Messenger RNA turnover is a critical determinant of eukaryotic gene expression. The stability of different mRNAs within the same cell can vary by orders of magnitude and thus contribute greatly to differential expression levels. Moreover, the stability of individual mRNAs can be regulated in response to a variety of stimuli, allowing for rapid alterations in gene expression. But how does eukaryotic mRNA turnover work, and how is it controlled? In this issue of PNAS, Wang et al. (1) provide a piece to the puzzle as to how eukaryotic mRNAs are degraded.
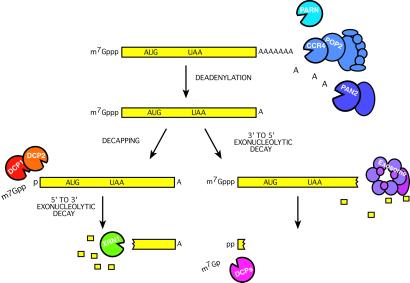
Early experiments in several eukaryotic systems revealed that the 5′ m7G cap and the 3′ poly(A) tail are critical protective features of mRNAs. Work in the yeast Saccharomyces cerevisiae has uncovered two general mRNA decay pathways that act on these protective ends (Fig. 1; reviewed in ref. 2). These pathways are general in that they appear to degrade most, if not all, normal mRNAs. The first step in both of these pathways is shortening of the poly(A) tail (3–5), which can be catalyzed by one of two different enzymes. The major deadenylase appears to be a large ≈1-mDa complex consisting of one known catalytic subunit, Ccr4p (6–8), a member of the ExoIII/AP endonuclease family (9), and several other proteins, most notably, Pop2, Not1, Not2, Not3, Not4, and Not5 (10). Alternatively, a complex of Pan2p/Pan3p can also function as a cytoplasmic deadenylase (6, 11, 12).
Figure 1.
Eukaryotic mRNA decay mechanisms and enzymes. Two general mRNA decay pathways. Both pathways are initiated by deadenylation by the Ccr4/Pop2/Not complex or possibly by the alternative deadenylases, Pan2/Pan3 and PARN. Poly(A) tail shortening can lead to either 3′-5′ exonucleolytic digestion by the exosome or decapping by the Dcp1/Dcp2 complex. Decapping is followed by 5′-3′ exonuclease digestion by Xrn1. The residual cap structure resulting from exosome digestion is cleaved by the scavenger decapping enzyme DcpS.
Deadenylation of the 3′ tail to a length that is too short to bind the major poly(A)-binding protein, Pabp1, can lead to two consequences in yeast. First, it can leave the 3′ end susceptible to a complex of multiple 3′-5′ exonucleases called the exosome (5, 13). Alternatively, loss of Pabp1 is thought to bring about a transition in the structure of the proteins associated with the mRNA, the mRNP, that disposes the 5′ end of the mRNA to decapping (14, 15). Decapping requires two proteins, Dcp1p and Dcp2p, that physically associate with each other (16, 17). Removal of the 5′ protective cap leaves the mRNA subject to 5′-3′ exonucleolytic degradation by the exoribonuclease, XRN1 (4, 13, 18). In yeast, the deadenylation-dependent decapping pathway degrades mRNAs more rapidly than the exosome-mediated decay pathway. The deadenylation-dependent decapping pathway and the exosome-mediated pathway apparently are the only general decay mechanisms in yeast. This conclusion is based on the observation that mRNAs are extremely stable when both pathways are inactivated (5). In fact, when both decay pathways are inactivated, the yeast cells die, which underscores the critical importance of the proper control of mRNA turnover for cell metabolism and division.
An unresolved issue is whether these mRNA decay enzymes and pathways are common in eukaryotes or are unique to yeast. Several lines of evidence suggest that at least some aspects of these pathways are conserved in eukaryotes. First, deadenylation precedes the decay of several mammalian mRNAs in vivo, and particular elements that stimulate the decay of mammalian mRNAs promote rapid deadenylation (e.g., refs. 19 and 20). Second, both the Ccr4p/Pop2p/Not protein complex (21–23) and the Pan2p/Pan3p deadenylase (24) are conserved in eukaryotic genomes. Moreover, human Ccr4p, like yeast Ccr4p, contains the exonuclease domain in its C terminus and can function as a deadenylase in vitro (7). An additional poly(A) ribonuclease, PARN, is present in vertebrates (25, 26). It is interesting to note that the three deadenylases have distinct substrate specificities. The Ccr4/Pop2/Not complex and PARN are inhibited by Pab1p in vitro (8, 26), whereas the Pan2p/Pan3p complex requires Pab1p for activity (11–12). In addition, PARN is stimulated by the presence of a 5′ cap structure (27), whereas CCR4 is not affected (8). These differences in specificity suggest that, in vivo, mRNAs with different mRNP structures may be preferentially deadenylated by alternative deadenylases, allowing for differential control of the poly(A) shortening process.
The deadenylation-dependent decapping pathway is indeed a common decay mechanism in eukaryotes.
Decay of mRNAs in a 3′ to 5′ direction after deadenylation also appears to be conserved. The individual components of the exosome and their ability to form a complex are conserved in eukaryotes (reviewed in ref. 28). Recent work in mammalian in vitro decay systems demonstrates that the exosome degrades RNAs after deadenylation (29–31). Interestingly, degradation by the exosome in vitro is stimulated by AU-rich elements (AREs) (29, 31). AREs are destabilizing elements found in many unstable mammalian mRNAs, especially those involved in growth control. Thus, the exosome may play a role in regulated mRNA turnover in mammals. Decay in a 3′ to 5′ direction may also be conserved in plants based on the structure of in vivo decay intermediates of a particular mRNA in oat (32).
The evidence has been less clear whether the decapping pathway is functionally conserved. mRNA decay intermediates that are trimmed from the 5′ end or lack the 5′ cap have been detected in mammals, chlamydomonas, and oat (32–34), although the mechanism of production of these intermediates has not been resolved. Apparent homologs to XRN1 (35–37), DCP1 (38), and DCP2 (17) have been identified in several eukaryotic genomes, and the Arabidopsis and Drosophila XRN1 homologs can functionally complement the absence of the yeast protein, arguing that they naturally play a role in mRNA turnover (36, 37). However, so far there has been no demonstration that proteins with similarity to DCP1 or DCP2 function in decapping. The identification of a functional human homolog to DCP2 by Wang et al. (1) provides evidence that the deadenylation-dependent decapping pathway is indeed a common decay mechanism in eukaryotes.
Wang et al. (1) identified the human DCP2 gene through its Nudix motif. Yeast Dcp2p contains a nucleotide diphosphate linked to an X moiety (Nudix) domain, or MutT motif, which is found in a particular class of pyrophosphatases (17). The human and yeast proteins are most similar at their N termini, which contain the Nudix motif in a larger Nudix fold domain as well as additional domains that are shared by other apparent homologs of DCP2 (1). Most significant is the demonstration by Wang et al. that recombinant human DCP2 protein has intrinsic decapping activity in vitro (1). This decapping activity depends on residues within the human DCP2 Nudix domain similar to this domain being required for Dcp2p activity in yeast. As described below, the biochemical role of Dcp2p in decapping in yeast has been unresolved for several years. Thus, the finding that DCP2 is indeed a decapping enzyme and therefore likely to have a direct role in decapping of mRNAs is an important advance. Human DCP2 activity shares many similarities to yeast decapping activity (1). For example, recombinant hDCP2 releases m7GDP from RNA substrates and is specific for N7 methylated cap structures but requires more than just the m7GTP moiety for efficient substrate recognition. Wang et al. find that hDCP2 is enriched in polysome fractions (1). Interestingly, DCP2-like decapping activity in polysome extracts is inhibited by the presence of a poly(A) tail. This finding suggests that, like yeast, decapping by hDCP2 may depend on deadenylation.
Parallel work by Jens Lykke-Andersen at the University of Colorado, Boulder, suggests that DCP1 is also conserved in mammals. By sequence comparison, he has identified two human homologs to human DCP1 as well as human DCP2 (39). At least one of the DCP1 proteins appears to be functionally related to the yeast DCP1 protein in that it coimmunoprecipitates with decapping activity, and conserved residues in the protein required for Dcp1p function in yeast are also required for decapping activity of the protein. This DCP1 homolog also coimmunoprecipitates with human DCP2, indicating that the interaction between DCP1 and DCP2 is also conserved in mammals. In summation, these results demonstrate that a Dcp1p/Dcp2p complex forms in both yeast and mammals and is capable of decapping mRNAs.
In both yeast and mammals, this Dcp1p/Dcp2p complex also shows an interaction with Upf proteins either by two-hybrid interactions (40) or by coimmunoprecipitation (ref. 39; F. Lejeune, M. Kiledjian, and L. E. Maquat, personal communication). Upf proteins function in the process of nonsense-mediated decay, wherein mRNAs with aberrant translation termination codons are recognized and rapidly degraded. In yeast, aberrant mRNAs recognized by this form of mRNA surveillance are degraded quickly, because they are rapidly decapped by Dcp1p/Dcp2p without requiring the normal slow deadenylation step (41). The conservation of the Dcp1p/Dcp2p–Upf interaction suggests that nonsense-containing mRNAs might also be rapidly decapped in mammalian cells. Although the idea that this interaction could promote rapid decapping is mechanistically appealing, it should be noted that there is no evidence that the interaction between Dcp1/Dcp2p and Upf proteins is functionally significant in any system.
It is surprising that mammalian DCP2 has decapping activity, given that in yeast Dcp1p is thought to be a decapping enzyme. Several pieces of evidence argue that Dcp1p has catalytic activity: (i) it is required for decapping in vivo and for decapping activity in yeast extracts (16); (ii) highly purified Dcp1p from yeast copurifies with decapping activity (42); (iii) decapping activity colocalizes with Dcp1p renatured in gels (42); and (iv) separation of Dcp1p and Dcp2p by high salt yielded preparations of Dcp1p with activity and no detectable Dcp2p (43). In addition, recombinant Dcp1p is reported to have decapping activity (44); however, it should be noted that efforts in our laboratory to obtain active recombinant Dcp1p have been unsuccessful. In contrast to Dcp1p, no biochemical activity has yet been ascribed to yeast Dcp2p. However, recent results suggest that recombinant yeast Dcp2p can have decapping activity (M. Steiger, A. Carr-Schmid, M. Kiledjian, and R.P., unpublished work). Given this result, either the experiments with yeast Dcp1p were somehow misleading, or the Dcp1p/Dcp2p complex contains two different decapping proteins. If there are two different decapping enzymes, why are both Dcp1p and Dcp2p required for decapping in vivo, when each may be sufficient for decapping in vitro? One idea is that their association may be necessary structurally to form active proteins in yeast. Consistent with this idea, under conditions where expression of either protein alone in Escherichia coli is insufficient for decapping activity, coexpression of Dcp1p and Dcp2p together allows for robust decapping (M. Steiger, A. Carr-Schmid, M. Kiledjian, and R.P., unpublished work).
The possibility that the decapping complex may contain two catalytic subunits is very intriguing, because other enzymes involved in eukaryotic mRNA decay exist in complexes of multiple proteins with similar enzymatic activities. The most dramatic example of this phenomenon is the exosome, which contains 10 different potential 3′-5′ exonucleases (45). In addition, in the Ccr4p/Pop2p/Not complex, both Cc4p and Pop2p contain exonuclease motifs (9, 46), and it appears they both can function as a mRNA deadenylase in vitro (7, 8, 47). Why are eukaryotic mRNA decay enzymes clustered in these complexes? One idea is that the association of nucleases with similar activity allows for the control of substrate specificity. That is, these complexes may allow the cell to limit multiple nucleases simultaneously from acting on inappropriate substrates as well as coordinate their activity with correct substrates. Future work aimed at understanding how mRNA turnover is controlled may shed light on this puzzling phenomenon.
The new findings of Wang et al. (1) and the recent demonstration that the exosome degrades RNAs after deadenylation in mammalian extracts (29–31) are exciting, because they suggest that the enzymes and mechanisms of mRNA turnover are conserved throughout the eukaryotic kingdom. The limitation, of course, is that functional studies in mammalian systems need to be conducted to verify whether these enzymes do indeed act to degrade mRNAs in vivo. If these two pathways turn out to be common general decay mechanisms in eukaryotes, the relative importance of the two pathways, i.e., which pathway is responsible for degrading the majority of mRNAs, may very well differ between eukaryotic species. Moreover, with the increased need for regulation, we should expect that complex organisms will have additional decay mechanisms that are specific to particular classes of mRNAs or to specific cellular responses (e.g., ref. 48). In any case, with the knowledge of the enzymes and pathways involved in decay, the pieces now appear to be in place for us to begin to understand how mRNA turnover is regulated by using a combination of biochemical and genetic approaches in multiple eukaryotic systems.
Footnotes
See companion article on page 12663.
References
- 1.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. Proc Natl Acad Sci USA. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tucker M, Parker R. Annu Rev Biochem. 2000;69:571–595. doi: 10.1146/annurev.biochem.69.1.571. [DOI] [PubMed] [Google Scholar]
- 3.Decker C J, Parker R. Genes Dev. 1993;8:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 4.Muhlrad D, Decker C J, Parker R. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs-Anderson J S, Parker R. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tucker M, Valencia-Sanchez M A, Staples R R, Chen J, Denis C L, Parker R. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Chiang Y C, Denis C L. EMBO J. 2002;21:1414–1426. doi: 10.1093/emboj/21.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tucker M, Staples R R, Valencia-Sanchez M A, Muhlrad D, Parker R. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dlakic M. Trends Biochem Sci. 2000;25:272–273. doi: 10.1016/s0968-0004(00)01582-6. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Rappsilber J, Chiang Y C, Russell P, Mann M, Denis C L. J Mol Biol. 2001;314:683–694. doi: 10.1006/jmbi.2001.5162. [DOI] [PubMed] [Google Scholar]
- 11.Boeck R, Tarun S, Jr, Rieger M, Deardorff J A, Muller-Auer S, Sachs A B. J Biol Chem. 1996;271:432–438. doi: 10.1074/jbc.271.1.432. [DOI] [PubMed] [Google Scholar]
- 12.Brown C E, Tarun S Z, Jr, Boeck R, Sachs A B. Mol Cell Biol. 1996;16:5744–5753. doi: 10.1128/mcb.16.10.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muhlrad D, Decker C J, Parker R. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caponigro G, Parker R. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 15.Tharun S, Parker R. Mol Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- 16.Beelman C A, Stevens A, Caponigro G, LaGrandeur T E, Hatfield L, Fortner D M, Parker R. Nature (London) 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- 17.Dunckley T, Parker R. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu C L, Stevens A. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson T, Treisman R. Nature (London) 1988;336:396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- 20.Shyu A B, Belasco J G, Greenberg M E. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 21.Draper M P, Salvadore C, Denis C L. Mol Cell Biol. 1995;15:3487–3495. doi: 10.1128/mcb.15.7.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dupressoir A, Barbot W, Loireau M P, Heidmann T. J Biol Chem. 1999;274:31068–31075. doi: 10.1074/jbc.274.43.31068. [DOI] [PubMed] [Google Scholar]
- 23.Dupressoir A, Morel A P, Barbot W, Loireau M P, Corbo L, Heidmann T. BMC Genom. 2001;2:9–22. doi: 10.1186/1471-2164-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuo Y, Deutscher M P. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korner C G, Wahle E. J Biol Chem. 1997;272:10448–10456. doi: 10.1074/jbc.272.16.10448. [DOI] [PubMed] [Google Scholar]
- 26.Korner C G, Wormington M, Muckenthaler M, Schneider S, Dehlin E, Wahle E. EMBO J. 1998;17:5427–5437. doi: 10.1093/emboj/17.18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dehlin E, Wormington M, Korner C G, Wahle E. EMBO J. 2000;19:1079–1086. doi: 10.1093/emboj/19.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butler J S. Trends Cell Biol. 2002;12:90–96. doi: 10.1016/s0962-8924(01)02225-5. [DOI] [PubMed] [Google Scholar]
- 29.Chen C Y, Gherzi R, Ong S E, Chan E L, Raijmakers R, Pruijn G J, Stoecklin G, Moroni C, Mann M, Karin M. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, Kiledjian M. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- 31.Mukherjee D, Gao M, O'Connor J P, Raijmakers R, Pruijn G, Lutz C S, Wilusz J. EMBO J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Higgs D C, Colbert J T. Plant Cell. 1994;6:1007–1019. doi: 10.1105/tpc.6.7.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Couttet P, Fromont-Racine M, Steel D, Pictet R, Grange T. Proc Natl Acad Sci USA. 1997;94:5628–5633. doi: 10.1073/pnas.94.11.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gera J F, Baker E J. Mol Cell Biol. 1998;18:1498–1505. doi: 10.1128/mcb.18.3.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bashkirov V I, Scherthan H, Solinger J A, Buerstedde J M, Heyer W D. J Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Till D D, Linz B, Seago J E, Elgar S J, Marujo P E, Elias M L, Arraiano C M, McClellan J A, McCarthy J E, Newbury S F. Mech Dev. 1998;79:51–55. doi: 10.1016/s0925-4773(98)00173-7. [DOI] [PubMed] [Google Scholar]
- 37.Kastenmayer J P, Green P J. Proc Natl Acad Sci USA. 2000;97:13985–13990. doi: 10.1073/pnas.97.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tharun S, Parker R. Genetics. 1999;151:1273–1285. doi: 10.1093/genetics/151.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lykke-Andersen, J. (2002) Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 40.He F, Jacobon A. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 41.Muhlrad D, Parker R. Nature (London) 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 42.LaGrandeur T E, Parker R. EMBO J. 1998;17:1487–1496. doi: 10.1093/emboj/17.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunckley T, Tucker M, Parker R. Genetics. 2001;157:27–37. doi: 10.1093/genetics/157.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vilela C, Velasco C, Ptushkina M, McCarthy J E. EMBO J. 2000;19:4372–4382. doi: 10.1093/emboj/19.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moser M J, Holley W R, Chatterjee A, Mian I S. Nucleic Acids Res. 1997;25:5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daugeron M C, Mauxion F, Seraphin B. Nucleic Acids Res. 2001;29:2448–2455. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dompenciel R E, Garnepudi V R, Schoenberg D R. J Biol Chem. 1995;270:6108–6118. doi: 10.1074/jbc.270.11.6108. [DOI] [PubMed] [Google Scholar]