Abstract
Force generation in several types of cell motility is driven by rapidly elongating cytoskeletal filaments that are persistently tethered at their polymerizing ends to propelled objects. These properties are not easily explained by force-generation models that require free (i.e., untethered) filament ends to fluctuate away from the surface for addition of new monomers. In contrast, filament end-tracking proteins that processively advance on filament ends can facilitate rapid elongation and substantial force generation by persistently tethered filaments. Such processive end-tracking proteins, termed here filament end-tracking motors, maintain possession of filament ends and, like other biomolecular motors, advance by means of 5′-nucleoside triphosphate (NTP) hydrolysis-driven affinity-modulated interactions. On-filament NTP hydrolysis/phosphate release yields substantially more energy than that required for driving steady-state assembly/disassembly of free filament ends (i.e., filament treadmilling), as revealed by an energy inventory on the treadmilling cycle. The kinetic and thermodynamic properties of two simple end-tracking mechanisms (an end-tracking stepping motor and a direct-transfer end-tracking motor) are analyzed to illustrate the advantages of an end-tracking motor over free filament-end elongation, and over passive end-trackers that operate without the benefit of NTP hydrolysis, in terms of generating force, facilitating rapid monomer addition, and maintaining tight possession of the filament ends. We describe an additional cofactor-assisted end-tracking motor to account for suggested roles of cofactors in the affinity-modulated interactions, such as profilin in actin-filament end-tracking motors and EB1 in microtubule end-tracking motors.
INTRODUCTION
Actin polymerization produces the protrusive forces needed for cell crawling and the intracellular propulsion of organelles and certain microbial pathogens (Bray, 2001). Microtubule polymerization/depolymerization is likewise responsible for chromosome alignment and locomotion. The thermodynamic driving force for force-generation in these processes has been widely thought to be the free energy change of monomer addition to free filament ends (Hill, 1981; Theriot, 2000), an assumption which underlies the well-known elastic-Brownian ratchet model for force generation (Mogilner and Oster, 1996, 2003b; Peskin et al., 1993). Essential features of the elastic-Brownian ratchet are 1), the free energy change of monomer addition to free filament ends is the energy source for force generation; 2), thermal fluctuations of the free filament end away from the surface is required for monomer addition; and 3), elongation of filaments oriented normal to the surface is kinetically prohibited. The Brownian ratchet mechanism therefore does not allow rapid elongation of nearly perpendicular filaments that are tethered to the object being pushed by their elongating (+)-ends, or substantial force generation from elongation in low free monomer concentrations near the critical concentration of monomer addition to free filament (+)-ends.
One important example of actin-based motility where elongating filaments appear to be tethered is the intracellular propulsion of Listeria monocytogenes, which requires only a single protein, ActA, on the bacterial surface. ActA binds to vasodilator-stimulated phosphoprotein (VASP), which forms a linkage between ActA and the actin filaments (Niebuhr et al., 1997). Several lines of evidence suggest the association between ActA and filaments ends is persistent and capable of supporting strong tensile or torsional forces over many cycles of monomer addition. Cameron et al. (2001) showed that sufficiently small (50-nm) beads grew actin tails consisting of a single filament attached to the bead, which, if untethered or cyclically detaching during monomer addition, would have quickly diffused away in the time required to add another monomer. Kuo and McGrath (2000) observed Listeria trajectories consisting of monomer-sized steps and very small fluctuations, a finding that was interpreted as evidence for persistent association of the filament-VASP interaction on taut filaments under high tension over several consecutive cycles of monomer addition. Consistent with this interpretation are observations of vesicle deformation by Upadhyaya et al. (2003) and Giardini et al. (2003), who both interpreted the teardrop shape of ActA-coated vesicles undergoing actin-based motility as resulting from simultaneous growth of actin filaments in regions of both high compression (up to ∼10 pN/filament) and high tension (up to ∼20 pN/filament). Both groups reported that the vesicle-surface-bound ActA co-localized with filament ends during motility, suggesting that ActA maintains its association with filament ends during filament elongation. Finally, Robbins and Theriot (2003) explained longitudinal rotational motion of Listeria as consequence of torque generated by adding monomers to (+)-ends of (helical) filaments, again requiring some persistent association with rotationally noncompliant surface-bound ActA for the torque to be sustained.
Mogilner and Oster (2003a) recently offered the idea that filament tethering results from transient ActA binding to filament-bound Arp2/3 complex during Arp2/3-mediated nucleation of new filament branches. To our knowledge, however, no experimental evidence has been offered to support tethering by Arp2/3. Moreover, Brieher et al. (2004) clearly demonstrated that Arp2/3 complex is unnecessary for persistent association of elongating filament ends with motile Listeria. Tethering by Arp2/3-ActA binding also does not explain persistent monomer-sized steps or rotational motion observed in Listeria trajectories, which are observations that suggest the tethering interaction is maintained during cycles of monomer addition.
Very similar (+)-end-binding behavior has been demonstrated for formins, the cytoskeletal proteins involved in stress-fiber and contractile-ring formation (Zigmond et al., 2003). Because of formin's ability to block capping protein yet allow polymerization, it has been described as a “leaky cap” which can maintain possession of filament (+)-ends over many cycles of monomer (Zigmond et al., 2003). The tracking of single actin filament (+)-ends by formins during polymerization of long actin filaments has been recently observed directly (Higashida et al., 2004; Zigmond, 2004). Similar processive properties are observed for the bacterial actin-like protein, ParM, which is a member of a superfamily of ATPases that includes actin and MreB (Bork et al., 1992; van den Ent et al., 2001, 2002). ParM forms actin-like filamentous structures that extend the length of bacterial cells and the resulting filament dynamics are necessary for segregation of DNA by binding protein ParR and parC, a gene sequence required for ParR binding. Analogous to tethered actin elongation, polymerization of ATP-ParM generates the mechanical force that drives separation and subsequent movement of plasmid molecules in bacteria, while maintaining a continual attachment with the plasmid at the elongating filament end.
In a manner similar to actin, microtubule plus-ends are capable of pushing while tightly bound to the kinetochore during cell mitosis. Kinetochores bind specifically to GTP-containing MT (+)-ends (Severin et al., 1997), and several proteins have been identified in the kinetochore to bind microtubule (+)-ends and to participate in force generation (Schuyler and Pellman, 2001). Of particular interest is EB1 a kinetochore-associated protein, which concentrates at elongating microtubule (+)-ends and promotes microtubule assembly and stabilization (Bu and Su, 2001; Tirnauer and Bierer, 2000; Tirnauer et al., 2002a). EB1 readily associates with the elongating GTP-rich (+)-ends (Schroer, 2001) but dissociates uniformly along the length of assembled microtubules (Tirnauer et al., 2002b). How GTP hydrolysis microtubules regulates EB1 localization remains unclear; co-polymerization of EB1-tubulin-GTP complexes from solution to (+)-ends appears unlikely given the much lower (∼100×) intracellular concentration of EB1 relative to tubulin (Tirnauer et al., 2002a,b). EB1 may be recruited to (+)-ends by adenomatous polyposis coli (APC) protein (Fodde et al., 2001a; Mimori-Kiyosue et al., 2000a) (other kinetochore proteins), a behavior that resembles the concentration of profilin near actin filament ends by ActA/VASP at Listeria surfaces (Kang et al., 1997; Southwick and Purich, 1996). Notably, APC is a multimeric protein (Joslyn et al., 1993) that binds both to EB1 (Mimori-Kiyosue et al., 2000b) and to microtubule (+)-ends (Kaplan et al., 2001).
To account for the rapid elongation and substantial force generation by tethered filaments—properties not easily explained by the Brownian ratchet model—we define and analyze the thermodynamic and kinetic properties of cytoskeletal filament end-tracking motors, which use available energy of 5′-nucleoside triphosphate (NTP) hydrolysis to maintain processive association with filament ends. This article expands and generalizes ideas we previously offered (Dickinson and Purich, 2002) in the actoclampin model for clamped-filament elongation, which explained stepwise motion and small fluctuations observed during Listeria motility (Kuo and McGrath, 2000) in terms of an affinity-modulated sliding clamp whose release and advancement on filament ends is driven by filament-bound ATP hydrolysis. We now show that modulating monomer binding in actin-filament and microtubule assembly/disassembly reactions (and likewise treadmilling) requires only a small fraction of the overall free energy of filament-bound NTP hydrolysis. We also analyze simple mechanisms to illustrate how this energy could be transduced into work by two simple types of enzymatic cycles that involve hydrolysis-induced affinity modulation of tracking protein interactions with the filament end in a manner that facilitates rapid filament elongation and force generation, while maintaining tight possession of the filament end to the surface. These mechanisms share several advantageous properties for force generation, which include 1), maintaining a strong continuous possession of the filament end to the propelled object during cycles of monomer addition; 2), harnessing a portion of the energy of filament-bound NTP hydrolysis to yield substantially more energy for work than provided by the free energy of monomer addition alone; and 3), allowing elongation and force generation, even when the filament is oriented perpendicular to the surface.
BIOENERGETICS OF FILAMENT ELONGATION AND TREADMILLING
In the Elastic Brownian Ratchet Model (Mogilner and Oster, 1996) or similar models relying on force generation by free filament ends (Hill, 1981), the pushing filament ends must be unattached to allow them to fluctuate away from a surface to incorporate new NTP-bound monomers (MT), thereby incrementally increasing their flexural force on the surface. However, since the monomer concentration above the (+)-end critical concentration ([MT](+)-crit) is the driving force for force generation, the mechanical work performed by monomer addition cannot exceed the free energy of monomer addition, 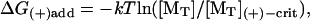 where kT = 4.1 pN/nm is the thermal energy (k is the Boltzmann constant, T is the absolute temperature) and [MT](+)-crit is ∼0.1 μM for actin and 0.03 μM for tubulin (Howard, 2001). See Appendix B for definitions of symbols. Under typical intracellular (unsequestered) monomer concentrations (∼0.1–0.5 μM for actin-ATP; Weber et al., 1992, and ∼10 μM for tubulin-GTP; Howard, 2001),
where kT = 4.1 pN/nm is the thermal energy (k is the Boltzmann constant, T is the absolute temperature) and [MT](+)-crit is ∼0.1 μM for actin and 0.03 μM for tubulin (Howard, 2001). See Appendix B for definitions of symbols. Under typical intracellular (unsequestered) monomer concentrations (∼0.1–0.5 μM for actin-ATP; Weber et al., 1992, and ∼10 μM for tubulin-GTP; Howard, 2001), 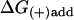 is ∼1–2 kT for actin-ATP and ∼5–6 kT for tubulin-GTP. Importantly, an upper bound on the intracellular [MT] is set by its (–)-end critical concentration ([MT](−)-crit ∼0.6 μM for actin), above which, NTP-containing subunits would accumulate on the (−)-end, thereby stabilizing the (−)-ends against disassembly and restricting the recycling of monomers needed for sustained filament assembly. When monomer addition to (+)-ends is the sole energy source for force generation, the thermodynamic maximum on the force that could be generated in the direction of elongation is ∼2–3 pN for actin filaments (two subunits added per 5.4-nm step) and ∼40 pN for microtubules (13 subunits added per 8-nm step). Importantly, the maximum achievable work by actin·ATP monomer addition to free filament ends has been commonly overestimated in published models (Abraham et al., 1999; Mogilner and Edelstein-Keshet, 2002; Mogilner and Oster, 2003a,b; Theriot, 2000) by assuming profilin-actin is equivalent to G-actin in its interactions with filament ends, thereby neglecting the capping of profilin and/or the weaker affinity of profilin·actin for filament ends (Kang et al., 1999; Kinosian et al., 2002; Pring et al., 1992). Profilin's catalytic role in filament elongation is considered in more detail below.
is ∼1–2 kT for actin-ATP and ∼5–6 kT for tubulin-GTP. Importantly, an upper bound on the intracellular [MT] is set by its (–)-end critical concentration ([MT](−)-crit ∼0.6 μM for actin), above which, NTP-containing subunits would accumulate on the (−)-end, thereby stabilizing the (−)-ends against disassembly and restricting the recycling of monomers needed for sustained filament assembly. When monomer addition to (+)-ends is the sole energy source for force generation, the thermodynamic maximum on the force that could be generated in the direction of elongation is ∼2–3 pN for actin filaments (two subunits added per 5.4-nm step) and ∼40 pN for microtubules (13 subunits added per 8-nm step). Importantly, the maximum achievable work by actin·ATP monomer addition to free filament ends has been commonly overestimated in published models (Abraham et al., 1999; Mogilner and Edelstein-Keshet, 2002; Mogilner and Oster, 2003a,b; Theriot, 2000) by assuming profilin-actin is equivalent to G-actin in its interactions with filament ends, thereby neglecting the capping of profilin and/or the weaker affinity of profilin·actin for filament ends (Kang et al., 1999; Kinosian et al., 2002; Pring et al., 1992). Profilin's catalytic role in filament elongation is considered in more detail below.
A long-held view is that NTP hydrolysis only drives “treadmilling,” the steady-state monomer flux observed when F-actin or microtubules undergo hydrolysis-dependent, opposite-end assembly/disassembly, which arises from monomer·NTP's high affinity for the (+)-end and monomer·NDP's low affinity for the (−)-end (Wegner and Engel, 1975). However, an inventory on the free energy changes in the assembly/disassembly cycle (illustrated for actin in Fig. 1) shows that most of the free energy of net NTP-to-NDP conversion is released upon or after filament-bound NTP hydrolysis, rather than in the treadmilling cycle where the free energy of monomer addition, release, and nucleotide exchange is relatively small. The additivity principle requires that the free energy changes of each step (i.e., MT addition to (+)-ends, hydrolysis of filament-bound nucleotide, phosphate release, loss of NDP-monomer (or MD) from (−)-ends) sum to the net free energy of NTP hydrolysis:
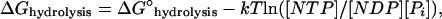 |
where 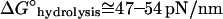 or 11–13 kT (Howard, 2001) is the standard-state free energy change under intracellular conditions, such that
or 11–13 kT (Howard, 2001) is the standard-state free energy change under intracellular conditions, such that
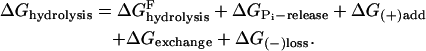 |
(1) |
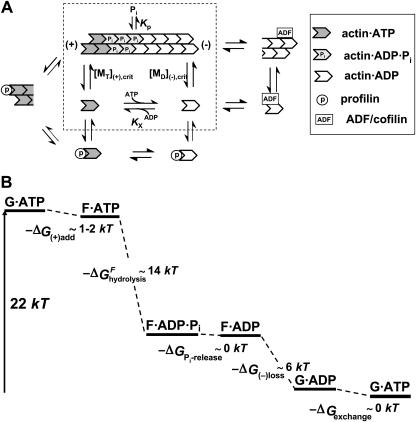
FIGURE 1.
Bioenergetics of actin filament dynamics and treadmilling. (A) The key reactions in actin filament dynamics. Actin·ATP binds to actin filament (+)-ends (critical concentration [MT](+)-crit). Key reactions are enclosed in the dotted box: Filament-bound ATP hydrolyzes to form ADP·Pi. Phosphate dissociates reversibly (KP) from the filament, and Actin·ADP dissociates from the (−)-end (critical concentration, [MD](−)-crit). Also shown are the roles of profilin in catalyzing monomer addition and ATP/ADP exchange, and of ADF/cofilin in catalyzing depolymerization at (−)-ends. Consistent with the principle of detailed balance, the known catalyzing properties of these actin-binding proteins do not alter the net free energy change in the cycle going from (−)-end bound actin·ADP to (+)-end bound actin·ATP. (B) The free energy changes in the treadmilling steps, with one ATP molecule consumed per monomer (ΔGhydrolysis ∼ −2 kT). The largest energy decrease occurs at ATP hydrolysis to form ADP·Pi on the filament end, a reaction releasing ∼14 kT, but that plays no role in treadmilling of filaments in solution. This available energy is proposed to facilitate monomer addition and force generation when (+)-end is coupled to a filament end-tracking motor.
Each of the contributing free energy changes are listed below:
Addition of MT to (+)-end: ΔG(+)add = −kT ln([MT]/[MT](+)-crit).
Hydrolysis of filament-bound NTP:
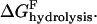
Phosphate release:
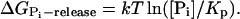
Loss of MD from (−)-end: ΔG(−)loss = kT ln([MD]/[MD](−)-crit).
Nucleotide exchange: ΔGexchange = kT ln([MT][NDP]/[MD][NTP])−kTln Kx.
Isolating the hydrolysis ΔGhydrolysis on the left-hand side and combining the other terms yields
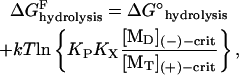 |
(2) |
for the energy released at the hydrolysis step, where KP is the equilibrium dissociation constant of reversible phosphate binding to NDP·filament subunits, and Kx is the equilibrium constant for the nucleotide exchange reaction. Based on literature values in Table 1, the combined hydrolysis and phosphate-release steps therefore yield ∼14 kT per monomer for actin and ∼11 kT for tubulin, accounting for a large fraction of the total ΔGhydrolysis ∼ −22 kT in each case, and which is substantially greater than the free energies of monomer addition to (+)-ends (∼1 kT for actin and ∼5.8 kT for tubulin). The magnitudes of the free energy changes of each step in actin's treadmilling cycle are illustrated in Fig. 1 B, assuming typical intracellular conditions ([Pi] ∼1 mM; [ATP]/[ADP] ∼20).
TABLE 1.
Values of equilibrium constants used in energy inventory
| Symbol | Reaction | Value | Reference |
|---|---|---|---|
| Actin | |||
| Kx | Nucleotide exchange | 6* | Kinosian et al. (1993) |
| Kp | Pi binding to filaments | 1.5 μM | Carlier and Pantaloni (1988) |
| [MT](+)-crit | MT addition to (+)-ends | 0.1 μM | Pollard et al. (2000) |
| [MD](−)-crit | MT addition to (−)-ends | 1.7 μM | Pollard et al. (2000) |
| Tubulin | |||
| Kx | Nucleotide exchange | 3* | Zeeberg and Caplow (1979) |
| Kp | Pi binding to filaments | 25 mM | Carlier et al. (1988) |
| [MT](+)-crit | MT addition to (+)-ends | 0.03 μM | Howard (2001) |
| [MD](−)-crit | MT addition to (−)-ends | 100 μM† | Howard (2001) |
Calculated from the ratio of measured equilibrium dissociation constants of nucleotide binding to the monomer, i.e., Kx = KNDP/KNTP.
Assumed equal to (+)-end critical concentration for tubulin·GDP.
Significantly, the actions of other protein species in catalyzing the treadmilling reactions (i.e., without altering the free energy changes) do not change the above conclusions. Examples illustrated in Fig. 1 A are profilin, which catalyzes AT P↔ADP exchange on actin monomers and increases the rate of actin assembly by increasing the net amount of ATP-actin in polymerization-competent form, and ADF/cofilin, which catalyzes disassembly at (−)-ends by binding ADP·actin subunits. Pantaloni and Carlier (1993) suggested that the profilin pathway uses some ATP hydrolysis energy to lower the critical concentration by a factor of 14 (a free energy reduction of 2.6 kT). On the other hand, other analyses (Kang et al., 1999; Pring et al., 1992) showed that both pathways are energetically equivalent, indicating that ATP hydrolysis has little or no effect. Kinosian et al. (2002) reported that filaments polymerized from nonmuscle actin·ATP had a more negative ΔG (by −3 kT per monomer) in the profilin-mediated pathway than in the direct pathway, but their conclusions are weakened by their additional finding that profilin-mediated assembly of actin·ADP (i.e., without ATP hydrolysis) also had a significantly more negative ΔG (by −2 kT). In any event, the profilin pathway appears to yield at most 2–3 kT per monomer, if any, additional energy in the assembly of free filament (+)-ends.
P↔ADP exchange on actin monomers and increases the rate of actin assembly by increasing the net amount of ATP-actin in polymerization-competent form, and ADF/cofilin, which catalyzes disassembly at (−)-ends by binding ADP·actin subunits. Pantaloni and Carlier (1993) suggested that the profilin pathway uses some ATP hydrolysis energy to lower the critical concentration by a factor of 14 (a free energy reduction of 2.6 kT). On the other hand, other analyses (Kang et al., 1999; Pring et al., 1992) showed that both pathways are energetically equivalent, indicating that ATP hydrolysis has little or no effect. Kinosian et al. (2002) reported that filaments polymerized from nonmuscle actin·ATP had a more negative ΔG (by −3 kT per monomer) in the profilin-mediated pathway than in the direct pathway, but their conclusions are weakened by their additional finding that profilin-mediated assembly of actin·ADP (i.e., without ATP hydrolysis) also had a significantly more negative ΔG (by −2 kT). In any event, the profilin pathway appears to yield at most 2–3 kT per monomer, if any, additional energy in the assembly of free filament (+)-ends.
ADF/cofilin also accelerates the treadmilling cycle by catalyzing actin·ADP disassembly from (−)-ends, but, consistent with its catalytic role without changing the free energy of net reaction (F-actin·ADP to G-actin·ADP), it does so without raising the actin·ATP concentration above its critical concentration for the filament's (−)-end (i.e., 0.6 μM) even at saturating concentrations of ADF/cofilin (Carlier et al., 1997). That is, solely catalytic factors like ADF/cofilin may increase the [MT] by accelerating depolymerization of NDP monomers, but not above the [MT](−)-crit where depolymerization is no longer energetically favorable.
The important conclusions from this energy inventory is that the free energy of monomer-NTP addition to (+)-ends for generating force is relatively small compared to the energy of NTP-to-NDP conversion on the filaments, especially for actin, and that there is a large fraction of the net free energy of NTP hydrolysis unused by treadmilling. Substantial force generation on the (∼10 pN) by rapidly elongating filaments requires a mechanism for capturing some of the hydrolysis energy. We show in the next section how a filament end-tracking motor can fulfill this function while also strongly tethering the elongating filament end to the motile surface.
FILAMENT END-TRACKING MOTORS
The two essential features of a filament end-tracking motor are 1), affinity-modulated interaction driven by hydrolysis of NTP on filament ends, and 2), multiple or multivalent interactions with the filament end to maintain its possession to the motile surface. A protein that binds preferentially to filament NTP subunits and releases from NDP (or NDP·Pi) subunits captures a portion of the available hydrolysis energy to stabilize NTP-bound terminal subunits, thereby increasing the net free energy change of monomer addition. For example, a 10,000× reduction in affinity upon hydrolysis would capture kT ln(104) = 9.2 kT of the available hydrolysis energy. If the end-binding protein is multivalent or multiple copies are bound to the propelled object (or motile surface) and capable of simultaneously operating upon multiple filament subunits, then a net strong interaction with the filament can be maintained even as individual end-tracking units release and rebind, thereby allowing the motor to advance processively with the elongating filament end. Such processive motion driven by NTP hydrolysis is a characteristic of a molecular motor, similar to actomyosin or kinesin, except that hydrolysis occurs on the filament rather than on the filament-binding protein.
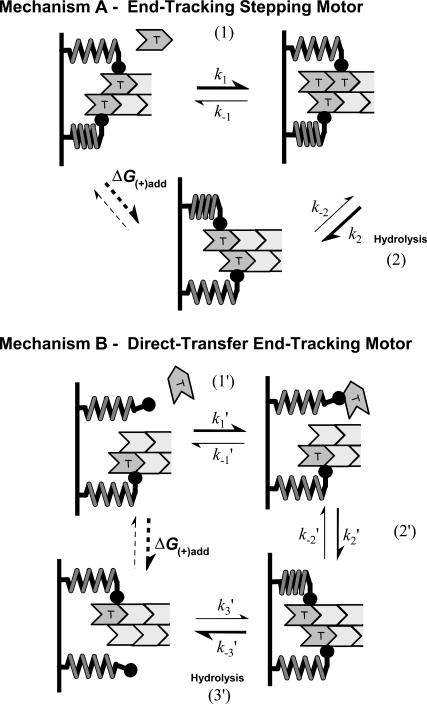
The two hypothetical reaction mechanisms in Fig. 2 demonstrate how end-tracking motors would facilitate elongation and force generation of an actin filament. These mechanisms are intended for conceptualization and may not accurately apply to any specific actin-filament (or microtubule) end-tracking motor. For simplicity, we assume that each protofilament interacts with a single tracking unit. Whether the end-tracking units are multimeric or simply bound independently to the motile surface does not alter the key properties of the end-tracking motor, which requires only that the units are co-localized on a filament end. Irreversible tethering of the end-tracking proteins to the motile surface is not essential for function of the end-tracking motor, but it does facilitate transduction of the force to the surface, as discussed below and shown in Appendix A. Moreover, reversible association/dissociation of the tracking proteins with the surface is not precluded by the reaction mechanisms, and an unbound multivalent end-tracking motor could even facilitate monomer addition to an untethered filament end (and simultaneously protect it from capping proteins).
FIGURE 2.
Hypothetical reaction schemes for cytoskeletal filament end-tracking motors. Each simple scheme shown requires two end-tracking units operating on an actin filament end. Actin·ATP subunits are shown in dark shading and actin·ADP (or actin·ADP·Pi) subunits are shown in light shading. The motor end-tracking units may be coupled together in a multimer or bound separately to the motile surface, as shown. Mechanism-A: end-tracking stepping motor (moving clockwise from the upper left). Step 1: The actin monomer binds to the filament end from solution. Step 2: The end-tracking unit advances to the new terminal subunit, in a manner that can be facilitated by hydrolysis. Mechanism-B: direct-transfer end-tracking motor (moving clockwise from the upper left). Step 1′: The actin monomer binds to the end-tracking unit. Step 2′: The end-tracking unit transfers the monomer to the filament end. Step 3′: Another end-tracking unit releases, in a manner that can be facilitated by ATP hydrolysis, which returns the system to the original state. In both Mechanisms, if hydrolysis were without effect, the principle of detailed balance would require that the net reaction would exhibit the same free energy change monomer addition to free filament ends, with critical concentration [MT](+)-crit and net free energy change, ΔG(+),add = −kT ln([MT]/[MT](+)-crit). With hydrolysis energy of the amount ɛ going to attenuate the affinity by the factor e−ɛ/kT, the net free energy change per assembled monomer is ΔG(+),add −ɛ.
Mechanism-A: End-tracking Stepping Motor
In Mechanism-A, the end-tracking motor consists of two tracking units that operate by releasing from a penultimate protofilament subunit after monomer addition to the filament end, followed by rebinding to the newly incorporated terminal subunit. One tracking unit remains bound during the release of the other unit, thereby maintaining at least one interaction at any instant. Because the net reaction is the addition of one monomer, the principle of detailed balance requires
 |
(3) |
where K1 ≡ k−1/k1, and K2 ≡ k−2/k2, when NTP hydrolysis has no effect on the reactions. As a consequence, strong binding of the tracking unit on terminal subunits also requires either 1), similarly strong binding interaction with penultimate subunits (i.e., K2 ∼1), or 2), weaker binding to the penultimate subunits (K2 ≪ 1) but a correspondingly weaker monomer binding to filament ends (K1 ≫ [MT](+)-crit). Detailed balance also requires that, absent another energy source, this reaction has the same free energy change as that of uncoupled monomer addition to a free filament end (i.e., ΔG1 + ΔG2 = ΔG(+)add = − kT ln[MT]/[MT](+)-crit). We refer to end-trackers operating without energetic coupling with hydrolysis under these constraints as “passive” end-trackers. The constraints can be circumvented, however, if hydrolysis of NTP on the penultimate subunit (triggered by monomer addition) attenuates the affinity of tracking unit to the penultimate subunit, thereby releasing and allowing it to quickly advance to the terminal subunit where it can rebind strongly. With this benefit of hydrolysis, the free energy of the net monomer-addition cycle is ΔG1 + ΔG2 = ΔG(+)add −ɛ, where ɛ is the portion of the hydrolysis energy going to attenuate the binding affinity by a factor of e−ɛ/kT. The constraints that the principle of detailed balance places on the thermodynamic favorability of the end-tracking reactions are summarized in Table 2 for the passive end-tracking stepping motor and an active motor that captures an amount ɛ of the hydrolysis energy in the net cycle of monomer addition.
TABLE 2.
Thermodynamic constraints on passive and active end-tracking reactions
| Mechanism-A: end-tracking stepping motor | |||
| Steps: | |||
| 1. Monomer binding from solution, ΔG1 = − kT ln([MT]/K1). | |||
| 2. Step of tracking unit to terminal subunit, ΔG2 = kT ln(K2). | |||
| Active motor: Hydrolysis energy ɛ facilitates Step 2. | |||
| Type | Energy | Equilibrium constants | Implications |
| Passive | ΔG1 + ΔG2 = ΔG(+),add ∼ O(−kT) | K1K2 = [MT](+)-crit | Favorable monomer binding ⇒ unfavorable stepping |
| Favorable stepping ⇒ unfavorable monomer binding | |||
| Active | ΔG1 + ΔG2 = ΔG(+),add−ɛ ≪ −kT | K1K2 = [MT](+)-crite−ɛ/kT | Monomer binding and stepping may both be favorable. |
| Mechanism-B: direct-transfer end-tracking motor | |||
| Steps: | |||
1′. Monomer binding from solution, 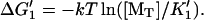
| |||
2′. Transfer of monomer to filament end, 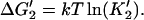
| |||
3′. Release of tracking unit, 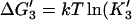 ). ). | |||
| Active motor: Hydrolysis energy ɛ facilitates Step 3 | |||
| Type | Energy | Equilibrium constants | Implications |
| Passive | 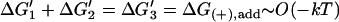 |
K1K2/K3 = [MT](+)-crit | Favorable monomer binding ⇒ unfavorable monomer transfer and/or tracker release |
| Favorable monomer transfer ⇒ unfavorable monomer binding and/or tracker release | |||
| Favorable tracking-unit release ⇒ unfavorable monomer binding and/or monomer transfer | |||
| Active | 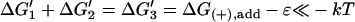 |
K1K2/K3 = [MT](+)-crite−ɛ/kT | Monomer binding, transfer, and tracking unit release may all be favorable. |
The following kinetic analysis of the end-tracking stepping motor (Mechanism-A) provides a quantitative perspective. Let ρ be the probability of the end-tracking protein being bound to the terminal subunit and (1–ρ) be the probability of it being bound to the penultimate subunit. For simplicity, we assume that binding to more distal subunits from the surface is sterically forbidden, and that the end-tracking unit is held in proximity to the filament end, such that the end-tracking protein must be bound to either the terminal or penultimate subunit. The steady-state balance on ρ is
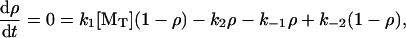 |
(4) |
which has the solution
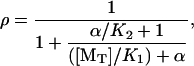 |
(5) |
where α ≡ k−2/k−1. The steady-state elongation rate is
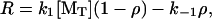 |
(6) |
which can be obtained explicitly by combining Eqs. 5 and 6 to find
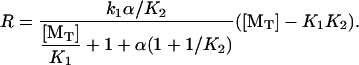 |
(7) |
This expression is general for cases with or without an affinity-modulation step. In the limit of rapid dissociation from the penultimate subunit (α ≫ 1, and K2 ≪ [MT] /K1), R approaches the monomer binding-rate-limited maximum, 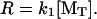 Absent an affinity-modulating effect from hydrolysis (i.e., “passive” end-tracker proteins), the equilibrium dissociation constants are constrained by the principle of detailed balance, i.e., [MT](+)-crit = K2K1, such that
Absent an affinity-modulating effect from hydrolysis (i.e., “passive” end-tracker proteins), the equilibrium dissociation constants are constrained by the principle of detailed balance, i.e., [MT](+)-crit = K2K1, such that
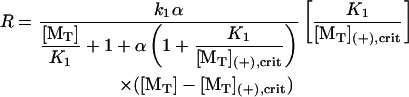 |
(8) |
for the elongation rate. Note that R is clearly negative when the monomer concentration is below the critical concentration ([MT] < [MT](+)-crit ), as expected. If, however, hydrolysis reduces the affinity of the tracking unit for the penultimate subunit, K2 could be greatly reduced by a factor, e−ɛ/kT, by capturing an amount of energy ɛ from the available hydrolysis energy (e.g., from the ∼14 kT available from hydrolysis on F-actin). Again, to attenuate affinity by a factor of 104 (i.e., K2 K1/[MT](+)-crit = 10−4), for example, ɛ would be 9.2 kT of the available hydrolysis energy.
Force generation by the End-tracking Stepping Motor
Now we consider polymerization of the filament against a force, F, exerted on the filament end. The net cycle to add one subunit of length d requires the amount of work Fd/2 (for two protofilaments). How the force would alter the kinetics depends on the force-dependent step. When the end-tracking units are sufficiently stiff, such that the monomer-addition step is unhindered by the force, K2 should increase with force by the factor, eFd/2kT (i.e., K2 = K2,0eFd/2kT where K2,0 ≡ K2(F = 0)), which accounts for the higher energy of the final state by the work increment Fd/2. Conversely, when the end-tracking units are very compliant, then the monomer addition step would require work and K1 would increase by the factor eFd/2kT. In either case (or a combination of the two), absent a hydrolysis effect, Eq. 6 implies the elongation by the passive end-tracking pathway is favored only when
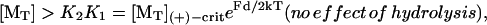 |
(9) |
such that the maximum work per monomer added (Fd/2) equals kT ln([MT]/[MT](+)-crit) for the passive end-tracking pathway, which is the same thermodynamic constraint limiting other Hill-type polymerization-force models (Hill, 1981). However, if an energy amount ɛ is captured from hydrolysis and used to modulate affinity (such that K2,0K1,0/[MT](+)-crit = e−ɛ/kT), the thermodynamic limit on elongation is instead
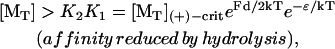 |
(10) |
such that now the maximum work per monomer added is kT ln([MT]/[MT](+)-crit) + ɛ for the end-tracking motor. Hence, elongation would be favored thermodynamically at lower monomer concentrations, and the maximum work increases by the amount of captured hydrolysis energy.
To compare the effect of force on the performance of this simple end-tracking motor, the force-dependent elongation rate is plotted in Fig. 3, shown with or without the benefit of hydrolysis. These rates are also compared to a Hill-type (or Brownian ratchet) rate equation for force-dependent monomer addition to filament ends, which is
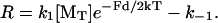 |
(11) |
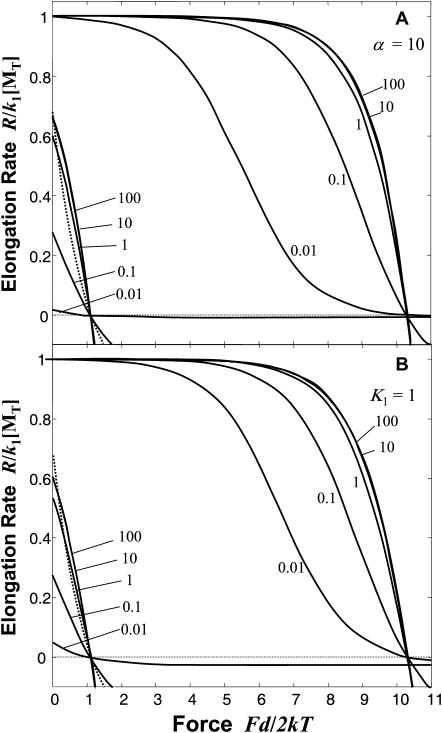
FIGURE 3.
Dimensionless elongation rate for the end-tracking stepping motor (Mechanism-A) versus dimensionless force. The elongation rate R, relative to the maximum rate k1[MT], is plotted as a function of dimensionless force, Fd/2kT, where k1 is the forward association rate constant in Step 1 of Mechanism-A (Fig. 2), [MT] is the actin monomer concentration, F is the force opposing the elongation, kT is the thermal energy, and d is the subunit length (5.4 nm for actin). For each plot, [MT]/[MT](+)-crit is set equal to 3, and 9.2 kT of the hydrolysis energy is captured to provide a 10−4 reduction in affinity for Step 2 in the Mechanism-A reaction cycle. The solid lines that intersect the zero line at Fd/2kT = ln(3) = 1.1 correspond to Mechanism-A without the benefit of hydrolysis (i.e., where K2,0 = [MT](+)-crit/K1), and those that intersect at 1.1 + 9.2 = 10.3 are for Mechanism-A with hydrolysis modulating the affinity (i.e., where K2,0 = 10−4[MT](+)crit/K1). (A) Curves are shown for various values of the equilibrium dissociation constant K1 at a fixed value of α = k−2/k−1 = 10. (B) Curves are shown for the various values of α at a fixed value of K1 = 1.0.
In the plots in Fig. 3, only K2 is assumed affected by force, and nominal parameters (i.e., ɛ = 9.2 kT, [MT] = 0.3 μM, and [MT](+)-crit = 0.1 μM) are assumed. Curves are shown with either K1 or α varied, with K2 set by Eq. 9 or Eq. 10. The advantages of the affinity attenuation step are clear from these plots: a much larger force can be generated by the end-tracking motor over the passive pathway, and its elongation rate is uninhibited over a larger range of forces, because the force-dependent step (Step 2) only becomes rate-limiting at large forces. Also, note that the thermodynamic stall forces are independent of the kinetic parameters, being determined only by [MT]/[MT](+)-crit and ɛ.
Mechanism-B: Direct-transfer End-tracking Motor
We now examine a second type of end-tracking motor, shown as Mechanism-B of Fig. 2. Here, monomers bind to the filament end-tracking unit and are then transferred to the filament end, with subsequent release of the end-tracking unit induced by hydrolysis. Let ρ again be the probability of the end-tracking unit being bound to the terminal filament subunit, and u be the probability of it being bound only to a monomer (i.e., not on the filament). The steady-state balance equations on ρ and u are
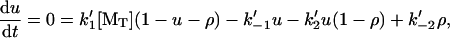 |
(12) |
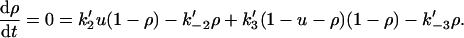 |
(13) |
The combined algebraic equation is cubic in ρ, but can be represented explicitly in u,
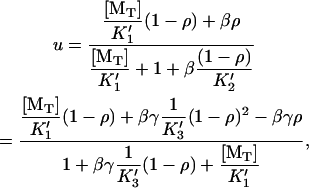 |
(14) |
where 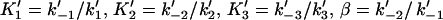 , and γ = k′−3/ k′−2. The elongation rate can be calculated from
, and γ = k′−3/ k′−2. The elongation rate can be calculated from
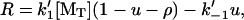 |
(15) |
where u and ρ are determined from Eq. 14, using a numeric solution for ρ. If this mechanism operates without the benefit of hydrolysis, the principle of detailed balance requires ΔG′1 + ΔG′2 + ΔG′3 = ΔG(+)add, such that K′3[MT](+)-crit = K′1K′2 for the passive end-tracking pathway. This condition disallows high affinity of the monomer for the tracking unit and for the filament end to be accompanied by low affinity of the tracking unit for the terminal subunit. However, if the hydrolysis energy accelerates the release step, then K′3 ≫ K′1K′2/[MT](+)-crit, and the net monomer-addition cycle becomes much more kinetically and energetically favorable. The thermodynamic constraints on passive and active direct-transfer end-tracking motors are also summarized in Table 2.
Force generation by Direct-transfer End-tracking Motor
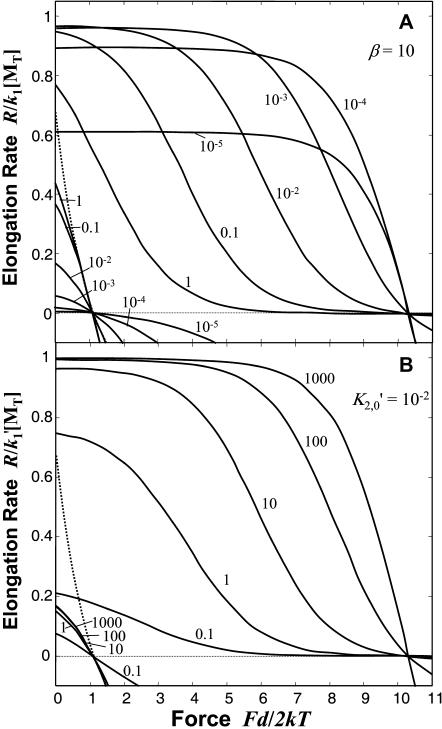
Calculation of the force effect on elongation rate again requires an assumption about the force-dependent kinetic step. For the direct-transfer pathway (Mechanism-B), it is reasonable to assume monomer binding to the tracking unit (Step 1′) is independent of force, such that force affects only the transfer and/or release steps (Steps 2′ and 3′) of the net cycle. The ratio K′2/K′3 must then increase by the factor eFd/2kT assuming K′1 is constant with F. Here we limit our examination to the case where only K′2 increases as K′2 = K′2,0eFd/2kT, which reflects inhibition of only the monomer-transfer step, and note that instead decreasing K′3 with F leads to similar results without altering the key conclusions. The resulting force-dependent kinetic behavior is plotted in Fig. 4 for various values of K′2,0 and β (again with ɛ = 9.2 kT, [MT] = 0.3 μM, and [MT](+)-crit = 0.1 μM), illustrating the advantage imparted by hydrolysis. The direct-transfer end-tracking pathway (Mechanism-B) has the same thermodynamic limits on the maximum work as the end-tracking stepping motor (Mechanism-A); that is, Fd/2 < kT ln([MT]/[MT](+)-crit) without affinity modulation, and Fd/2 < kT ln([MT]/[MT](+)-crit)+ɛ with affinity modulation). As illustrated in Fig. 4, the direct-transfer end-tracking motor also offers the same important advantages for force generation over free filament elongation, particularly when the force-dependent step is fast (reflected in small K′2,0 or large β): the maximum force is greater; the elongation rate is uninhibited by force over a large range; and at least one strong interaction is maintained on the other protofilament(s) while the tracking unit advances. The additional mechanical advantage of tethering the elongating working filament to the surface is considered in the Discussion and Appendix A.
FIGURE 4.
Dimensionless elongation rate for the direct-transfer end-tracking motor (Mechanism-B) versus dimensionless force. The elongation rate R, relative to the maximum rate 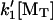 , is plotted as a function of dimensionless force, Fd/2kT, where k′1 is the forward association rate constant of Step 1′ in Mechanism-B (Fig. 2). (Other parameters are defined in the Fig. 3 caption.) Step 2′ is assumed the only force-dependent step (K′2 = K′2,0 e−Fd/2kT). As in Fig. 3, [MT]/[MT](+)-crit is set equal to 3, and the 9.2 kT of the hydrolysis energy is captured to provide a 10−4 reduction in affinity for Step 3 of Mechanism-B. The solid lines that intersect the zero line at Fd/2kT = ln(3) = 1.1 are for Mechanism-B without the benefit of hydrolysis (i.e., where K′3 = K′1K′2,0/[MT](+)-crit), and those that intersect at 1.1 + 9.2 = 10.3 are for Mechanism-A with hydrolysis modulating the affinity (i.e., where K′3 = 104 K′1K′2,0/[MT](+)crit). (A) Curves are for various shown values of the equilibrium constant K′20 with a fixed value of β = k′−2/k′−1 =10 and K′1 = 1.0. (B) Curves are for the various shown values of β with a fixed values of K′2,0 = 10−2 and K′1 = 1.0.
, is plotted as a function of dimensionless force, Fd/2kT, where k′1 is the forward association rate constant of Step 1′ in Mechanism-B (Fig. 2). (Other parameters are defined in the Fig. 3 caption.) Step 2′ is assumed the only force-dependent step (K′2 = K′2,0 e−Fd/2kT). As in Fig. 3, [MT]/[MT](+)-crit is set equal to 3, and the 9.2 kT of the hydrolysis energy is captured to provide a 10−4 reduction in affinity for Step 3 of Mechanism-B. The solid lines that intersect the zero line at Fd/2kT = ln(3) = 1.1 are for Mechanism-B without the benefit of hydrolysis (i.e., where K′3 = K′1K′2,0/[MT](+)-crit), and those that intersect at 1.1 + 9.2 = 10.3 are for Mechanism-A with hydrolysis modulating the affinity (i.e., where K′3 = 104 K′1K′2,0/[MT](+)crit). (A) Curves are for various shown values of the equilibrium constant K′20 with a fixed value of β = k′−2/k′−1 =10 and K′1 = 1.0. (B) Curves are for the various shown values of β with a fixed values of K′2,0 = 10−2 and K′1 = 1.0.
Compared to the end-tracking stepping motor (Mechanism-A), a motor operating by the direct-transfer pathway (Mechanism-B) would be at a disadvantage only if the linkage between the terminal subunit and the rest of the filament were to maintain the filament end's association with the motile surface. This disadvantage could be partially alleviated by additional binding interactions between the end-tracking unit and other subunits on filament. This problem could also be reduced if a greater number of end-tracking units were available for each protofilament, thus allowing prompt insertion of new monomers from other nearby end-tracker units after the release of a tracking unit from the filament. For example, VASP's tetrameric structure could supply a greater number of tracking units than two F-actin protofilaments.
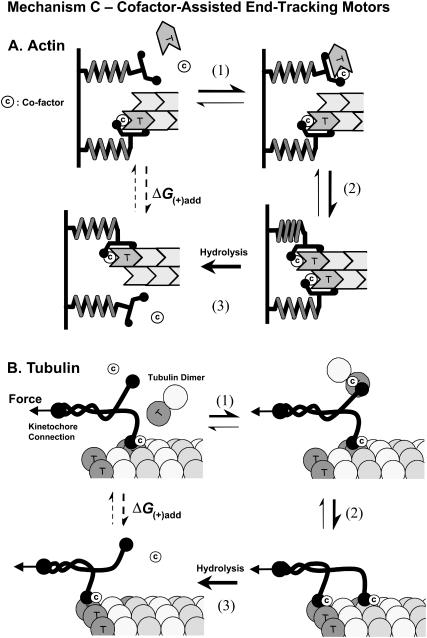
Mechanism-C: Cofactor-assisted End-tracking Motor
Considering the important but incompletely understood role of cofactors such as profilin in actin-based motility and EB1 in microtubule elongation in the kinetochore, we examine an additional mechanism that has a monomer-transfer step as in Mechanism-B, but which also involves a cofactor in the filament end-tracking complex (Fig. 1). Profilin and EB1 are similar in that both proteins attached to the motile object (e.g., profilin binds to formins and to VASP/ActA on the Listeria surface, and EB1 to proteins such as APC in the kinetochore), and both are associated with ends (+)-end only during elongation, yet they dissociate after monomer addition. In the latter sense, both EB1 and profilin act as soluble end-tracking proteins, even though EB1 can remain transiently bound to non-terminal subunits (Tirnauer et al., 2002b), whereas profilin apparently cannot. Profilin has also been found to “gate” monomer addition to actin filament ends tracked by yeast formin Cdc12p (Kovar et al., 2003), and inhibition of binding of profilin binding to VASP inhibits Listeria motility in vivo (Kang et al., 1997). Such properties suggest a potentially critical role of cofactors such as profilin or EB1 in end-tracking mechanisms beyond simply providing a higher concentration of monomers at filament ends.
The cofactor-assisted direct-transfer pathway (Mechanism-C) is shown in Fig. 5 for both actin-filament and microtubule polymerization (Fig. 5, parts A and B, respectively). In both cases, the cofactor and monomer bind the surface-bound end-tracking protein (either as a complex or independently), and the resulting ternary complex then ushers the monomer to the filament end. The additive bond energy between the three components stabilizes the complex until hydrolysis attenuates one or more of the bonds (i.e., between cofactor and monomer, between the filament and the end-tracker, and/or between the cofactor and the end-tracker). Note that the processive end-tracking could be maintained if motor monomer addition triggered NTP hydrolysis and release from the adjacent subunits (as illustrated in Fig. 5 A for actin) or from non-terminal subunits on the same protofilament (as illustrated in Fig. 5 B for tubulin). The cofactor may return immediately to solution, or remain transiently bound to the tracking unit or to the filament before dissociating later (as EB1 apparently does).
FIGURE 5.
Mechanism-C: cofactor assisted end-tracking motor filament. Cofactor assisted pathways are shown for (A) actin polymerization, and (B) tubulin polymerization. Step 1: The soluble cofactor and monomer bind together to the end-tracking unit. Step 2: The cofactor and monomer add to the filament end. Step 3: Another tracking unit and cofactor dissociate from the adjacent protofilament, in a step that can be facilitated by ATP hydrolysis energy to modulate the affinity of the cofactor and/or the tracking unit for the filament. The cofactor may return immediately to solution, or remain transiently bound to the tracking unit or to the filament.
Mechanism-C shows how a soluble cofactor such as profilin or EB1 can be critical in the monomer addition step and resultant force generation of an end-tracking complex without staying bound to the filament. Because Mechanism-C is functionally similar to Mechanism-B and has similar kinetic and thermodynamic properties, it will not be analyzed separately. Each critical step in the reaction cycle (1, 2, 3) has a counterpart in Mechanism-B (1′, 2′, 3′), but with ternary complexation/dissociation reactions in steps 1 and 3 of Mechanism-C replacing the corresponding binary reactions in steps 1′ and 3′ of Mechanism-B.
DISCUSSION
NTP hydrolysis-driven affinity modulation is a recurring theme in the action of molecular motors, solute-transporters, and even enzyme catalysis. In all known molecular motors (e.g., actomyosin, kinesin, dynein, etc.), the free energy liberated during myosin-bound ATP hydrolysis modulates the affinity between protein components to permit efficient and processive action. Likewise, active solute transporters utilize ATP hydrolysis to transit between high- and low-affinity transporter-solute interactions on opposite membrane faces to permit binding at low solute concentration and release at higher solute concentration. This theme also extends to enzyme catalysis, as exemplified by the case of ATP sulfurylase (ATP + 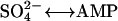 -sulfate + diphosphate), an enzyme that also hydrolyzes GTP (at a site other than the active site) to modulate the enzyme's affinity for sulfate ion, thus promoting catalysis even at low sulfate concentration (Leyh, 1999; Wei and Leyh, 1999). We now extend these principles to cytoskeletal filament assembly by filament end-tracking proteins.
-sulfate + diphosphate), an enzyme that also hydrolyzes GTP (at a site other than the active site) to modulate the enzyme's affinity for sulfate ion, thus promoting catalysis even at low sulfate concentration (Leyh, 1999; Wei and Leyh, 1999). We now extend these principles to cytoskeletal filament assembly by filament end-tracking proteins.
Driven by the small free energy of monomer addition to free filament ends (Hill, 1981), the Brownian ratchet mechanism (Mogilner and Oster, 1996, 2003b; Peskin et al., 1993) cannot explain rapid elongation and substantial force generation by tethered filaments. We propose that end-tracking motors, consisting of multivalent, modulated binding interactions between surface-tethered proteins (tracking units) and filament ends, define a new class of processive NTP-driven molecular motors. To illustrate this concept, two simple mechanoenzymatic reaction mechanisms were analyzed to show how affinity-modulated interactions with subunits at filament ends offer several advantages for filament elongation and force generation over monomer addition to free filament ends (i.e., the elastic-Brownian ratchet model).
There is a clear analogy between facilitated elongation with or without the aid of hydrolysis and facilitated diffusion by active versus passive membrane transporters. Active transporters utilize ATP to drive molecules against concentration (hence free energy) gradients, whereas passive transporters may provide a kinetic advantage to membrane transport, but they have the same thermodynamic limit (i.e., they cannot transport molecules up a concentration gradient).
Although there has been no definitive test of whether actin-based motility operates by a Brownian ratchet model or a filament end-tracking motor model, several observations and the properties of certain required proteins appear to be more consistent with the end-tracking motor hypothesis. First, filament end-tracking motors yield a substantially greater force per filament than possible by mechanisms driven by the free energy of monomer addition to free filament ends. For actin, we estimate up to 14 kT additional energy is available per monomer, potentially yielding up to 21 pN additional force per filament. In actin-based motility, the actin monomer concentration is constrained by the (−)-end critical concentration (<0.6 μM), which, without another energy source, constrains the maximum force that could be generated by Brownian ratchet mechanism in vivo to <2.7 pN, a value which is lower than many experimental estimates. (In principle, somewhat higher forces can be generated if the Brownian ratchet operates at larger angles of incidence, θ0, because the distance over which the work is performed is reduced by a factor cosθ0. However, as shown in Appendix A, free filaments oriented at glancing angles to the surface (large θ0) would be expected to buckle mechanically under relatively small forces.) Upadhyaya et al. (2003) estimated 10 pN/filament generated by filaments on the surface of ActA-coated vesicle during motility (i.e., away from equilibrium). The observed tight force balance implied by small fluctuations of motile Listeria (Kuo and McGrath, 2000) also suggests that large forces accumulate between pushing and pulling filaments. Noireaux et al. (2000) found that a force-generation model using ΔG(+)add = −14 kT yielded reasonably good comparison to measurements of the steady-state thickness of an actin gel growing on ActA-coated microspheres. Moreover, several applications of the Brownian ratchet model treated profilin-actin (p·MT) as equivalent to actin (e.g., Abraham et al., 1999; Mogilner and Edelstein-Keshet, 2002; Mogilner and Oster, 2003a), without accounting for profilin capping or the lower affinity for profilin-actin for filament ends, thereby making the equilibrium condition [p·MT] = [MT](+)-crit rather than [MT] = [MT](+)-crit. Such a treatment largely overestimates the maximum work of free filament elongation (which becomes ΔG(+)add = − kT ln([p·MT]/[MT](+)-crit) rather than ΔG(+)add = − kT ln([MT]/[MT](+)-crit)), because the affinity of profilin for actin (hence the ratio [p·MT]/[MT]) can be quite large. Thus, the reasonable experimental agreement claimed by models that treated [p·MT] as equivalent to [MT] was achieved by overestimating the driving force of monomer addition to free filament ends.
Second, filament end-tracking motors could maintain tight possession of the filament end to the surface, which keeps filament ends localized to the surface for force generation. Elongating kinetochore microtubules are held tightly to the kinetochore during mitosis, and actin-like-filament end-tracking proteins such as formin (Higashida et al., 2004) and ParR (van den Ent et al., 2002) have been directly shown to maintain possession of the elongating filament ends. Moreover, several independent observations also attest to this property in Listeria motility; i.e., (+)-ends elongate while continually tethered to surface-bound ActA without fluctuating away or detaching from the surface after each monomer-addition cycle. These observations include 1), monomer-sized steps of Listeria trajectories (Kuo and McGrath, 2000; McGrath et al., 2003); 2), single-filament tails observed by Cameron et al. (2001); 3), colocalization of membrane-bound ActA with filaments on motile vesicles (Giardini et al., 2003; Upadhyaya et al., 2003); 4), the inability of capping protein to inhibit intracellular Listeria motility (Laine et al., 1998), attributed to VASP's persistent association with filament (+)-ends (Bear et al., 2002); and 5), the apparent torque generated by elongating filament ends on the motile surface, as implied by Listeria rotation (Robbins and Theriot, 2003) and their clockwise helical trajectories (W. Zeile, F. Zhang, R. B. Dickinson, and D. L. Purich, unpublished findings). Such observations are at odds with a mechanism whereby possession of the motile surface is maintained by a subpopulation of filaments that are transiently attached to ActA via Arp2/3 complex only during filament nucleation/branching (Mogilner and Oster, 2003a; Samarin et al., 2003), or by cyclical binding/unbinding of the filament from VASP with each monomer addition to filament ends (as proposed by Laurent et al., 1999). Similarly, the fact that lamellipodial actin filaments appear to be consistently branched toward the leading edge (Verkhovsky et al., 2003), and not growing backward away from the leading edge, is difficult to explain by any model requiring elongation of only free filament ends.
Third, unlike the elastic-Brownian ratchet model, which requires filament ends to make excursions from the motile surface to free up space for monomer addition, processive end-tracking motors can allow unhindered addition of monomers to the filament end, by relying instead on spatial fluctuations of the tracking units to advance from subunit to subunit. The Brownian ratchet model anticipates that nonperpendicular orientation angles are optimal, because fluctuations away from the surface by stiff perpendicular filament ends are kinetically prohibited (Mogilner and Oster, 1996). However, filaments in filipodial extensions and Listeria tails are typically oriented normal to the surface, yet filipodial extension proceeds at rates up to 1 μm/s. Moreover, the end-tracking motors could allow the force-dependent reaction steps to be fast relative to the slower steps, such as monomer diffusion to the surface, thus allowing the filament elongation rate to remain unhindered by force up to several picoNewtons. This property may explain how a balance of large forces can accumulate between “pushing” and “pulling” filaments in Listeria motility (Kuo and McGrath, 2000) and with motility of phospholipid vesicles (Giardini et al., 2003; Upadhyaya et al., 2003).
Elongation of surface-tethered filament ends also has a mechanical advantage in terms of converting filament elongation into mechanical work, in that an untethered filament segment spanning the distance between the motile surface and the actin network will buckle at much lower forces than will a tethered filament. It is well known that the buckling force of a perpendicular filament free to slide laterally on a surface is much lower (by factor of ∼8; Howard, 2001) than that of a filament unable to slide laterally. Moreover, as shown in Appendix A, the buckling force falls sharply with the angle of incidence. For example, an elongating actin filament (with ∼10-μM persistence length) spanning a ∼70-nm gap between the surface and an anchored position (e.g., in the filament network) should buckle at only ∼2-pN force when initially oriented at 45° to the surface, whereas a corresponding tethered filament can exert force with a component normal to the surface of up to ∼60 pN before buckling (neglecting, of course, other kinetic and thermodynamic limits) (Fig. 7).
We have considered only simple reaction mechanisms that illustrate the advantages of force generation by filament end-tracking motors over force generation by similar mechanisms operating with the benefit of hydrolysis or force generation relying on only monomer addition to free filament ends. The end-tracking stepping motor illustrated in Mechanism-A uses penultimate subunit·NTP hydrolysis to facilitate processive end-tracker advancement on the same protofilament, whereas the direct-transfer end-tracking motors (Mechanism-B and Mechanism-C) uses the end-tracking unit to usher monomers to the filament end, which then triggers hydrolysis and end-tracker release. There are several possible variations of these mechanisms that do not alter the following general properties: strong binding to NTP-bound subunits and weaker binding of NDP-bound (or NDP·Pi-bound) subunits, and multivalent interaction to maintain possession of the filament end. For example, there may be some level of cooperativity between advancement of end-tracking proteins on adjacent protofilaments, which could explain how elongation of the tautmost actin filament resulted in predominantly 5.4 nm steps (as opposed to 2.7-nm steps) in Listeria motility (Dickinson and Purich, 2002). There may also be multiple additional tracking units (above one per protofilament), ready to rapidly replace the dissociating units (i.e., ready to bind to the terminal filament subunit in Mechanism-A after monomer addition, or to bind and feed additional monomers in Mechanism-B and Mechanism-C). The reservoir of monomer-binding sites on surface-bound tracking units that transfer monomers to the filament ends can explain how high rates of elongation can occur, despite the lower diffusion-limited rate of monomer binding directly to filament ends (Dickinson et al., 2002).
End-tracking proteins need not be multimeric to achieve a multivalent interaction with filament ends. For example, the end-tracking proteins may be independently bound to the motile object (e.g., in the kinetochore, on the membrane, or on Listeria surface) without being bound to each other. Moreover, there is a clear kinetic and functional advantage to having a degree of flexibility and independent action of the different protein subunits: one end-tracking unit can release and advance independently, whereas the others remain bound to maintain possession of the filament end to the surface, without requiring all tracking units to release simultaneously for the motor to advance.
Mechanism-C was motivated by the potential role of such soluble cofactors as EB1 and profilin, respectively, in microtubule and actin filament end-tracking motors. EB1 and profilin are known to bind to elongating (+)-ends as well as to the motile surface-bound filament-binding proteins such as APC found at the kinetochores and Ena/VASP proteins on Listeria or host-cell membranes. In addition to their potential participation in the affinity-modulated interaction, these cofactors may offer kinetic advantages by recruiting and concentrating monomers for transfer to the elongating (+)-ends (Dickinson, et al., 2002). An attractive way to reconcile conflicting ideas about profilin-mediated filament elongation (Kang et al., 1999; Pantaloni and Carlier, 1993; Pring et al., 1992) is to postulate that profilin's binding affinity may be affected by the ATP hydrolysis state only when it is bound to the lagging protofilament (as in state 2 of Mechanism-C, but uncoupled from the tracking proteins). Noting its ability to bind profilin, actin monomers, and filaments, one role of VASP may be to transiently stabilize an otherwise energetically unfavorable addition of the second p·A complex (Step 2 in Fig. 5) in a manner that triggers ATP hydrolysis. If so, in vitro profilin-mediated elongation in the absence of VASP would be attended by less frequent p·A occupancy of the higher-energy binding site, causing sporadic affinity-modulation by ATP hydrolysis and resulting in a slightly more negative measured ΔG of monomer addition through the profilin pathway (and in a manner that may be sensitive to slight variation in experimental conditions).
In view of the high complexity and numerous molecular players involved in the kinetochore, we are hesitant to suggest that EB1/APC end-tracking motors represent the predominant mechanism generating a pushing force. Nevertheless, GTP subunits at microtubule ends are required for kinetochore capture of microtubules, and EB1 clearly accumulates at (+)-ends and detaches from microtubule sides (Tirnauer et al., 2002b), indicating that there is a thermodynamic driving force for accumulation on the end. Because both APC and EB1 bind to microtubule ends (Mimori-Kiyosue et al., 2000b) and apparently to each other (Fodde et al., 2001b) in the kinetochore, it is possible that EB1 and APC (or another kinetochore protein) bind to GTP-containing protofilament subunits in a ternary complex, which is subsequently disrupted by the GTP hydrolysis. If tracking units operated on all or several of the 13 microtubule protofilaments simultaneously, Mechanism-C would account for how the kinetochore facilitates rapid monomer addition and force generation during tight possession of the GTP-rich filament end.
Hill (1985) treated a type of microtubule end-tracking mechanism in which the kinetochore remains tethered to a depolymerizing microtubule by means of an enclosing “sleeve” that translates with a shrinking (+)-end to maintain maximum contact with microtubule subunits. In contrast to “pushing” by affinity-modulated end-tracking motors as described here, the sleeve mechanism is not driven by NTP hydrolysis, and it is restricted to kinetochore “pulling,” because addition of new terminal subunits provides no way for increasing the already-maximized contact between the sleeve and the microtubule end.
Hill's sleeve model and the end-tracking motors are therefore distinct mechanisms that account for different phenomena and as such are not mutually exclusive. However, the same molecular players may participate in both mechanisms; i.e., end-tracking proteins may push during GTP-tubulin addition, but then pull the kinetochore by the sleeve mechanism during GDP-tubulin release from depolymerizing (+)-ends.
In summary, the affinity-modulated end-tracking mechanisms presented here describe a new class of molecular motors that enjoy kinetic and functional advantages in facilitating monomer addition and generating force. The properties of end-tracking motors provide attractive explanations for a number of puzzling phenomena that cannot be readily explained by models requiring free filament ends. An intriguing possibility is that filament assembly by end-tracking motors (whether tethered or untethered) is the norm in vivo, with assembly of free (+)-ends contributing only modestly (if at all) to synthesis of the cytoskeletal filaments. For these reasons, affinity-modulated filament end-tracking motors merit closer consideration in efforts to explain cell motility.
Acknowledgments
The authors thank William Zeile and Robert Cohen for helpful suggestions in preparing this manuscript.
This work was partially supported by the National Science Foundation Engineering Research Center for Particle Science and Technology at the University of Florida (EEC-94-0289) and by the National Institutes of Health (NIGMS-R01 GM067828).
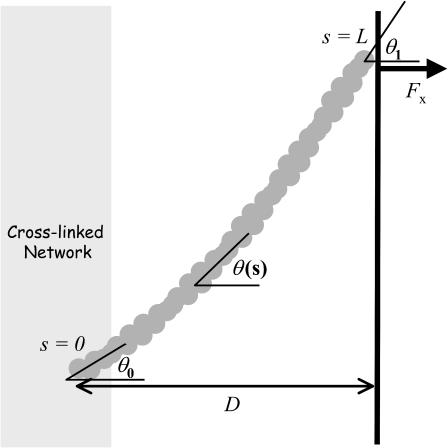
APPENDIX A: BUCKLING FORCE ESTIMATION FOR AN ELONGATING FILAMENT
In this Appendix, we derive the mechanical buckling force limit of an elongating free filament and compare it to that of a tethered filament. Following the approach of Mogilner and Oster (1996), we consider a filament (Fig. 6) of contour length, 0 < s < L, spanning a gap distance, D, between fixed position (x(s),y(s)) = (0,0) at fixed angle θ0, with the equilibrium end position constrained on the surface (x(L) = D). At mechanical equilibrium, the orientation angle tangent to the filament, θ(s), is governed by the differential equation (Landau and Lifshitz, 1986),
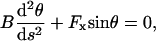 |
(A-1) |
where B is the bending modulus and Fx is the magnitude force on the filament end (the y component of the force is 0 for the freely sliding end). The appropriate boundary conditions are θ(0) ≡ θ0 at the fixed end, and 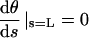 at the free end, corresponding to a zero moment of internal stress. Integrating Eq. A-1 yields
at the free end, corresponding to a zero moment of internal stress. Integrating Eq. A-1 yields
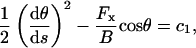 |
(A-2) |
with 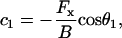 where θ1 ≡ θ(L) is determined by applying the boundary conditions at L. Separating the variables and integrating a second time yields
where θ1 ≡ θ(L) is determined by applying the boundary conditions at L. Separating the variables and integrating a second time yields
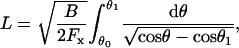 |
(A-3) |
which can be rearranged to determined the force parameterized by θ1,
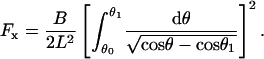 |
(A-4) |
The x position on the filament is obtained from
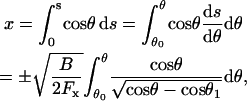 |
(A-5) |
which, from Eq. A-3, can be written as
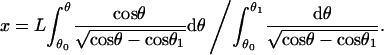 |
(A-6) |
Eq. A-4 is used to express the filament end distance, D, also parameterized by θ1,
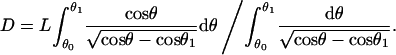 |
(A-7) |
Although not needed here, the y position can be obtained from
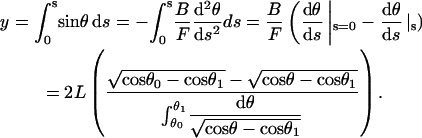 |
(A-8) |
Also relevant is the mechanical energy of the filament, which is given by
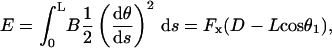 |
(A-9) |
which was obtained by substituting Eq. A-2 into the integrand. The corresponding dimensionless force, f ≡ FxL2/B, dimensionless end position, δx ≡ D/L, and dimensionless energy ɛ ≡ EL/B, are all parameterized by θ1 and expressible in terms of tabulated elliptic functions:
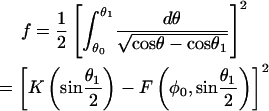 |
(A-10) |
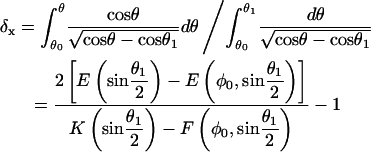 |
(A-11) |
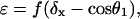 |
(A-12) |
where 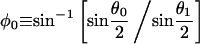 ; K(k) and F(z|k) are complete and incomplete elliptic integrals of first kind, respectively; and E(k) and E(z|k) are the complete and incomplete elliptic integrals of second kind, respectively. The maximum force that an elongating filament achieves before buckling, Fx,max, is calculated by solving
; K(k) and F(z|k) are complete and incomplete elliptic integrals of first kind, respectively; and E(k) and E(z|k) are the complete and incomplete elliptic integrals of second kind, respectively. The maximum force that an elongating filament achieves before buckling, Fx,max, is calculated by solving
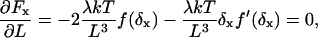 |
(A-13) |
which is equivalent to minimizing the function, ɛ(δx) + δxf(δx) with respect to δx(θ1), then substituting the solution for θ1 into Eq. A-10.
FIGURE 6.
Schematic of filament with a free end flexed against a surface. The relevant parameters and variables for a flexed filament analyzed in Appendix A are shown, including the gap distance D, filament angle θ(s), arc length s, fixed initial angle of incidence, θ0, final angle of incidence θ1, and total length, L.
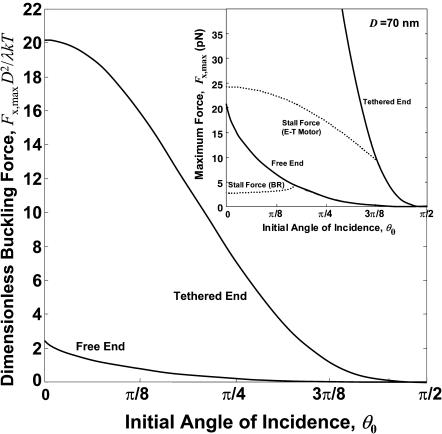
The calculated dimensionless buckling force FxD2/B is plotted versus initial angle of incidence θ0 in Fig. 7. The curve shows the well-known buckling instability of π2/4 for θ0 = 0, and declines rapidly to 0 with increasing θ0. For comparison, the much larger x component of buckling force for a tethered filament, one that is unable to slide laterally on the surface, is also shown, using the known formula, FL2/B = 20.19 (Howard, 2001), together with Fx = F cos θ0 and D2 = L2 cos2 θ0 for a straight filament (tethered, before buckling). The corresponding dimensional solutions for the case of an actin filament with persistence length, λ = B/kT = 10 μm and D = 70 nm, are shown in the Fig. 7 inset. Also plotted in the inset by dotted lines are the thermodynamic maximum forces achievable for the Brownian ratchet and end-tracking motors, assuming [MT]/[MT](+)-crit = 6 and ɛ = 14 kT, which simultaneously highlights the thermodynamic and mechanical advantages of surface-tethered end-tracking motors in terms of maximum force generated. For the dotted lines, filaments were assumed capable of elongating and generating force only until dE/dL reaches the thermodynamic limit of [dE/dL]max = kT ln(6)/2.7 nm = 2.7 pN for the Brownian ratchet model, or [dE/dL]max = kT(ln(6)+14)/2.7 nm = 24 pN for the end-tracking motor, or until the buckling limit was reached. For the free filament case, this maximum force was obtained by numerically solving
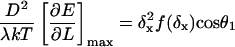 |
(A-14) |
for θ1, using Eqs. A-10 and A-11, while noting that 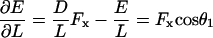 from Eq. A-9.
from Eq. A-9.
FIGURE 7.
Maximum force versus initial angle of incidence for tethered and untethered filaments. The dimensionless buckling force for both cases is shown in the main plot, and the corresponding dimensional forces are shown for the case of a persistence length λ = 10 μm and a gap distance, D = 70 nm. The thermodynamic stall forces (dotted lines) for the Brownian ratchet model (BR Model) and the end-tracking motor (E-T motor) are also shown, assuming a factor of six greater monomer concentration above the critical concentration and 14 kT additional energy from ATP hydrolysis captured by the end-tracking motor.
The low-force buckling limit illustrates why the linear spring-model for free filament flexure assumed by Mogilner and Oster (1996) should be used with caution, especially at larger angles of incidence and gap distances, D. It should also be noted their spring constant, κ = −dFx/dD ≅ 4B/L3sin2 θ0 for small deflections, was derived assuming a uniform filament curvature, which cannot be valid because the moment (hence the curvature) necessarily approaches zero at the free filament end (Landau and Lifshitz, 1986). The spring constant for the small deflection limit can be correctly obtained by expanding θ and θ1 around θ0 and keeping only the first-order terms in the integrands in Eqs. A-10 and A-11,
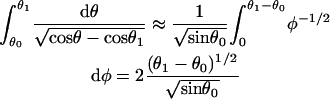 |
(A-15) |
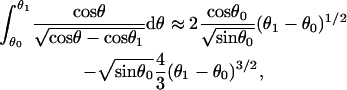 |
(A-16) |
such that
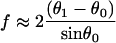 |
(A-17) |
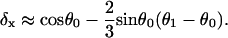 |
(A-18) |
Therefore, noting that 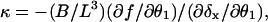 the linear filament spring constant for the small-deflection limit of a free filament is
the linear filament spring constant for the small-deflection limit of a free filament is
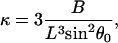 |
(A-19) |
which differs by the integer factor of 3 instead of the factor of 4 in stiffness formula derived by Mogilner and Oster by assuming constant filament curvature.
APPENDIX B: DEFINITIONS
TABLE 3.
Definition of parameters and variables
| Symbol | Definition |
|---|---|
| d | Length of one protofilament subunit (5.4 nm for actin, 8 nm for tubulin) |
| F | Force on filament end, opposing elongation |
| ΔG(+)add | Free energy change upon monomer·NTP binding to filament (+)-end |
| ΔGexchange | Free energy change upon NTP exchange with monomer-bound NDP |
| ΔGhydrolysis | Free energy change of NTP hydrolysis to form NDP and Pi |
| ΔG(−)loss | Free energy change upon monomer·NDP dissociation from filament (−)-end |
Δ
|
Free energy change upon hydrolysis of filament-bound NTP |
Δ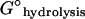
|
Standard free energy change of NTP hydrolysis to form NDP and Pi |
| ΔGPi-release | Free energy change upon reversible release of filament-bound Pi |
| k1, k−1, K1 | Forward and reverse rate constants and equilibrium dissociation constant (K1 = k−1/k1) for monomer binding to filament (+)-end (Mechanism-A) |
| k2, k−2, K2 | Forward and reverse rate constants and equilibrium dissociation constant (K2 = k−2/k2) for tracking unit translocation to terminal subunit (Mechanism-A) |
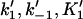 |
Forward and reverse rate constants and equilibrium dissociation constant (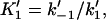 ) for monomer binding to tracking protein (Mechanism-B) ) for monomer binding to tracking protein (Mechanism-B) |
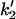 , , 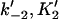
|
Forward and reverse rate constants and equilibrium dissociation constant (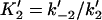 ) for monomer-tracking protein complex binding to filament end (Mechanism-B) ) for monomer-tracking protein complex binding to filament end (Mechanism-B) |
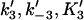 |
Forward and reverse rate constants and equilibrium dissociation constant (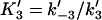 ) for tracking protein binding to terminal subunit (Mechanism-B) ) for tracking protein binding to terminal subunit (Mechanism-B) |
| kT | Thermal energy (Boltzmann constant, k, × absolute temperature, T) |
| KP | Equilibrium dissociation constant for Pi binding to filament subunits |
| Kx | Equilibrium constant for nucleotide exchange (Kx = [MT]eq[NDP] /[MD]eq[NTP]) |
| MT | NTP-bound monomer |
| MD | NDP-bound monomer |
| [MT](+)-crit | Critical concentration of NTP-bound monomers for filament (+)-ends |
| [MD](−)-crit | Critical concentration of NDP-bound monomers for filament (−)-ends |
| u | Probability of the end-tracking protein bound only to monomer (Mechanism-B) |
| α | Ratio of rate constants (k−2/k−1) in Mechanism-A |
| β | Ratio of rate constants (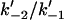 ) in Mechanism-B ) in Mechanism-B |
| γ | Ratio of rate constants (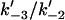 ) in Mechanism-B ) in Mechanism-B |
| ɛ | Energy captured from hydrolysis that is used for affinity modulation |
| ρ | Probability of the end-tracking protein being bound to the terminal subunit (Mechanism-A or Mechanism-B) |
References
- Abraham, V. C., V. Krishnamurthi, D. L. Taylor, and F. Lanni. 1999. The actin-based nanomachine at the leading edge of migrating cells. Biophys. J. 77:1721–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear, J. E., T. M. Svitkina, M. Krause, D. A. Schafer, J. J. Loureiro, G. A. Strasser, I. V. Maly, O. Y. Chaga, J. A. Cooper, G. G. Borisy, and F. B. Gertler. 2002. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 109:509–521. [DOI] [PubMed] [Google Scholar]
- Bray, D. 2001. Cell Movements: From Molecules to Motility. Garland Publishing, New York.
- Bork, P., C. Sander, and A. Valencia. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA. 89:7290–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieher, W. M., M. Coughlin, and T. J. Mitchison. 2004. Fascin-mediated propulsion of Listeria monocytogenes independent of frequent nucleation by the Arp2/3 complex. J. Cell Biol. 165:233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu, W., and L. K. Su. 2001. Regulation of microtubule assembly by human EB1 family proteins. Oncogene. 20:3185–3192. [DOI] [PubMed] [Google Scholar]
- Cameron, L. A., T. M. Svitkina, D. Vignjevic, J. A. Theriot, and G. G. Borisy. 2001. Dendritic organization of actin comet tails. Curr. Biol. 11:130–135. [DOI] [PubMed] [Google Scholar]
- Carlier, M. F., D. Didry, R. Melki, M. Chabre, and D. Pantaloni. 1988. Stabilization of microtubules by inorganic phosphate and its structural analogues, the fluoride complexes of aluminum and beryllium. Biochemistry. 27:3555–3559. [DOI] [PubMed] [Google Scholar]
- Carlier, M. F., V. Laurent, J. Santolini, R. Melki, D. Didry, G. X. Xia, Y. Hong, N. H. Chua, and D. Pantaloni. 1997. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136:1307–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier, M. F., and D. Pantaloni. 1988. Binding of phosphate to F-ADP-actin and role of F-ADP-Pi-actin in ATP-actin polymerization. J. Biol. Chem. 263:817–825. [PubMed] [Google Scholar]
- Dickinson, R. B., and D. L. Purich. 2002. Clamped-filament elongation model for actin-based motors. Biophys. J. 82:605–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, R. B., F. S. Southwick, and D. L. Purich. 2002. A direct-transfer polymerization model explains how the multiple profilin-binding sites in the actoclampin motor promote rapid actin-based motility. Arch. Biochem. Biophys. 406:296–301. [DOI] [PubMed] [Google Scholar]
- Fodde, R., J. Kuipers, C. Rosenberg, R. Smits, M. Kielman, C. Gaspar, J. H. van Es, C. Breukel, J. Wiegant, R. H. Giles, and H. Clevers. 2001a. Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat. Cell Biol. 3:433–438. [DOI] [PubMed] [Google Scholar]
- Fodde, R., R. Smits, and H. Clevers. 2001b. APC, signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer. 1:55–67. [DOI] [PubMed] [Google Scholar]
- Giardini, P. A., D. A. Fletcher, and J. A. Theriot. 2003. Compression forces generated by actin comet tails on lipid vesicles. Proc. Natl. Acad. Sci. USA. 100:6493–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashida, C., T. Miyoshi, A. Fujita, F. Oceguera-Yanez, J. Monypenny, Y. Andou, S. Narumiya, and N. Watanabe. 2004. Actin polymerization-driven molecular movement of mDia1 in living cells. Science. 303:2007–2010. [DOI] [PubMed] [Google Scholar]
- Hill, T. L. 1981. Microfilament or microtubule assembly or disassembly against a force. Proc. Natl. Acad. Sci. USA. 78:5613–5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, T. L. 1985. Theoretical problems related to the attachment of microtubules to kinetochores. Proc. Natl. Acad. Sci. USA. 82:4404–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, J. 2001. Mechanics of Motor Proteins and the Cytoskeleton. Sinauer, Sunderland, MA.
- Joslyn, G., D. S. Richardson, R. White, and T. Alber. 1993. Dimer formation by an N-terminal coiled coil in the APC protein. Proc. Natl. Acad. Sci. USA. 90:11109–11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, F., R. O. Laine, M. R. Bubb, F. S. Southwick, and D. L. Purich. 1997. Profilin interacts with the Gly-Pro-Pro-Pro-Pro-Pro sequences of vasodilator-stimulated phosphoprotein (VASP): implications for actin-based Listeria motility. Biochemistry. 36:8384–8392. [DOI] [PubMed] [Google Scholar]
- Kang, F., D. L. Purich, and F. S. Southwick. 1999. Profilin promotes barbed-end actin filament assembly without lowering the critical concentration. J. Biol. Chem. 274:36963–36972. [DOI] [PubMed] [Google Scholar]
- Kaplan, K. B., A. A. Burds, J. R. Swedlow, S. S. Bekir, P. K. Sorger, and I. S. Nathke. 2001. A role for the adenomatous polyposis coli protein in chromosome segregation. Nat. Cell Biol. 3:429–432. [DOI] [PubMed] [Google Scholar]
- Kinosian, H. J., L. A. Selden, J. E. Estes, and L. C. Gershman. 1993. Nucleotide binding to actin. Cation dependence of nucleotide dissociation and exchange rates. J. Biol. Chem. 268:8683–8691. [PubMed] [Google Scholar]
- Kinosian, H. J., L. A. Selden, L. C. Gershman, and J. E. Estes. 2002. Actin filament barbed end elongation with nonmuscle MgATP-actin and MgADP-actin in the presence of profilin. Biochemistry. 41:6734–6743. [DOI] [PubMed] [Google Scholar]
- Kovar, D. R., J. R. Kuhn, A. L. Tichy, and T. D. Pollard. 2003. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 161:875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo, S. C., and J. L. McGrath. 2000. Steps and fluctuations of Listeria monocytogenes during actin-based motility. Nature. 407:1026–1029. [DOI] [PubMed] [Google Scholar]
- Laine, R. O., K. L. Phaneuf, C. C. Cunningham, D. Kwiatkowski, T. Azuma, and F. S. Southwick. 1998. Gelsolin, a protein that caps the barbed ends and severs actin filaments, enhances the actin-based motility of Listeria monocytogenes in host cells. Infect. Immun. 66:3775–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau, L. D., and E. M. Lifshitz. 1986. Theory of Elasticity. Pergamon Press, Oxford, UK.
- Laurent, V., T. P. Loisel, B. Harbeck, A. Wehman, L. Grobe, B. M. Jockusch, J. Wehland, F. B. Gertler, and M. F. Carlier. 1999. Role of proteins of the Ena/VASP family in actin-based motility of Listeria monocytogenes. J. Cell Biol. 144:1245–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyh, T. S. 1999. On the advantages of imperfect energetic linkage. Methods Enzymol. 308:48–70. [DOI] [PubMed] [Google Scholar]
- McGrath, J. L., N. J. Eungdamrong, C. I. Fisher, F. Peng, L. Mahadevan, T. J. Mitchison, and S. C. Kuo. 2003. The force-velocity relationship for the actin-based motility of Listeria monocytogenes. Curr. Biol. 13:329–332. [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue, Y., N. Shiina, and S. Tsukita. 2000a. Adenomatous polyposis coli (APC) protein moves along microtubules and concentrates at their growing ends in epithelial cells. J. Cell Biol. 148:505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue, Y., N. Shiina, and S. Tsukita. 2000b. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr. Biol. 10:865–868. [DOI] [PubMed] [Google Scholar]
- Mogilner, A., and L. Edelstein-Keshet. 2002. Regulation of actin dynamics in rapidly moving cells: a quantitative analysis. Biophys. J. 83:1237–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner, A., and G. Oster. 1996. Cell motility driven by actin polymerization. Biophys. J. 71:3030–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner, A., and G. Oster. 2003a. Force generation by actin polymerization II: the elastic ratchet and tethered filaments. Biophys. J. 84:1591–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner, A., and G. Oster. 2003b. Polymer motors: pushing out the front and pulling up the back. Curr. Biol. 13:R721–R733. [DOI] [PubMed] [Google Scholar]
- Niebuhr, K., F. Ebel, R. Frank, M. Reinhard, E. Domann, U. D. Carl, U. Walter, F. B. Gertler, J. Wehland, and T. Chakraborty. 1997. A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO J. 16:5433–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noireaux, V., R. M. Golsteyn, E. Friederich, J. Prost, C. Antony, D. Louvard, and C. Sykes. 2000. Growing an actin gel on spherical surfaces. Biophys. J. 78:1643–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaloni, D., and M. F. Carlier. 1993. How profilin promotes actin filament assembly in the presence of thyroxin β-4. Cell. 75:1007–1014. [DOI] [PubMed] [Google Scholar]
- Peskin, C. S., G. M. Odell, and G. F. Oster. 1993. Cellular motions and thermal fluctuations: the Brownian ratchet. Biophys. J. 65:316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T. D., L. Blanchoin, and R. D. Mullins. 2000. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29:545–576. [DOI] [PubMed] [Google Scholar]
- Pring, M., A. Weber, and M. R. Bubb. 1992. Profilin-actin complexes directly elongate actin filaments at the barbed end. Biochemistry. 31:1827–1836. [DOI] [PubMed] [Google Scholar]
- Robbins, J. R., and J. A. Theriot. 2003. Listeria monocytogenes rotates around its long axis during actin-based motility. Curr. Biol. 13:R754–R756. [DOI] [PubMed] [Google Scholar]
- Samarin, S., S. Romero, C. Kocks, D. Didry, D. Pantaloni, and M. F. Carlier. 2003. How VASP enhances actin-based motility. J. Cell Biol. 163:131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer, T. A. 2001. Microtubules don and doff their caps: dynamic attachments at plus and minus ends. Curr. Opin. Cell Biol. 13:92–96. [DOI] [PubMed] [Google Scholar]
- Schuyler, S. C., and D. Pellman. 2001. Microtubule “plus-end-tracking proteins”: the end is just the beginning. Cell. 105:421–424. [DOI] [PubMed] [Google Scholar]
- Severin, F. F., P. K. Sorger, and A. A. Hyman. 1997. Kinetochores distinguish GTP from GDP forms of the microtubule lattice. Nature. 388:888–891. [DOI] [PubMed] [Google Scholar]
- Southwick, F. S., and D. L. Purich. 1996. Intracellular pathogenesis of Listeriosis. N. Engl. J. Med. 334:770–776. [DOI] [PubMed] [Google Scholar]
- Theriot, J. A. 2000. The polymerization motor. Traffic. 1:19–28. [DOI] [PubMed] [Google Scholar]
- Tirnauer, J. S., and B. E. Bierer. 2000. EB1 proteins regulate microtubule dynamics, cell polarity, and chromosome stability. J. Cell Biol. 149:761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer, J. S., J. C. Canman, E. D. Salmon, and T. J. Mitchison. 2002a. EB1 targets to kinetochores with attached, polymerizing microtubules. Mol. Biol. Cell. 13:4308–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer, J. S., S. Grego, E. D. Salmon, and T. J. Mitchison. 2002b. EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol. Biol. Cell. 13:3614–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya, A., J. R. Chabot, A. Andreeva, A. Samadani, and A. van Oudenaarden. 2003. Probing polymerization forces by using actin-propelled lipid vesicles. Proc. Natl. Acad. Sci. USA. 100:4521–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent, F., L. A. Amos, and J. Lowe. 2001. Prokaryotic origin of the actin cytoskeleton. Nature. 413:39–44. [DOI] [PubMed] [Google Scholar]
- van den Ent, F., J. Moller-Jensen, L. A. Amos, K. Gerdes, and J. Lowe. 2002. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 21:6935–6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhovsky, A. B., O. Y. Chaga, S. Schaub, T. M. Svitkina, J. J. Meister, and G. G. Borisy. 2003. Orientational order of the lamellipodial actin network as demonstrated in living motile cells. Mol. Biol. Cell. 14:4667–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, A., V. T. Nachmias, C. R. Pennise, M. Pring, and D. Safer. 1992. Interaction of thymosin β-4 with muscle and platelet actin: implications for actin sequestration in resting platelets. Biochemistry. 31:6179–6185. [DOI] [PubMed] [Google Scholar]
- Wegner, A., and J. Engel. 1975. Kinetics of the cooperative association of actin to actin filaments. Biophys. Chem. 3:215–225. [DOI] [PubMed] [Google Scholar]
- Wei, J., and T. S. Leyh. 1999. Isomerization couples chemistry in the ATP sulfurylase-GTPase system. Biochemistry. 38:6311–6316. [DOI] [PubMed] [Google Scholar]
- Zeeberg, B., and M. Caplow. 1979. Determination of free and bound microtubular protein and guanine nucleotide under equilibrium conditions. Biochemistry. 18:3880–3886. [DOI] [PubMed] [Google Scholar]
- Zigmond, S. H. 2004. Formin-induced nucleation of actin filaments. Curr. Opin. Cell Biol. 16:99–105. [DOI] [PubMed] [Google Scholar]
- Zigmond, S. H., M. Evangelista, C. Boone, C. Yang, A. C. Dar, F. Sicheri, J. Forkey, and M. Pring. 2003. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr. Biol. 13:1820–1823. [DOI] [PubMed] [Google Scholar]