule from the left to the right. The vertical position of the microtubule in the most left pixel column was first assigned manually. Next, the exact position in this pixel column was estimated within a window of 60 pixels centered on this initially chosen point. For this purpose, the intensity profile of the 60 pixels (e.g., Fig. 1 b) was convoluted with one period of a sine function (Fig. 1 c). This function was chosen to mimic the shadow-cast appearance of an imaged microtubule. The position of the maximum in the convoluted data was assigned as the microtubule's vertical position (Fig. 1 d). For all other pixel columns, an initial point was assigned automatically based on the microtubule position in preceding columns and the convolution procedure was repeated. Typically, when available, 40 points on the left were extrapolated to the right to obtain the initial point. The convolution procedure was repeated for every column. By using extrapolation, the 60-point window remained centered around the microtubule. Furthermore, in case the maximum in the convoluted signal deviated more than four pixels from the extrapolated value, the extrapolated value was chosen as the microtubule position. This prevented the algorithm from assigning false values caused by low local contrast or “dirt” particles on the coverslip that obscure locally the microtubule (e.g., Fig. 2 b). The width of the sine period that was used for convolution was taken equal to 13 pixels as this yielded the smoothest traces.
FIGURE 1.
Shape digitization by semiautomated image tracing. (a) Example of a partly traced image; the black line corresponds to points found by the tracing algorithm. The white horizontal bar equals 5 μm or 100 pixels. (b) Intensity line scan of the pixel column indicated by the vertical white line in Fig. 1 a. The lower left pixel in Fig. 1 a has pixel coordinates [0,0] . (c) The single period of a sine functions that was used to model the characteristic shadow-cast appearance of a microtubule. (d) Convolution results. The shaded line corresponds to the convolution of the raw data of Fig. 1 b, with the kernel of Fig. 1 c. Black lines are convolutions of nine neighboring pixel columns on the left and right side of the white line in Fig. 1 a.
FIGURE 2.
(a) DIC image of an elongating microtubule. The growing plus end is on the right and the seed is on the left. The scale bar corresponds to 5 μm. (b) Same microtubule as in Fig. 2 a imaged 5 min later. The microtubule has grown out of the field of view and the shape has been changed by thermal fluctuations except for the left seed region. A digitized curve, found by automated image tracing (see Fig. 1), is superimposed on the image. The displayed coordinate system is chosen as explained in c. (c) Parameterization of microtubule shape. The path length s is chosen to be zero at the transition between the clamped (shaded) and the free (black plus white) part of the microtubule. The center of the coordinate system is located at s = 0 with the x axis along the direction of clamping. Shape can be parameterized either by y(s) or θ(s). In the experiment, only the length between s = 0 and s = 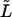 (black) is analyzed. During the course of the experiment, the microtubule is growing and its full length corresponds to s = L.
(black) is analyzed. During the course of the experiment, the microtubule is growing and its full length corresponds to s = L.
Shape parameterization
At the start of observation, most microtubules had grown already to a length of several micrometers after having been nucleated from a seed. The seed's attachment end, i.e., the location at which the microtubule becomes free from the surface, was estimated by looking at the video image in real time. This allowed for a good observation of small thermal fluctuations close to the seed that indicated where the microtubule could still move. A new transformed set of coordinates [x,y] was obtained by rotation of the traced microtubule shape around the observed attachment end such that the new x axis was parallel to the direction of the seed (see Fig. 2, b and c). Finally the microtubule was parameterized as y(s), where s is the path length along the microtubule, with s = 0 at the seed's attachment end. To calculate the increase in path length ds between two neighboring traced points (typically 50 nm), we applied a 30 point moving average filter to the raw [x,y] data. Smoothing prevents uncertainty in the determination of y from having a severe effect on the determination of microtubule length. Nonsmoothed y-values were used when calculating mode amplitudes.
Shape analysis in the cantilevered beam geometry
Thermal fluctuations of a nongrowing microtubule. To analyze shape fluctuations on elongating microtubules, we first describe the thermally excited dynamics of a microtubule that is clamped at one side and does not change its total length (L) during the observation time (see Fig. 2 c). As a result of thermal motion, the tangent angle θ(s) along the microtubule deviates from the value θ0(s) that describes the relaxed shape of the microtubule, which is straight apart from a possible small intrinsic curvature. The flexural rigidity κ (Nm2) of a microtubule is related to its three-dimensional persistence length, Lp = κ/kBT, with kB Boltzmann's constant and T the absolute temperature (Landau and Lifshitz, 1986). In general, θ(s) will be small because Lp is expected to be much larger than L. Therefore, 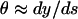 and the balance between elastic restoring forces inside the microtubule and the hydrodynamic drag experienced by a moving microtubule (Wiggins et al., 1998) can be written as
and the balance between elastic restoring forces inside the microtubule and the hydrodynamic drag experienced by a moving microtubule (Wiggins et al., 1998) can be written as
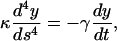 |
(1) |
where γ(kg m−1s−1) is the perpendicular drag coefficient for the microtubule. The solutions (n = 1,2,3..) of Eq. 1 are
 |
(2) |
The spatial part, Wn(s/L), that satisfies the boundary conditions for the clamped and free microtubule end, y(0) = 0, y′(0) = 0, y″(L) = 0, and y‴(L) = 0, was calculated previously (Wiggins et al., 1998) as
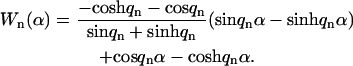 |
(3) |
Here 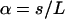 and the parameters qn are given by the subsequent solutions of
and the parameters qn are given by the subsequent solutions of
 |
(4) |
Solutions of Eq. 4 are q1 ≈ 1.875, q2 ≈ 4.695, q3 ≈ 7.855, and 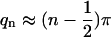 for n > 3. The first three eigenfunctions, Wn=1..3, are plotted in Fig. 3.
for n > 3. The first three eigenfunctions, Wn=1..3, are plotted in Fig. 3.
FIGURE 3.
Spatial part, Wn, of the first three solutions of the hydrodynamic beam equation plotted as a function of the scaled path length α = s/L. The functions are orthonormal on the interval [0,1].
Any arbitrary shape y(s) that the microtubule adopts due to thermal motions can be expressed in terms of the Wn series:
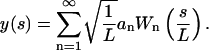 |
(5) |
The amplitudes an are calculated by
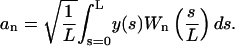 |
(6) |
The mode amplitudes will fluctuate in time with a correlation time that is given by (Wiggins et al., 1998)
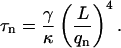 |
(7) |
One can now calculate the bending energy that is stored in a microtubule as
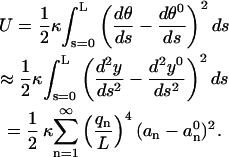 |
(8) |
Here we used the equality 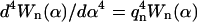 (Wiggins et al., 1998). The intrinsic, relaxed shape of the microtubule is represented by θ0, y0, and
(Wiggins et al., 1998). The intrinsic, relaxed shape of the microtubule is represented by θ0, y0, and 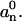 Equipartition theorem (Gittes et al., 1993; Reif, 1965) states that each quadratic term in Eq. 8 contributes on average ½kBT to the total bending energy. Therefore, the variance in each mode amplitude equals
Equipartition theorem (Gittes et al., 1993; Reif, 1965) states that each quadratic term in Eq. 8 contributes on average ½kBT to the total bending energy. Therefore, the variance in each mode amplitude equals
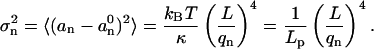 |
(9) |
This variance can be determined experimentally, from which then an estimate for Lp can be obtained as was described in Gittes et al. (1993) for the case of nonclamped microtubules.
Analysis of an elongating microtubule. The variance expressed by Eq. 9 is a strong function of L, and a priori it is not clear how a variance should be calculated if the microtubule grows significantly during the time needed to sample the fluctuating amplitudes. We can try to infer the rigidity of a microtubule by only analyzing a constant part of the growing microtubule (between s = 0 and 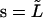 ; see Fig. 2 c). Let us project the shape
; see Fig. 2 c). Let us project the shape 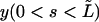 of this part of the microtubule onto the function set Wl (Eq. 3) with
of this part of the microtubule onto the function set Wl (Eq. 3) with 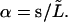 A tilde indicates a quantity for the analyzed part of the microtubule. The amplitudes of these new modes, which we shall call “analyzed-length” modes, are calculated as
A tilde indicates a quantity for the analyzed part of the microtubule. The amplitudes of these new modes, which we shall call “analyzed-length” modes, are calculated as
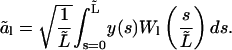 |
(10) |
From this we can reconstitute the shape of the microtubule between s = 0 and 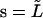 by
by
 |
(11) |
The use of the function set Wl implies that 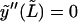 and
and 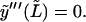 In reality, the microtubule satisfies these conditions, however, only at s = L (note that this problem also occurs when using cosine or sine modes, which in addition violates the boundary conditions y(0) = 0 or y′(0) = 0 at the clamped end). We will investigate the implications of this discrepancy later. If we use the reconstituted shape
In reality, the microtubule satisfies these conditions, however, only at s = L (note that this problem also occurs when using cosine or sine modes, which in addition violates the boundary conditions y(0) = 0 or y′(0) = 0 at the clamped end). We will investigate the implications of this discrepancy later. If we use the reconstituted shape 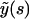 to calculate the bending energy in the analyzed part, we find
to calculate the bending energy in the analyzed part, we find
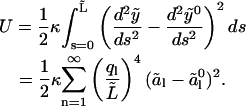 |
(12) |
Equipartition theorem may be applied to the energy of the analyzed part only. The variances of the new mode amplitudes are then calculated as
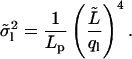 |
(13) |
This variance is independent of the actual length L of the microtubule.
Mode spectrum. The fluctuations of the full-length modes of a microtubule are governed by the correlation time  To understand the dynamics of the analyzed-length modes, we need to calculate how the amplitude fluctuations of each full-length mode contribute to the amplitude of analyzed-length modes. We will show, for example, that the slow fluctuations of the first full-length mode hardly change the amplitude of the second and higher analyzed-length modes. As a consequence, the fluctuations of these modes are fast. We consider a nongrowing microtubule with length L of which only a length
To understand the dynamics of the analyzed-length modes, we need to calculate how the amplitude fluctuations of each full-length mode contribute to the amplitude of analyzed-length modes. We will show, for example, that the slow fluctuations of the first full-length mode hardly change the amplitude of the second and higher analyzed-length modes. As a consequence, the fluctuations of these modes are fast. We consider a nongrowing microtubule with length L of which only a length 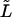 is analyzed and substitute the shape of the full-length microtubule (Eq. 5), into Eq. 10:
is analyzed and substitute the shape of the full-length microtubule (Eq. 5), into Eq. 10:
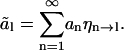 |
(14) |
Here we introduced coupling coefficients, ηn→l, which link modes of the full-length microtubule to the analyzed-length mode amplitude ãl, that are a function of the parameter 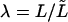 :
:
 |
(15) |
Because of the linearity in Eq. 14, the variance of the analyzed-length mode amplitudes is related to that of the full-length variances by
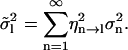 |
(16) |
The contributions can be normalized by dividing Eq. 16 by the total variance (Eq. 13):
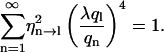 |
(17) |
The normalized contributions (calculated using numerical methods; Mathematica, Wolfram Research, Champaign, IL) of the first 11 full-length modes (n) to the variance of the first six analyzed-length modes (l) are plotted in Fig. 4 a for λ = 1.5. This λ-value represents the average ratio between full length and analyzed length as experimentally used in this study; λ-values >2 were not used. Fig. 4 a shows that a narrow band of full-length modes contributes to the variance in the analyzed-length modes. As an example, for l = 3, only n = 4 and n = 5 contribute significantly whereas other contributions are minor. As a consequence, two timescales will determine the correlation time of the third analyzed-mode: τ5 and the slower τ4. For λ = 3 (Fig. 4 b), the band of contributing modes becomes wider and the contributions come from higher order full-length modes. In general, the following picture arises: the correlation time of an analyzed-length mode l is dominated by the fluctuations of the full-length mode λ × l and we may therefore write
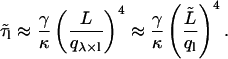 |
(18) |
FIGURE 4.
Normalized contributions of full-length modes (n = 1..11) to the variance of analyzed-length modes (l = 1..6) for λ = 1.5 (a) and λ = 3 (b). Plotted contributions equal 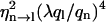 (Eq. 17).
(Eq. 17).
The sum of the first 11 contributions of full-length modes (n = 1..1) to the sixth analyzed-length mode (l = 6) is 0.995. For smaller values of l, this sum is even closer to 1 as expected by Eq. 17. This shows that for all practical purposes, Eq. 13 is a good approximation for 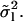 Therefore, we conclude that the chosen expansion of y(s) into Wl functions does not generate errors that are related to the mentioned discrepancy of higher order derivatives at
Therefore, we conclude that the chosen expansion of y(s) into Wl functions does not generate errors that are related to the mentioned discrepancy of higher order derivatives at  Fig. 4 also shows that mode spectra for different l have very little overlap. Because full-length modes fluctuate statistically independently, this finding implicates that rigidity estimates from different analyzed-length modes (l) are also statistically independent.
Fig. 4 also shows that mode spectra for different l have very little overlap. Because full-length modes fluctuate statistically independently, this finding implicates that rigidity estimates from different analyzed-length modes (l) are also statistically independent.
RESULTS
Growing microtubules
Variances in analyzed-length mode amplitudes were measured on microtubules in the cantilevered beam geometry. The microtubules were confined in between two coverslips spaced 1 μm apart. The samples were scanned for microtubule seeds that already nucleated a microtubule at their plus end, i.e., the faster growing end of a microtubule that can be identified because of its higher growth velocity (Walker et al., 1988). When found, the seed was positioned somewhere near the edge of the field of view. Some microtubules could be followed for 1–10 min before the tip of the microtubule left the visible part of the sample (Fig. 2, a and b). The straight distance between microtubule seed and tip was measured at discrete time points from which the average growth velocity, v, was obtained using a linear fit to the data.
We studied microtubule rigidity under two different growth conditions (A and B). For three samples (to which we added 28 μM tubulin and an oxygen-scavenging system; condition A) we measured v = 2.72 ± 0.53 μm/min (N = 9, mean ± SD, data were weighted with the observation time per microtubule) and for one sample (26 μM tubulin without oxygen-scavenging system; condition B) we measured v = 1.50 ± 0.39 μm/min (N = 5). Growth velocities measured on individual microtubules ranged between 1.84 and 3.51 μm/min for condition A (N = 9) and were between 1.11 and 1.73 μm/min for condition B (N = 5).The two growth conditions are therefore well separated based on their measured growth velocities, indicating that the tubulin concentration in the last sample was in fact lower than 26 μM (see Methods). Microtubule catastrophes were rare under both conditions and most microtubules grew up to lengths exceeding 60 μm without undergoing rapid shrinkage.
After the tip of a growing microtubule disappeared from the field of view (35 μm × 26 μm) the observation was continued for another 10–15 min or stopped earlier after the occurrence of a catastrophe. Some microtubules were pinned at a single point along their length during part of the observation time. Pinning, in contrast to seed binding, probably involves temporary nonspecific bonding between a microtubule and the surface. Pinned microtubules were not analyzed because pinning hinders thermal fluctuations. A few microtubules had a large intrinsic curvature and were also not analyzed. In total, 7 out of 12 observed microtubules were analyzed for condition A and 5 out of 5 for condition B.
Shape digitization
Once every 6 s a video frame was captured. Two snapshots are shown in Fig. 2, a and b. An algorithm was used to trace the shape of a microtubule using the intensity profile that is created by the DIC microscope (Fig. 1). The intensity variation of the pixels in the vertical pixel column that is indicated in Fig. 1 a is plotted in Fig. 1 b. It is impossible to judge directly the vertical coordinate of the microtubule from this intensity plot. The data was therefore convoluted with a profile that mimics the image of a microtubule (Fig. 1 c) to filter out the vertical coordinate (Fig. 1 d). The tracing algorithm that is described in the methods section follows the ridge that becomes visible when subsequent pixel columns along the length of the microtubule are treated in the same manner (Fig. 1 d). Small dirt particles or crossing microtubules as visible in Fig. 2 b do not hinder the algorithm. Microtubules did not move out of the plane of focus because the sample chamber had a thickness of only 1 μm. Therefore the algorithm worked on almost all digitized images. Incorrectly traced images were not analyzed. Traced curves were parameterized as y(s) as described in the methods section (Fig. 2 c).
Calculation of mode amplitudes
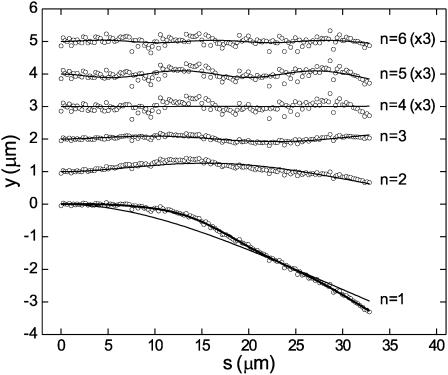
Of each microtubule, ∼30 μm was analyzed, corresponding to the field of view. Mode amplitudes were calculated every 6 s, using a discrete version of Eq. 10. An example of an analyzed shape is displayed at the bottom of Fig. 5. The first analyzed-length mode with its appropriate amplitude is plotted over the raw digitized data. This first mode describes the largest part of the observed microtubule's deflection. Next, the first mode is subtracted from the raw data and the result is plotted over the second analyzed-length mode with its corresponding amplitude. Subsequent modes are plotted similarly. Raw data points from which the first four modes were subtracted still showed significant undulation above the noise level (Fig. 5; n = 5). This indicates that the fifth analyzed-length mode amplitude still contains significant bending information. The sixth and higher analyzed-length modes often appeared to describe merely digitization noise (Fig. 5; n = 6).
FIGURE 5.
Decomposition in modes. Plotted are raw digitized points that represent a microtubule shape at a single time point (lower curve). Only 25% of all digitized points are plotted for clarity. Mode amplitudes were calculated using Eq. 10. The thick overlaid curve is reconstituted (Eq. 11) from the first five mode amplitudes. The thin overlaid line corresponds to a reconstitution using only the first mode. Reconstitutions using only the second, third, fourth, fifth, or sixth mode are plotted with an increasing vertical offset of 1 μm (thin lines). The fourth, fifth, and sixth modes are scaled by a factor 3 for clarity. Overlaid on each nth mode curve are the raw data points with mode reconstitutions 1 until n − 1 subtracted.
In Fig. 6, the analyzed-length mode amplitudes, ãl, are shown as a function of time. The second and higher modes behave stochastically, whereas a clear trend can be seen in the data for the first mode. This trend is indicative for a correlation time that is much larger than the 6 s between subsequent time points.
FIGURE 6.
Calculated mode amplitudes, ãl, for a single microtubule (condition A, analyzed length is 34.3 μm). Data are plotted as a function of time for the first eight mode numbers. The time between data points was 6 s, and the total measurement time was 8 min.
The variance in mode amplitudes is predicted to decrease with ql following an inverse fourth power law (Eq. 13): 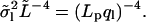 To test this relation experimentally, the measured
To test this relation experimentally, the measured  was multiplied by
was multiplied by 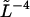 and plotted as a function of ql for all microtubules within each growth condition (Fig. 7, a and b). For both growth conditions, the second until approximately the fifth mode values obey the straight line that represents the expected fourth power dependence on a double log scale.
and plotted as a function of ql for all microtubules within each growth condition (Fig. 7, a and b). For both growth conditions, the second until approximately the fifth mode values obey the straight line that represents the expected fourth power dependence on a double log scale.
FIGURE 7.
Variance of the measured mode amplitudes, 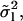 multiplied by
multiplied by 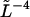 as a function of ql. The theoretical expectation,
as a function of ql. The theoretical expectation,  is plotted for our final estimates of the persistence length (Table 1). The analyzed length of the eight microtubules is given in the legend (μm). (a) Condition A. (b) Condition B.
is plotted for our final estimates of the persistence length (Table 1). The analyzed length of the eight microtubules is given in the legend (μm). (a) Condition A. (b) Condition B.
Rigidity estimates
From the observed variances in amplitude, we calculated for each individual microtubule five persistence length estimates based on mode numbers 1 until 5 (Eq. 13, Fig. 8). Statistical errors that are caused by a limited number of analyzed time points were calculated as in Gittes et al. (1993). Fig. 8 shows that the first order estimate is for most microtubules significantly larger than the second until fifth estimates. Estimates from mode 2 and 3 (see Discussion) were averaged to obtain a single rigidity estimate per microtubule (Table 1). To test the consistency of the two averaged values, P-values were calculated for each microtubule that express the probability that the two values can be drawn from a single parent distribution given the statistical errors (χ2 test d = 1, Table 1). The smallest value is 18%, indicating that modes 2 and 3 are consistent. A similar analysis was done by averaging estimates from mode 2, 3, and 4. Now 2 out of 12 P-values were found to be <2%, a situation that has a probability of occurrence that is 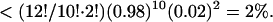 This analysis shows that the fourth mode estimate is not always consistent with the second and third mode estimates.
This analysis shows that the fourth mode estimate is not always consistent with the second and third mode estimates.
FIGURE 8.
Estimates for the persistence length for the first five modes are plotted in order of increasing filament length. Standard deviations are plotted as error bars (statistical error only). The straight line corresponds to our final estimate of the persistence length (Table 1). (a) Condition A. (b) Condition B. The first mode persistence length estimate for the microtubule with length 33.5 μm does not fit on the scale, but equals 104 mm.
TABLE 1.
Persistence length estimates (mean ± SE) per microtubule for experimental conditions A and B based on the 2nd and 3rd mode
| A: v = 2.72 ± 0.53 μm/min
|
B: v = 1.50 ± 0.39 μm/min
|
||||
|---|---|---|---|---|---|
| MT length (μm) | Lp,2&3 (mm) | P (%) | MT length (μm) | Lp,2&3 (mm) | P (%) |
| 28.5 | 2.8 ± 0.3 (2) | 72 | 27.6 | 5.8 ± 1.1 (2) | 67 |
| 30.8 | 5.0 ± 0.9 (2) | 35 | 29.3 | 8.3 ± 1.1 (2) | 21 |
| 32.0 | 3.9 ± 0.4 (2) | 34 | 30.7 | 5.0 ± 0.8 (2) | 18 |
| 32.6 | 4.5 ± 0.4 (2) | 98 | 31.2 | 4.5 ± 0.8 (2) | 33 |
| 32.8 | 4.8 ± 0.6 (2) | 31 | 33.5 | 9.3 ± 1.8 (2) | 88 |
| 34.3 | 4.4 ± 0.5 (2) | 81 | |||
| 34.5 | 3.6 ± 0.3 (2) | 88 | |||
| Average | 4.2 ± 0.3 (7) | Average | 6.6 ± 0.9 (5) | ||
The average microtubule (MT) growth velocity, v, is indicated for both growth conditions (mean ± SD). P-values were calculated for each microtubule (χ2 test with d = 1). The bottom row shows our final rigidity estimates obtained by averaging the Lp,2&3 values for individual microtubules (mean ± SE, weighted with the number of time points taken per microtubule). The number of values averaged is indicated in parentheses.
Persistence length values for individual microtubules, based on the second and third mode values, were averaged to obtain a single estimate per growth condition (Table 1). The obtained average values, 4.2 ± 0.3 mm for condition A (mean ± SD, data was weighted with the number of time points taken per microtubule) and 6.6 ± 0.9 mm for B, indicate a significant difference in rigidity between the two growth conditions. To check for rigidity differences between microtubules within a single growth condition, P-values were calculated for the averaged values: P = 0.05% (A, χ2 test with d = 6) and P = 0.2% (B, χ2 test with d = 4). These values suggest a nonconstant rigidity; however, the used errors are statistical errors (see Fig. 8 and Table 1) that do not include systematic errors. If we increase all standard errors by 5% of the persistence length to roughly account for errors introduced by an uncertainty in the location of the seed's attachment end (see Discussion and Appendix), we find P = 9% (A) and P = 4% (B). These probabilities are still small, which suggests the existence of differences in rigidity between individual microtubules within a single growth condition. Differences between the two growth conditions appear larger, however.
Correlation times
To investigate the dynamics of bending fluctuations, we measured the correlation time of the second until fifth mode. For this we increased our sampling time from 6 s to 40 ms. To ease automated shape digitization, we picked out a microtubule that remained particularly well in focus (condition A, previously analyzed with 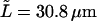 ) and which grew 12.7 μm at 2.9 μm/min before its growing tip left the field of view. This complete event was subdivided into 10 short fragments of 24 s. Fig. 9 a shows autocorrelation functions of mode amplitudes that were calculated for one of these 24 s fragments using the near constant full length of the microtubule during this time fragment. Correlation times were successively obtained by fitting an exponential to these curves. The curves were fitted until they dropped below 0.4 to minimize interference of the observed nonzero base levels. The fifth mode autocorrelation function decays very fast and a correlation time was derived only from the first point (t = 40 ms). During the time span of the 24 s fragment, the microtubule hardly changes its length and the measured correlation times thus correspond to the correlation times of the full-length modes, τn. Correlation times were obtained for all other 24 s fragments in a similar way and the results are plotted in Fig. 9 b as a function of microtubule length. To investigate the expected (q/L)4 dependency of τn (Eq. 7), we multiplied τn by κ*(q/L)4 using the value for κ that was measured earlier for this microtubule. The result is an estimate of the perpendicular drag coefficient γ, which is plotted in Fig. 9 c.
) and which grew 12.7 μm at 2.9 μm/min before its growing tip left the field of view. This complete event was subdivided into 10 short fragments of 24 s. Fig. 9 a shows autocorrelation functions of mode amplitudes that were calculated for one of these 24 s fragments using the near constant full length of the microtubule during this time fragment. Correlation times were successively obtained by fitting an exponential to these curves. The curves were fitted until they dropped below 0.4 to minimize interference of the observed nonzero base levels. The fifth mode autocorrelation function decays very fast and a correlation time was derived only from the first point (t = 40 ms). During the time span of the 24 s fragment, the microtubule hardly changes its length and the measured correlation times thus correspond to the correlation times of the full-length modes, τn. Correlation times were obtained for all other 24 s fragments in a similar way and the results are plotted in Fig. 9 b as a function of microtubule length. To investigate the expected (q/L)4 dependency of τn (Eq. 7), we multiplied τn by κ*(q/L)4 using the value for κ that was measured earlier for this microtubule. The result is an estimate of the perpendicular drag coefficient γ, which is plotted in Fig. 9 c.
FIGURE 9.
Analysis of the dynamics of full-length modes. (a) Autocorrelation function (n = 2..5) of mode amplitudes that were calculated over the full length of a microtubule (L = 33.3 μm) during 24 s with 40 ms sampling time. Dashed lines are exponential fits to the initial decrease. (b) Fitted correlation time as a function of microtubule length for four full-length modes (n = 2..5). (c) Estimates for γ as a function of qn/L. The plotted line is a fit of Eq. 20 to the data.
DISCUSSION
We have measured flexural rigidities on elongating microtubules by analyzing only a constant part of their continuously increasing full length. A mode analysis was applied based on the full-length modes of cantilevered microtubules. We will first discuss errors that are of methodical and experimental origin, then discuss the dynamics of bending fluctuations, and finally compare rigidity estimates.
Errors due to slow and fast dynamics
Our analysis in Fig. 6 shows that thermal fluctuations of the first analyzed-length mode progress slowly, suggesting a correlation time on the order of 100 s. Therefore we cannot get enough statistically independent estimates for mode amplitude 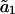 within the observation time of typically 10 min (limited by the occurrence of catastrophes). Rigidity estimates of the first mode are therefore inaccurate and often overestimated because only part of the accessible amplitude distribution is sampled (Fig. 8 and Table 2). The correlation time of the first analyzed-length mode is long because the slowly fluctuating first full-length mode couples strongly into this mode (Fig. 4). This effect is analyzed in detail in the Appendix, where it is also shown that higher order analyzed-length modes do not suffer from this long correlation time.
within the observation time of typically 10 min (limited by the occurrence of catastrophes). Rigidity estimates of the first mode are therefore inaccurate and often overestimated because only part of the accessible amplitude distribution is sampled (Fig. 8 and Table 2). The correlation time of the first analyzed-length mode is long because the slowly fluctuating first full-length mode couples strongly into this mode (Fig. 4). This effect is analyzed in detail in the Appendix, where it is also shown that higher order analyzed-length modes do not suffer from this long correlation time.
TABLE 2.
Persistence length estimates per mode
| Mode number l | Lp (mm) for condition A: v = 2.72 ± 0.53 μm/min | Lp (mm) for condition B: v = 1.50 ± 0.39 μm/min |
|---|---|---|
| 1 | 12.2 ± 2.6 (7) | 26.1 ± 16.4 (5) |
| 2 | 4.0 ± 0.3 (7) | 6.2 ± 0.8 (5) |
| 3 | 4.1 ± 0.3 (7) | 7.1 ± 1.2 (5) |
| 4 | 4.3 ± 0.6 (7) | 8.1 ± 1.2 (5) |
| 5 | 4.7 ± 0.3 (7) | 8.8 ± 1.1 (5) |
| 6 | 4.0 ± 0.5 (7) | 5.9 ± 0.7 (5) |
Shown is the average of the lth mode rigidity estimates for all microtubules within one experimental condition (mean ± SE, weighted with the number of time points taken per microtubule). The number of values averaged is indicated in parentheses.
Fast correlation times of higher order modes may also introduce errors. Fluctuations with a correlation time shorter than the image acquisition time of 40 ms will appear smoothed on the digitized images (Gittes et al., 1993). Some evidence for mode smoothing is obtained from Table 2, where the average fifth mode estimates appear somewhat larger than the second until fourth mode estimates. To further investigate the occurrence of mode smoothing, we measured the correlation time of the second until fifth full-length mode (Fig. 9, a and b; a correlation time for the first mode could not be measured because the lifetime of microtubules was limited by catastrophes). As expected, the correlation time of full-length modes increases with mode number and microtubule length (Fig. 9 b). For a 30 μm long microtubule, we approximately find τ2 = 1000 ms, τ3 = 200 ms, τ4 = 80 ms, and τ5 = 40 ms. Comparison of Eq. 7 and Eq. 18 predicts similar correlation times for the analyzed-length mode amplitudes of longer microtubules of which only 30 μm is analyzed. We verified this experimentally (data not shown). An image acquisition time of 40 ms will therefore smooth the amplitudes of the fifth mode to some degree, explaining why we found higher rigidity estimates for the fifth mode in Table 2.
In the Appendix, we discuss several other experimental sources of error. It is shown that positional noise, caused by a limited resolution of the microscope, causes additional variance in mode amplitudes that influences rigidity estimates. These noise contributions are relatively large for higher modes. We therefore based our final rigidity estimates only on the second and third mode, which were shown to be consistent with each other. In the Appendix, it is shown that the remaining source of experimental error is the uncertainty in the location of the seed's attachment end. This uncertainty can cause a relative uncertainty in the rigidity estimate of 5%. In size, this is comparable to the statistical error that is caused by a limited number of analyzed time points.
Dynamics of bending fluctuations
The measured correlation times for a 30 μm long microtubule are surprisingly long when compared to theoretical predictions that can be obtained using an estimate for the perpendicular drag coefficient γ0 of a long cylinder in the vicinity of two coverslips (Mickey and Howard, 1995):
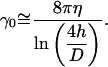 |
(19) |
Here η (kg m−1s−1) equals the viscosity of water, h the distance between microtubule and coverslips, and D the diameter of the microtubule. We find γ0 = 0.0057 s Nm−2 assuming the microtubule is halfway in between the two coverslips. This gives for a 30 μm long microtubule, τ1 ≈ 20 s, τ2 ≈ 0.5 s, τ3 ≈ 60 ms, τ4 ≈ 15 ms, and τ5 ≈ 6 ms (Eq. 7).
Experimentally we find that γ is not constant but increases with the scaled wavenumber q/L (Fig. 9 c). This observation can be explained by a more elaborate equation of motion for biofilaments, which includes a term that accounts for internal friction within the filament or flow of water through pores within a filament (Poirier and Marko, 2002). According to this model, the apparent drag, as plotted in Fig. 9 c, becomes mode number dependent:
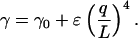 |
(20) |
Here ɛ depends on the amount of internal friction and the radius of the filament. When fitted to the data in Fig. 9 c, we find γ0 = 0.011 s Nm−2 and ɛ = 6.9?10−25 s Nm2. The value for γ0 is in reasonable agreement with the theoretical estimate, which itself is only an approximation. This extended model predicts a mode number-independent correlation time equal to ɛ/κ in the limit of large q/L (Poirier and Marko, 2002). For our microtubule, this value equals 33 ms. If true, the model would imply that the correlation time of the fifth and higher modes of a 30 μm long microtubule are all approximately equal to 33 ms. It is not clear what structural property of a microtubule may yield such a long correlation time. In any case, our analysis shows that additional friction on top of external hydrodynamic drag changes the dynamics of bending modes and explains why higher order modes are not severely affected by mode smoothing. Some evidence suggests that chromosomes show similar dynamics, which may be caused by internal friction (Poirier and Marko, 2002), but there are no reports for other biofilaments. Correlation times for microtubules were not previously reported except for the relaxation of the first mode (Felgner et al., 1996).
A relation between growth velocity and rigidity
We made rigidity estimates on microtubules under two different experimental conditions that displayed a different average microtubule growth velocity (2.72 μm/min and 1.50 μm/min). Our results indicate a significant difference in rigidity between the two experimental conditions (Table 1). In Fig. 10 and Table 3, we compare our data to earlier (less precise) rigidity measurements from our lab on elongating microtubules grown from GTP-tubulin using a simpler analysis of thermal fluctuations (Janson and Dogterom, 2004). There is no clear separation in rigidity between measurements done with and without an OXS system, which could be one explanation for rigidity differences (Mickey and Howard, 1995). However, there seems to be a consistent correlation between growth velocity and microtubule rigidity: slower-growing microtubules appear to be more rigid. Other published work on microtubule rigidity of elongating microtubules (Cassimeris et al., 2001; Kurz and Williams, 1995; Venier et al., 1994) does not document the growth velocity and one cannot check whether it is consistent with the observed trend in Fig. 10. A relation between growth velocity and rigidity may, however, provide part of an explanation for the large scatter in rigidity estimates that has been reported in the past.
FIGURE 10.
Persistence length estimates for five different growth conditions versus average growth velocity (see Table 3). Standard errors are plotted for the velocity and standard deviations (statistical error only) for Lp. Growth conditions with added OXS system are plotted as solid symbols and without OXS system as open symbols.
TABLE 3.
Summary of five persistence length estimates on elongating microtubules obtained with three different analysis methods of thermally driven fluctuations
| Source | Number of microtubules | Temp. (°C) | OXS system | Growth velocity (μm/min) (mean ± SE) | Lp (mm) (mean ± SD) |
|---|---|---|---|---|---|
| This work, condition A* | 7 | 23 | yes | 2.72 ± 0.18 | 4.2 ± 0.8 |
| This work, condition B* | 5 | 23 | no | 1.50 ± 0.18 | 6.6 ± 2.1 |
| Janson and Dogterom (2004)*† | 23 | 23 | yes | 1.88 ± 0.03 | 5.2 ± 1.9 |
| Janson and Dogterom (2004)*† | 16 | 23 | no | 2.40 ± 0.01 | 3.4 ± 1.6 |
| Dogterom and Yurke, (1997)‡ | 12 | 22 | yes | 1.21 ± 0.07 | 8.3 ± 1.7 |
All measurements are done in the cantilevered beam geometry. The number of microtubules that were analyzed, the sample temperature, and the addition of an oxygen scavenging system (Dogterom and Yurke, 1997) are indicated.
These four measurements were done using the same batch of tubulin.
Estimates are based on deflections of a single point of a microtubule near its end.
Estimates are based on the deflections of the middle point of the microtubule from the straight line connecting the seed and a point near the end.
Possible explanations
One may speculate about the reason for a relation between growth velocity and rigidity. Differences in rigidity between fast and slowly growing microtubules indicate either a difference in microtubule structure or chemical composition. Hydrolysis of GTP-tubulin to GDP-tubulin at the growing tip is a fast process (Desai and Mitchison, 1997) and therefore the analyzed length of microtubules should consist only out of GDP-tubulin. Rigidity differences, therefore, most likely have a structural origin. Faster growing microtubules could be made on average out of less protofilaments. The number of protofilaments inside a microtubule varies between 10 and 16, but values of 12, 13, or 14 are by far the most observed (Chretien and Fuller, 2000). If a microtubule is modeled by a homogeneous rod, one can show that the rigidity difference between a 13- and 14-filament microtubule should be 20% (Gittes et al., 1993). Although this can explain rigidity variations observed within one single growth condition (Table 1), the effect seems too small to explain the full range of observed rigidity values (Fig. 10). Furthermore, no structural data are available that suggests a relation between growth velocity and protofilament number.
Another explanation for the observed relation may be provided by defects in the microtubule structure that may give rise to microtubule segments with decreased rigidity. Transitions between different types of lattices and protofilament numbers were observed within individual microtubules and with higher frequency at increasing tubulin concentrations (Chretien and Fuller, 2000). Fast-growing microtubules may have less time to “repair” defects that occur during growth. If the spatial frequency of transitions is high and indeed growth velocity dependent, they might explain our data. Reported frequencies observed with electron microscopy are, however, low and range from 0.08 transitions per micrometer, averaged over many microtubules, (Chretien et al., 1992) to 2 μm−1 on one individual microtubule (Chretien and Fuller, 2000). Similarly, we recently observed that abrupt changes in growth velocity occur during microtubule growth that may be a signature of lattice changes (Janson and Dogterom, 2004). Up to two changes were seen per micron of growth. A mechanism based on defects is, however, very speculative. Recent work with an atomic force microscope suggests that shear or protofilament sliding may influence microtubule rigidity on short length scales (Kis et al., 2002). These kinds of deformations are reported to be of influence for other filament systems (Cohen and Mahadevan, 2003). Shear deformations together with growth velocity-dependent lattice defects could possibly create a complex bending behavior. However, structural electron microscopy work suggests that the bonds between protofilaments are in fact very strong and hard to deform (Chretien and Fuller, 2000).
Comparing the technique to other methods
The analysis of thermally induced fluctuations in microtubule shape is a convenient and often used method to study microtubule rigidity. A full analysis of bending modes is preferred over methods in which only the motion of the tip of a microtubule is analyzed, because multiple rigidity estimates are obtained that can be checked for consistency. To measure microtubules in their native state, i.e., growing and shrinking, we had to attach one side of a microtubule to the coverslip, and adapt an earlier bending analysis (Gittes et al., 1993) to a cantilevered beam geometry. The attached side functions as a fixed reference point along the length of a microtubule. For stabilized microtubules the ends can be used as a reference and unattached microtubules can be studied (Gittes et al., 1993). Attaching one side of a microtubule has the advantage that it prevents rotation around its long axis, which in the case of intrinsically bent microtubules can introduce errors (Gittes et al., 1993). We applied automated image analysis methods for digitizing microtubule shape yielding ∼600 points along the microtubule length. Due to the large amount of points used in the calculation of mode amplitudes, digitization errors averaged out to a large degree allowing for the characterization of five bending modes (mode 1 until 5) above the noise level set by positional noise in the quantification of microtubule shape (Appendix). In earlier work, ∼10 points were assigned manually (Gittes et al., 1993; Kurz and Williams, 1995) and the number of analyzed modes was limited to the first (for 30 μm long microtubules (Kurz and Williams, 1995)) or the first and second (for microtubules shorter than 60 μm (Gittes et al., 1993)). In our case, the first bending mode could not be used to measure rigidity because of its slow dynamics, and the fourth and fifth modes were discarded because they may be effected by mode smoothing and are more influenced by positional noise compared to the second and third modes. The first bending mode is very sensitive to flows of liquid in the sample, and therefore rejecting the first mode is not a disadvantage. Rigidity measurements that are based on deflections of the microtubule tip alone basically probe the first mode and are therefore more prone to errors. The accuracy of our rigidity estimates is limited by statistical errors due to a limited sample size and errors introduced during the estimation of the seed's attachment end. Because of the automated shape digitization, the statistics can in principal be improved by analyzing shape fluctuations at video rate. This approach is, of course, limited by the correlation time of the modes. The localization of the seed's attachment end may be improved by visualizing the biotin region of the seeds with fluorescent labels or other labels, e.g., gold-labeled streptavidin.
In conclusion, we have measured, with high accuracy, the flexural rigidity of elongating microtubules using an analysis of thermally induced bending modes. The bending amplitude of a constant part of the microtubule's length was analyzed in terms of “analyzed-length” modes. We found that the correlation time of bending modes levels off toward ∼30 ms for higher order modes, suggesting that internal friction within the microtubule plays an important role in shape fluctuations. By combining our current rigidity estimates on elongating microtubules with estimates we obtained by simpler methods, we find an apparent relation between average microtubule growth velocity and rigidity: the persistence length decreases by a factor of 2 (from 8 to 4 mm) between growth velocities of 1.2 and 2.7 μm/min. This finding suggests that the structural properties of microtubules, at least in vitro, may vary depending on their growth velocity. It is possible that the effect we observe is of transient nature and that fast-growing microtubules can “heal” their rigidity when left for some time in a background of tubulin dimers. Additional experiments are needed to test whether this is indeed the case.
Acknowledgments
We thank Cătălin Tănase, Marco Cosentino-Lagomarsino, and Dirk-Jan Spaanderman for helpful discussions and numerical calculations, and Dima Lukatsky for a critical reading of the manuscript.
This work is part of the research program of the Stichting voor Fundamenteel Onderzoek der Materie, which is financially supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek.
APPENDIX: SOURCES OF ERROR
Errors due to limited observation time
Both a theoretical estimate of τ1 and the experimentally observed slow dynamics of the first analyzed-length mode amplitude (Fig. 6) indicate that the experimentally used observation time of ∼10 min is insufficient to sample the complete variance of the first full length mode. Here, we will examine how a limited observation time affects rigidity estimates for different modes and different values of λ. A microtubule samples only a limited part of its phase space during a short observation time T. For a worst-case scenario, we will assume that the first full-length mode amplitude remains constant during observation, corresponding to 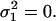 Higher order full-length modes are treated as being sampled completely, a situation that is roughly equivalent to τ1 < T < τ2 representing our experimental situation. For this situation, we calculate (Eq. 16) the anticipated measured analyzed-length variances over the observation time T as
Higher order full-length modes are treated as being sampled completely, a situation that is roughly equivalent to τ1 < T < τ2 representing our experimental situation. For this situation, we calculate (Eq. 16) the anticipated measured analyzed-length variances over the observation time T as
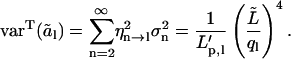 |
(A1) |
An inaccurate estimate of the persistence length (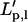 ) is obtained when the measured variance is used instead of the long-term variance
) is obtained when the measured variance is used instead of the long-term variance 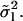 Comparison with Eq. 16 yields
Comparison with Eq. 16 yields
 |
(A2) |
This ratio is plotted for the first four modes for different values of λ in Fig. 11. As expected, the first mode estimate 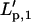 deviates as much as 100% from Lp. The second mode estimate, however, deviates at most 10% from Lp (for λ ≈ 3), and discrepancies are even smaller for higher modes. The second and higher modes are thus hardly affected by an incomplete sampling of the first mode for all experimentally used values of λ (1 < λ <2). In an experimental setting, the sampled first mode variance will be larger than zero and realistic errors will be even less. Second and higher mode values are thus valid rigidity estimates.
deviates as much as 100% from Lp. The second mode estimate, however, deviates at most 10% from Lp (for λ ≈ 3), and discrepancies are even smaller for higher modes. The second and higher modes are thus hardly affected by an incomplete sampling of the first mode for all experimentally used values of λ (1 < λ <2). In an experimental setting, the sampled first mode variance will be larger than zero and realistic errors will be even less. Second and higher mode values are thus valid rigidity estimates.
FIGURE 11.
The ratio 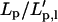 (Eq. A2), calculated for the first four mode numbers as a function of λ.
(Eq. A2), calculated for the first four mode numbers as a function of λ.
Errors caused by positional noise
Errors are made during the quantification of microtubule shape. This “positional noise” is expected to be only significant for higher modes for which thermally induced mode amplitudes are small and comparable in size to positional errors. A complete treatment of this kind of measurement error is given by Gittes et al. (1993). Here a slightly different treatment (not shown) is necessary because microtubule shape is parameterized as y(s) instead of θ(s) as in Gittes et al. (1993). The additional variance that is introduced by positional noise can be shown to be a mode number-independent constant value. The experimentally measured mode variance (Fig. 7) indeed levels off to a constant value for higher modes. This level is low enough to not have significantly influenced the second and third mode persistence length estimates, which were used for our final persistence length values. Modes 6 and higher are clearly influenced by noise, but modes 4 and 5 are still reasonably above the noise level. Possibly, the noise level can be decreased further if more advanced image tracing routines are developed. Subpixel resolution can in principal be achieved if the whole convoluted intensity curve is used to find the microtubule position with a fitting algorithm (Danuser et al., 2000). Currently only the position of the maximum in the curve is used. Additional positional noise can be caused by sample drift during observation, which was visible for 40% of the analyzed microtubules. For these cases we tracked the position of a fixed point in the sample to correct the drift. The residual shift after correction has a standard deviation on the order of half a pixel (25 nm). The additional mode variance that is caused by such sample shift along the y axis can be calculated using Eq. 10. The variances of mode 1 until 6 are increased by 0.1, 1, 3, 6, 9, and 14%, respectively (calculated for Lp = 5 mm). Persistence length estimates based on these modes will therefore be underestimated by a similar percentage for 40% of the analyzed microtubules. The use of a more stable microscope construction will help to further reduce measurement errors. The above analysis shows that positional noise has not significantly affected our final rigidity estimates that are based on the second and third mode.
Errors due to an uncertainty in the location of the seed's attachment end
The transition point between the part of the microtubule that is clamped and the free part, i.e., the seed's attachment end, could be estimated best by observing thermal fluctuations near the seed. Errors are, however, made in the assignment of this position causing an over- or underestimation of the analyzed length (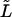 ) by
) by  and a miscalculation of the persistence length. We estimated this type of error using methods similar to Eqs. 10, 14, 15, and 16. The integral bounds in Eq. 10 were however changed by
and a miscalculation of the persistence length. We estimated this type of error using methods similar to Eqs. 10, 14, 15, and 16. The integral bounds in Eq. 10 were however changed by 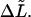 The calculated relative error was approximately
The calculated relative error was approximately 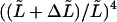 for the second mode estimate and slightly smaller than
for the second mode estimate and slightly smaller than 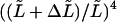 for higher modes. Errors on the seed's attachment end localization are not likely to be larger than 0.5 μm. Therefore, we take for the uncertainty (or standard deviation) on
for higher modes. Errors on the seed's attachment end localization are not likely to be larger than 0.5 μm. Therefore, we take for the uncertainty (or standard deviation) on 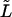 a value of 0.4 μm, which for a typical analyzed length of 30 μm results in a relative uncertainty in the persistence length of ∼5%.
a value of 0.4 μm, which for a typical analyzed length of 30 μm results in a relative uncertainty in the persistence length of ∼5%.
Errors induced by background flow
Small, undetected flows of liquid in the sample may induce nonthermal bending, especially if one side of the microtubule is fixed to the surface (Mickey and Howard, 1995). By closing the sides of our samples, we tried to avoid currents related to the evaporation of fluid. Here we try to estimate the consequences of possible currents by looking at the effect of a uniform flow field with velocity vflow in a direction perpendicular to microtubule. The deformation introduced by such a field was estimated previously (Venier et al., 1994) in the small angle limit,
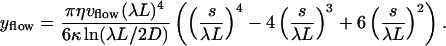 |
(A3) |
This shape can be projected onto the functions Wl(s/L) to obtain the induced mode amplitudes ãl, flow. Fluctuations in flow amplitude vvlow can be shown to induce additional variance in mode amplitudes that roughly follow a 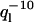 mode dependency. A mode analysis can thus discriminate between liquid currents and thermally induced bending (
mode dependency. A mode analysis can thus discriminate between liquid currents and thermally induced bending (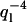 mode dependency). Although not all currents are necessarily uniform, it is clear that the observed mode dependency of variances in Fig. 7, a and b, were primarily caused by thermal bending and not background flow.
mode dependency). Although not all currents are necessarily uniform, it is clear that the observed mode dependency of variances in Fig. 7, a and b, were primarily caused by thermal bending and not background flow.
Marcel E. Janson's present address is Dept. of Cell and Developmental Biology, University of Pennsylvania School of Medicine, Philadelphia, PA 19104-6058 USA.
References
- Alberts, B., A. Johnson, J. Lewis, M. Raff, K. Roberts, and P. Walter. 2002. Molecular Biology of the Cell, 4th ed. Garland Science, New York.
- Ashkin, A. 1997. Optical trapping and manipulation of neutral particles using lasers. Proc. Natl. Acad. Sci. USA. 94:4853–4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris, L., D. Gard, P. T. Tran, and H. P. Erickson. 2001. XMAP215 is a long thin molecule that does not increase microtubule stiffness. J. Cell Sci. 114:3025–3033. [DOI] [PubMed] [Google Scholar]
- Chretien, D., and S. D. Fuller. 2000. Microtubules switch occasionally into unfavorable configurations during elongation. J. Mol. Biol. 298:663–676. [DOI] [PubMed] [Google Scholar]
- Chretien, D., F. Metoz, F. Verde, E. Karsenti, and R. H. Wade. 1992. Lattice-defects in microtubules—protofilament numbers vary within individual microtubules. J. Cell Biol. 117:1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, A. E., and L. Mahadevan. 2003. Kinks, rings, and rackets in filamentous structures. Proc. Natl. Acad. Sci. USA. 100:12141–12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danuser, G., P. T. Tran, and E. D. Salmon. 2000. Tracking differential interference contrast diffraction line images with nanometre sensitivity. J. Microsc.-Oxf. 198:34–53. [DOI] [PubMed] [Google Scholar]
- de Pablo, P. J., I. A. T. Schaap, and C. F. Schmidt. 2003. Observation of microtubules with scanning force microscopy in liquid. Nanotechnology. 14:143–146. [Google Scholar]
- Desai, A., and T. J. Mitchison. 1997. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13:83–117. [DOI] [PubMed] [Google Scholar]
- Dogterom, M., and B. Yurke. 1997. Measurement of the force-velocity relation for growing microtubules. Science. 278:856–860. [DOI] [PubMed] [Google Scholar]
- Felgner, H., R. Frank, J. Biernat, E. M. Mandelkow, E. Mandelkow, B. Ludin, A. Matus, and M. Schliwa. 1997. Domains of neuronal microtubule-associated proteins and flexural rigidity of microtubules. J. Cell Biol. 138:1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felgner, H., R. Frank, and M. Schliwa. 1996. Flexural rigidity of microtubules measured with the use of optical tweezers. J. Cell Sci. 109:509–516. [DOI] [PubMed] [Google Scholar]
- Fygenson, D. K., M. Elbaum, B. Shraiman, and A. Libchaber. 1997. Microtubules and vesicles under controlled tension. Phys. Rev. E. 55:850–859. [Google Scholar]
- Gittes, F., E. Meyhofer, S. Baek, and J. Howard. 1996. Directional loading of the kinesin motor molecule as it buckles a microtubule. Biophys. J. 70:418–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittes, F., B. Mickey, J. Nettleton, and J. Howard. 1993. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J. Cell Biol. 120:923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, H., J. Howard, and V. Vogel. 2002. A piconewton forcemeter assembled from microtubules and kinesins. Nano Lett. 2:1113–1115. [Google Scholar]
- Howard, J. 2001. Mechanics of Motor Proteins and the Cytoskeleton. Sinauer Associates, Sunderland, MA.
- Hunt, A. J., F. Gittes, and J. Howard. 1994. The force exerted by a single kinesin molecule against a viscous load. Biophys. J. 67:766–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson, M. E., and M. Dogterom. 2004. Scaling of microtubule force-velocity curves obtained at different tubulin concentrations. Phys. Rev. Lett. 92:248101. [DOI] [PubMed] [Google Scholar]
- Janson, M. E., M. E. de Dood, and M. Dogterom. 2003. Dynamic instability is regulated by force. J. Cell Biol. 161:1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson, M. E. 2002. Force generation by growing microtubules. PhD thesis. Leiden University, Leiden, The Netherlands.
- Kis, A., S. Kasas, B. Babic, A. J. Kulik, W. Benoit, G. A. D. Briggs, C. Schonenberger, S. Catsicas, and L. Forro. 2002. Nanomechanics of microtubules. Phys. Rev. Lett. 89:248101. [DOI] [PubMed] [Google Scholar]
- Kurachi, M., M. Hoshi, and H. Tashiro. 1995. Buckling of a single microtubule by optical trapping forces - direct measurement of microtubule rigidity. Cell Motil. Cytoskeleton. 30:221–228. [DOI] [PubMed] [Google Scholar]
- Kurz, J. C., and R. C. Williams. 1995. Microtubule-associated proteins and the flexibility of microtubules. Biochemistry. 34:13374–13380. [DOI] [PubMed] [Google Scholar]
- Landau, L. D., and E. M. Lifshitz. 1986. Theory of elasticity. Pergamon, New York.
- Mickey, B., and J. Howard. 1995. Rigidity of microtubules is increased by stabilizing agents. J. Cell Biol. 130:909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier, M. G., and J. F. Marko. 2002. Effect of internal friction on biofilament dynamics. Phys. Rev. Lett. 88:228103. [DOI] [PubMed] [Google Scholar]
- Reif, F. 1965. Statistical Physics. McGraw-Hill, New York.
- Takasone, T., S. Juodkazis, Y. Kawagishi, A. Yamaguchi, S. Matsuo, H. Sakakibara, H. Nakayama, and H. Misawa. 2002. Flexural rigidity of a single microtubule. Jpn. J. Appl. Phys. 41:3015–3019. [Google Scholar]
- Venier, P., A. C. Maggs, M.-F. Carlier, and D. Pantaloni. 1994. Analysis of microtubule rigidity using hydrodynamic flow and thermal fluctuations. J. Biol. Chem. 269:13353–13360. [PubMed] [Google Scholar]
- Walker, R. A., E. T. Obrien, N. K. Pryer, M. F. Soboeiro, W. A. Voter, H. P. Erickson, and E. D. Salmon. 1988. Dynamic instability of individual microtubules analyzed by video light-microscopy - rate constants and transition frequencies. J. Cell Biol. 107:1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggins, C. H., D. Riveline, A. Ott, and R. E. Goldstein. 1998. Trapping and wiggling: elastohydrodynamics of driven microfilaments. Biophys. J. 74:1043–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]