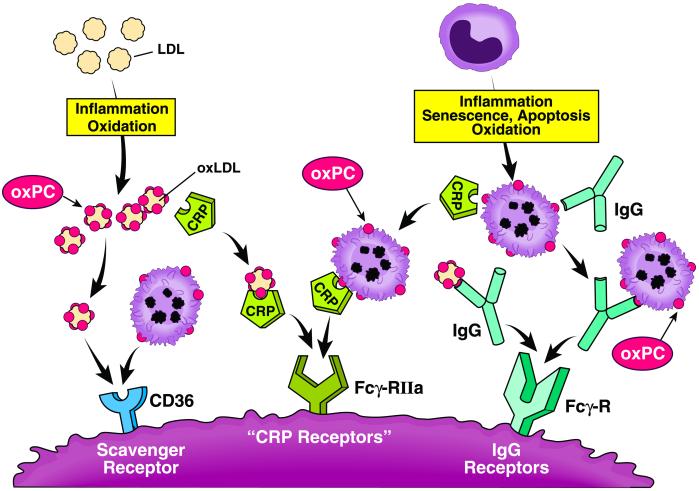
One consequence of living in an aerobic environment is an inexorable oxidative modification of molecular targets in vivo. At the cellular level the effects of the oxidized lipid and lipoprotein by-products can include leukocyte recruitment, activation, and apoptosis. Accumulating at sites of inflammation, these oxidation products can have profound pathological consequences, such as in the case of developing arterial lesions (1–5). Formation of oxidized lipids and lipoproteins in inflammation has thus been linked to the advancement of atherosclerosis and other degenerative diseases of aging.
Evolution has provided us with various means to cope with oxidative insults and challenges. A growing body of evidence suggests that distinct host defense mechanisms have evolved to dispose of damaged molecular complexes and hopelessly injured cells by means of recognition of oxidized phosphatidylcholine (oxPC) species. One such pathway involves recognition of modified lipoproteins and apoptotic or senescent cells by specific scavenger receptors involved in innate immunity, such as CD36 and SR-B1. An alternative pathway involves recognition of oxPC species and protein-oxPC adducts on lipoproteins through autoantibodies and subsequent Fc-γ receptor-mediated endocytosis. In this issue of PNAS, Chang et al. (6) expand upon this theme. These authors show evidence that modifications to phosphatidylcholine, the principal phospholipid present in low density lipoprotein (LDL) and cell membranes, render lipoprotein particles and apoptotic cells recognizable by C-reactive protein (CRP).
A growing body of evidence suggests that distinct host defense mechanisms have evolved to dispose of damaged molecular complexes.
CRP is produced by the liver in large quantities in response to varied stimuli, especially those emanating from trauma, infection, or other inflammatory diseases. This primitive member of the innate immune response serves as an acute-phase reactant, binding to cell wall C-lipopolysaccharide of pathogens such as Streptococcus pneumoniae, and facilitating complement recognition and receptor-mediated clearance. Although the functional consequences of CRP recognition were not directly examined by Chang et al. (6), their findings with those of others (7, 8) suggest that, under certain conditions, CRP binding may mark or opsonize LDL for macrophage or smooth muscle cell recognition. CRP-mediated enhancement in the uptake of modified forms of LDL could then lead to cholesterol accumulation and formation of foam cells, the characteristic cells of the early atherosclerotic lesions termed “fatty streaks.”
The findings of Chang et al. (6) shed light on probable targets for CRP recognition of LDL and apoptotic cells—i.e., oxPC species. Opsonization of oxidized LDL (oxLDL) through oxPC recognition by autoantibodies has similarly been described as a clearance pathway of aged or modified lipoproteins and cells by this group. Palinski and colleagues (9) initially reported natural autoantibodies to oxLDL in atherosclerosis-prone apolipoprotein E (apoE)-deficient mice, and subsequent studies have shown that they recognize oxPC as their cognate epitope (10). Detection of such autoantibodies within the first weeks of life, even in mice raised under germ-free conditions, supports the hypothesis that they serve as germ-line antibodies selected during development by production of oxidized phospholipids present within cellular debris, apoptotic cells, and/or oxLDL (10, 11). EO6 is a clonospecific IgM autoantibody isolated from apoE-null mice that has been extensively characterized and shown to bind specifically to the phosphocholine moiety of microbial capsular polysaccharide, as well as oxPC, oxLDL and apoptotic cells, but not native LDL or viable cells (10, 11). Initial studies aimed at characterizing the specific oxPC recognized by EO6 suggested that 1-palmitoyl-2-(5-oxovaleroyl)-sn-glycero-3-phosphocholine (POV-PC), both free and after reductive alkylation to proteins, mimicked the endogenous epitopes recognized by EO6 (12, 13). Subsequent studies revealed that several POV-PC-related structures are recognized by EO6, including both the reduced Schiff base adduct between POV-PC and the ɛ-amino group of lysine residues, and the initial aldol condensation products of POV-PC. The present studies by Chang et al. (6) suggest that the structural patterns formed by oxPC that are recognized by CRP are similar to those recognized by EO6.
As Chang et al. (6) point out, information published to date regarding CRP's binding to lipoproteins has been inconsistent, and the impact of that binding on function remains uncertain. Using either immobilized CRP or immobilized lipoproteins, a number of investigators have shown that native LDL or native very low density lipoprotein (VLDL) binds CRP in a calcium-dependent and phosphocholine-inhibitable manner (8, 14–16). These findings would seem to be contradictory to the concept that LDL must be modified before CRP will bind. However, Chang et al. (6) show that the process of immobilization of native LDL enhances CRP binding. They speculate that this occurs by means of structural changes accompanying LDL immobilization that unveil “cryptic” phosphocholine epitopes. They further suggest that some accounts of native LDL binding to CRP may be the result of undetected oxidation of LDL. Others have shown that certain types of enzymatic treatment of LDL facilitates CRP binding. Trypsin and cholesterol esterase treatment of LDL enhanced CRP binding that was inhibited by phosphocholine or phospholipase C (17, 18). Chang et al. (6) speculate that such enzymatic treatment could unmask phosphocholine on the LDL preparations. These inferences provide a beginning hypothesis to explain the binding of CRP to lipoproteins by means of a common ligand; however, not all of the published data on lipoprotein–CRP associations can be readily reconciled. For example, Taskinen et al. (18) recently reported that CRP binding to LDL modified by trypsin and cholesterol esterase was dependent on unesterified cholesterol on the lipoprotein's surface, but neither oxidation nor phospholipase-A2 treatment of LDL enhanced CRP binding. These seeming inconsistencies may be the result of CRP's recognition of multiple lipid ligands, but this needs further resolution.
The nature of the modifications of LDL that pertain in vivo, the ligand on the lipoprotein that binds CRP, and the Fc-γ receptors or other receptor systems that recognize and ingest CRP–lipoprotein complexes all need further elucidation. Chang et al. (6) offer data on the second of these three steps in a plausible in vitro system and present a hypothesis that can be tested further for the relevance of this process in vivo. There are some published data on the third step. For example, it has been reported that macrophages can take up CRP–LDL complexes via the “CRP receptor” (CD32, Fc-γ-IIR) (7, 8), but this uptake mechanism may need to be revisited as the precise nature of the CRP–lipoprotein complexes that accumulate in vivo becomes known.
As noted above, oxidized phospholipids also serve as ligands for members of the scavenger receptor class B, such as CD36 (19–21). CD36, the prototypic member of class B scavenger receptors, is a multiligand receptor that participates in macrophage recognition of oxLDL and apoptotic cells (20, 22–24). Studies with CD36-null mice confirm a primary role for this receptor in macrophage foam cell formation and atherosclerosis progression (24). Through use of cross-competition assays and antibody-based studies using EO6, Witztum and colleagues (10, 21) previously attributed CD36 recognition of oxLDL to oxPC such as POV-PC and its protein adducts on apoB-100.
The studies of Chang et al. in this issue (6) provide evidence for a critical role for the phosphocholine moiety of oxidized lipids in CRP-mediated recognition, similar to prior conclusions by this group for EO6 and CD36 recognition. While a wealth of data clearly support the concept that oxidized phospholipids play a major role in the binding of oxLDL forms by CRP, CD36, and autoantibodies to oxLDL, the precise nature of the lipid ligands within oxLDL for these respective protein binding partners have not yet been definitively elucidated at the molecular level. Identification of lipid ligands is difficult because of the large number and complexity of products generated during LDL oxidation and the daunting challenges of their isolation, structural and biochemical characterization, and synthesis. Moreover, the potential contributions of alterations in lipid “presentation” through changes in macromolecular structures of lipids (i.e., the mesomorphic form of the lipid) are not easily investigated.
Recently, a systematic attempt has been made to define at the structural level the oxidized lipids of oxLDL that serve as ligands for the scavenger receptor CD36 (25, 26). A highly conserved family of oxidized choline glycerophospholipids that support high-affinity recognition of CD36 was structurally defined. These were shown to be enriched in atherosclerotic lesions and to be formed in various oxLDL preparations in parallel with increased receptor recognition. The oxPC ligands identified for CD36 were shown to support CD36-specific recognition when incorporated into particles even at trace levels (e.g., 0.3 mol %, equivalent to only a few molecules per LDL particle), and to promote CD36-specific cholesterol deposition and foam cell formation (25, 26). Structures of the specific lipid ligands were identified by using a combination of binding studies to recombinant expressed CD36, multiple distinct chromatographic and mass spectrometric methods in conjunction with chemical derivatization strategies, NMR, inference of plausible structures based on known mechanisms of lipid oxidation and fragmentation, and de novo synthesis of each lipid (25, 26). Consistent with the results of Chang et al. for CRP (6), a critical role of the phosphocholine head group for CD36 binding to oxidized lipids was observed. In addition, a remarkably conserved structural motif was required for high-affinity receptor recognition: a γ-hydroxy (or oxo)-α,β-unsaturated carbonyl (terminal aldehyde or carboxylic acid) tethered to the sn-2 position of lysophosphatidylcholine (26). Based on the parallel binding patterns noted between oxPC and oxLDL vs. CRP, CD36, or EO6, the insights gained into the molecular patterns of oxidized lipids recognized by CD36 may shed light on the potential oxPC structures that bind with high affinity to CRP and oxLDL autoantibodies.
In light of the cumulative results of Witztum's group and the apparent parallel nature of oxPC and oxLDL recognition by members of three distinct arms of innate host defenses (i.e., CRP, autoantibodies such as EO6, and CD36) (6, 10–13, 21, 27, 28), it is tempting to speculate that the novel family of oxPCs recently defined for CD36 (25, 26) may confer enhanced recognition of modified lipoproteins and senescent or apoptotic cells by CRP. It should be noted, however, that glycerophospholipids can adopt alternative polymorphic and mesomorphic forms (29). Even though CD36 recognition could be conferred by the addition of only trace levels of specific oxPCs to a particle, one cannot exclude the possibility that it is thermodynamically favorable to form microdomains of oxPC molecules. The “pattern recognition” for oxPC species that has been exploited through evolution for CRP, CD36, and anti-oxLDL recognition thus may not be a single (monomeric) oxPC ligand, but rather a motif or “patch” of lipids presenting as a “raft” of oxPC species.
In recent years the plasma level of CRP has become a clinical diagnostic for assessing risk of atherosclerosis development, progression, and cardiovascular events, because it provides additive predictive benefit beyond that gleaned from conventional lipoprotein-associated risk factors (30, 31). Interest in CRP has catalyzed an awareness of the prominent role of inflammation in coronary artery disease pathogenesis (32–35). The recent growing interest in CRP as a marker for vascular disease risk has sparked both research and speculation as to CRP's possible roles in disease processes. It is imperative to understand the significance of CRP elevations—i.e., whether CRP (i) exacerbates the severity of inflammation and the progression of arterial lesions, (ii) reflects attempts by the body to protect itself, or (iii) accumulates as an inconsequential epiphenomenon. There are indications that CRP's role is more than passive (31, 32). Exogenous CRP, for example, enhanced complement activation and worsened myocardial damage in a rat coronary artery ligation infarction model (36). While there are dozens of papers confirming and furthering the value of CRP as a predictor of disease progression, there is less secure information about potential mechanistic links to atherosclerosis risk or protection. The findings of Chang et al. (6) provide insights into potential pathways linking CRP to lipoprotein and cholesterol handling by cells of the artery wall.
The in vivo roles played by CRP, autoantibodies to oxPC, and scavenger receptors, seemingly redundant protective pathways of the innate immune system, are matters of considerable interest. An approach used to assess the importance of the scavenger receptors has been to test their role in the progression of atherosclerosis in mice. Genetically engineered deficiencies in CD36 have been shown to reduce the progression of arterial disease in mouse models of atherosclerosis (24). The consequences of disrupting the CRP-mediated clearance mechanism inferred from the study by Chang et al. (6) are largely unknown. It will be particularly informative to assess directly the role of CRP–lipoprotein complexes on models of atherosclerosis and other inflammatory processes as more tools to block specific pathways for CRP–lipoprotein complex formation and uptake become available.
Figure.
Oxidized phosphatidylcholines as pattern recognition ligands for multiple pathways of innate immunity.
Acknowledgments
We thank Mr. David Schumick for graphic arts.
Footnotes
See companion article on page 13043.
References
- 1.Witztum J L, Berliner J A. Curr Opin Lipidol. 1998;9:441–448. doi: 10.1097/00041433-199810000-00008. [DOI] [PubMed] [Google Scholar]
- 2.Navab M, Hama S Y, Ready S T, Ng C J, Van Lenten B J, Laks H, Fogelman A M. Curr Opin Lipidol. 2002;13:363–372. doi: 10.1097/00041433-200208000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Glass C K, Witztum J L. Cell. 2001;104:503–516. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 4.Chisolm G M, Steinberg D. Free Radical Biol Med. 2000;28:1815–1826. doi: 10.1016/s0891-5849(00)00344-0. [DOI] [PubMed] [Google Scholar]
- 5.Podrez E A, Abu-Soud H M, Hazen S L. Free Radical Biol Med. 2000;28:1717–1725. doi: 10.1016/s0891-5849(00)00229-x. [DOI] [PubMed] [Google Scholar]
- 6.Chang M-K, Binder C J, Torzewski M, Witztum J L. Proc Natl Acad Sci USA. 2002;99:13043–13048. doi: 10.1073/pnas.192399699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zwaka T P, Hombach V, Torzewski J. Circulation. 2001;103:1194–1197. doi: 10.1161/01.cir.103.9.1194. [DOI] [PubMed] [Google Scholar]
- 8.Fu T, Borensztajn J. Biochem J. 2002;366:195–201. doi: 10.1042/BJ20020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palinski W, Horkko S, Miller E, Steinbrecher U P, Powell H C, Curtiss L K, Witztum J L. J Clin Invest. 1996;98:800–814. doi: 10.1172/JCI118853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horkko S, Bird D A, Miller E, Itabe H, Leitinger N, Subbanagounder G, Berliner J A, Friedman P, Dennis E A, Curtiss L K, et al. J Clin Invest. 1999;103:117–128. doi: 10.1172/JCI4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw P X, Horkko S, Chang M K, Curtiss L K, Palinski W, Silverman G J, Witztum J L. J Clin Invest. 2000;105:1731–1740. doi: 10.1172/JCI8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman P, Horkko S, Steinberg D, Witztum J L, Dennis E A. J Biol Chem. 2002;277:7010–7020. doi: 10.1074/jbc.M108860200. [DOI] [PubMed] [Google Scholar]
- 13.Chang M K, Bergmark C, Laurila A, Horkko S, Han K H, Friedman P, Dennis E A, Witztum J L. Proc Natl Acad Sci USA. 1999;96:6353–6358. doi: 10.1073/pnas.96.11.6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Beer F C, Soutar A K, Baltz M L, Trayner I M, Feinstein A, Pepys M B. J Exp Med. 1982;156:230–242. doi: 10.1084/jem.156.1.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rowe I F, Soutar A K, Trayner I M, Baltz M L, de Beer F C, Walker L, Bowyer D, Herbert J, Feinstein A, Pepys M B. J Exp Med. 1984;159:604–616. doi: 10.1084/jem.159.2.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nunomura W, Hatakeyama M. Hokkaido J Med Sci. 1990;65:474–480. [PubMed] [Google Scholar]
- 17.Bhakdi S, Torzewski M, Klouche M, Hemmes M. Arterioscler Thromb Vasc Biol. 1999;19:2348–2354. doi: 10.1161/01.atv.19.10.2348. [DOI] [PubMed] [Google Scholar]
- 18. Taskinen, S., Kovanen, P. T., Jarva, H., Meri, S. & Pentikainen, M. O. (2002) Biochem. J., in press. [DOI] [PMC free article] [PubMed]
- 19.Terpstra V, Bird D A, Steinberg D. Proc Natl Acad Sci USA. 1998;95:1806–1811. doi: 10.1073/pnas.95.4.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Podrez E A, Febbraio M, Sheibani N, Schmitt D, Silverstein R L, Hajjar D P, Cohen P A, Frazier W A, Hoff H F, Hazen S L. J Clin Invest. 2000;105:1095–1108. doi: 10.1172/JCI8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boullier A, Gillotte K L, Horkko S, Green S R, Friedman P, Dennis E A, Witztum J L, Steinberg D, Quehenberger O. J Biol Chem. 2000;275:9163–9169. doi: 10.1074/jbc.275.13.9163. [DOI] [PubMed] [Google Scholar]
- 22.Nozaki S, Kashiwagi H, Yamashita S, Nakagawa T, Kostner B, Tomiyama Y, Nakata A, Ishigami M, Miyagawa J, Kameda-Takemura K, et al. J Clin Invest. 1995;96:1859–1865. doi: 10.1172/JCI118231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huh H Y, Pearce S F, Yesner L M, Schindler J L, Silverstein R L. Blood. 1996;87:2020–2028. [PubMed] [Google Scholar]
- 24.Febbraio M, Podrez E A, Smith J D, Hajjar D P, Hazen S L, Hoff H F, Sharma K, Silverstein R L. J Clin Invest. 2000;105:1049–1056. doi: 10.1172/JCI9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Podrez E A, Poliakov E, Shen Z, Zhang R, Deng Y, Sun M, Finton P J, Shan L, Febbraio M, Hajjar D P, et al. J Biol Chem. 2002;277:38517–38523. doi: 10.1074/jbc.M205924200. [DOI] [PubMed] [Google Scholar]
- 26.Podrez E A, Poliakov E, Shen Z, Zhang R, Deng Y, Sun M, Finton P J, Shan L, Gugiu B, Fox P L, et al. J Biol Chem. 2002;277:38503–38516. doi: 10.1074/jbc.M203318200. [DOI] [PubMed] [Google Scholar]
- 27.Gillotte-Taylor K, Boullier A, Witztum J L, Steinberg D, Quehenberger O. J Lipid Res. 2001;42:1474–1482. [PubMed] [Google Scholar]
- 28.Huber J, Vales A, Mitulovic G, Blumer M, Schmid R, Witztum J L, Binder B R, Leitinger N. Arterioscler Thromb Vasc Biol. 2002;22:101–107. doi: 10.1161/hq0102.101525. [DOI] [PubMed] [Google Scholar]
- 29.Koynova R, Caffrey M. Biochim Biophys Acta. 1998;1376:91–145. doi: 10.1016/s0304-4157(98)00006-9. [DOI] [PubMed] [Google Scholar]
- 30.Ridker P M, Cushman M, Stampfer M J, Tracy R P, Hennekens C H. N Engl J Med. 1997;336:973–979. doi: 10.1056/NEJM199704033361401. [DOI] [PubMed] [Google Scholar]
- 31.Taubes G. Science. 2002;296:242–245. doi: 10.1126/science.296.5566.242. [DOI] [PubMed] [Google Scholar]
- 32.Pepys M B, Hirschfield G M. Ital Heart J. 2001;2:196–199. [PubMed] [Google Scholar]
- 33.Libby P. Sci Am. 2002;286(5):46–55. doi: 10.1038/scientificamerican0502-46. [DOI] [PubMed] [Google Scholar]
- 34.Blake G J, Ridker P M. Circ Res. 2001;89:763–771. doi: 10.1161/hh2101.099270. [DOI] [PubMed] [Google Scholar]
- 35.Ridker P M, Hennekens C H, Buring J E, Rifai N. N Engl J Med. 2000;342:836–843. doi: 10.1056/NEJM200003233421202. [DOI] [PubMed] [Google Scholar]
- 36.Griselli M, Herbert J, Hutchinson W L, Taylor K M, Sohail M, Krausz T, Pepys M B. J Exp Med. 1999;190:1733–1740. doi: 10.1084/jem.190.12.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]