Abstract
A key step in the rational design of new DNA binding agents is to obtain a complete thermodynamic characterization of small molecule-DNA interactions. Ethidium bromide has served as a classic DNA intercalator for more than four decades. This work focuses on delineating the influence(s) of the 3- and 8-amino substituents of ethidium on the energetic contributions and concomitant fluorescent properties upon DNA complex formation. Binding affinities decrease by an order of magnitude upon the removal of either the 3- or 8-amino substituent, with a further order-of-magnitude decrease in the absence of both amino groups. The thermodynamic binding mechanism changes from enthalpy-driven for the parent ethidium to entropy-driven when both amino groups are removed. Upon DNA binding, fluorescence enhancement is observed in the presence of either or both of the amino groups, likely because of more efficient fluorescence quenching through solvent interactions of free amino groups than when buried within the intercalation site. The des-amino ethidium analog exhibits fluorescence quenching upon binding, consistent with less efficient quenching of the chromophore through interactions with solvent than within the intercalation site. Determination of the quantum efficiencies suggests distinct differences in the environments of the 3- and 8-amino substituents within the DNA binding site.
INTRODUCTION
The ability to understand the interactions of small molecules with specific DNA sequences is fundamental in any attempt to control gene expression. In designing novel chemotherapeutic agents, one of the major strategies is to develop novel DNA binding ligands that influence crucial cellular processes such as DNA topology, replication, transcription, and DNA repair (Chaires, 1998; Haq et al., 2001). Systematic modifications of clinically effective chemotherapeutic agents have the potential for positively influencing their activity and delivery. Fundamental to this approach is the rigorous description of the interactions of known DNA binding agents with their macromolecular target.
Ethidium bromide has served as a classic model for small molecule-DNA interactions for more than four decades. Although there has been considerable work directed toward the elucidation of the thermodynamic, kinetic, and structural parameters that underlie the physicochemical properties exhibited upon DNA complex formation, there remain a number of unanswered questions. One important question pertains to the understanding of the role of 3- and 8-amino substituents in directing the energetics of the DNA binding process. The binding of ethidium to DNA has been described as involving two distinct steps—an initial electrostatic interaction between the negatively charged phosphate oxygens and the positively charged phenanthridinium ring nitrogen, followed by intercalative stacking interactions that stabilize the ligand-DNA complex through hydrophobic interactions. At high ligand concentrations, a secondary binding mode was observed involving external stacking of ethidium molecules in the DNA grooves resulting from ionic interactions with the phosphate backbone (Laugaa et al., 1983; Waring, 1965).
Structure-based techniques, such as x-ray diffraction and nuclear magnetic resonance (NMR), have been employed to achieve understanding of the interaction of ethidium with nucleic acids. Fuller and Waring (1964) used x-ray diffraction and molecular modeling studies to describe a model wherein the geometry of the intercalated ethidium chromophore was such as to orient the amino groups in close proximity to the charged oxygen atoms of the DNA phosphate backbone, facilitating additional stabilization of the intercalated complex through hydrogen-bonding interactions. This model placed the phenyl and ethyl groups of ethidium in the major groove of DNA. In 1983, Laugaa et al. (1983) report an alternative model structure. Their study examines the binding of ethidium and its azido analogs to ribodinucleosides using NMR and visible spectroscopic methods. Their studies support earlier crystallographic data that describe an ethidium-DNA complex wherein the long axis of ethidium is oriented parallel to that of the hydrogen-bonded dinucleosides with the phenyl and ethyl side chains lying in the minor groove (Jain et al., 1977). It is evident, therefore, that the binding geometry is not clearly defined; indeed, it might reflect binding involving both orientations.
Although structural studies have clear importance, they alone cannot provide a complete description of the binding event. To achieve a complete understanding, the nature of the molecular forces involved in complex formation must be defined. The thermodynamic characterization of the binding of ethidium bromide to DNA has been well defined in terms of the intercalation of the phenanthridinium ring system (Fuller and Waring, 1964; Jain et al., 1977; LePecq and Paoletti, 1967; Tsai et al., 1977; Waring, 1965); however, no direct quantitative assessment of the role(s) played by the amino substituents has been presented. The objectives of this work were to delineate the contributions of the 3- and 8-amino substituents of ethidium on the thermodynamics of binding associated with DNA complex formation. These studies were carried out using three analogs of ethidium: 3-amino, 8-amino, and des-amino ethidium (Fig. 1), synthesized as described by Firth et al. (1983). The DNA binding of ethidium and its three amino analogs will be described by obtaining complete thermodynamic profiles from the application of high-sensitivity isothermal titration calorimetry and fluorescence spectroscopy. Employing the methodology of Qu and Chaires (2000), we have exploited the significant changes in the fluorescence properties of ethidium bromide upon binding to DNA (LePecq and Paoletti, 1967) in the determination of its DNA binding properties. Furthermore, a model independent excess-site isothermal titration calorimetry (ITC) method allowed the determination of an enthalpy of binding. Knowledge of 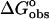 (from a binding constant) and
(from a binding constant) and 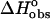 then yielded
then yielded 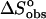 to permit the thermodynamics of the binding event to be fully described.
to permit the thermodynamics of the binding event to be fully described.
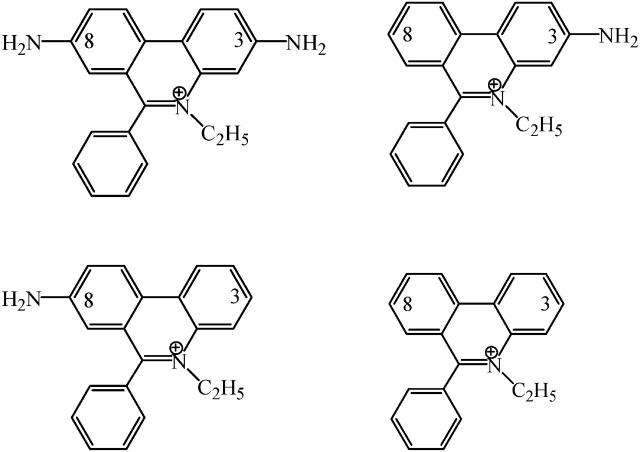
FIGURE 1.
Chemical structures of ethidium bromide and its amino analogs. (Top left) Ethidium, 3,8-diamino-5-ethyl-6-phenylphenanthridinium; (top right) 3-amino ethidium, 3-amino-5-ethyl-6-phenylphenanthridinium; (bottom left) 8-amino ethidium, 8-amino-5-ethyl-6-phenylphenanthridinium; and (bottom right) des-amino ethidium, 5-ethyl-6-phenylphenanthridinium.
MATERIALS AND METHODS
Sample preparation
All experiments were performed in BPES buffer (6 mM Na2HPO4, 4 mM NaH2PO4, 1 mM Na2EDTA, and 100 mM NaCl) at pH 7.00 ± 0.01. Ethidium bromide was purchased from Sigma-Aldrich (St. Louis, MO) and used without further purification. The ethidium analogs 3-amino-5-ethyl-6-phenylphenanthridinium chloride (3-amino ethidium), 8-amino-5-ethyl-6-phenylphenanthridinium chloride (8-amino ethidium), and 5-ethyl-6-phenylphenanthridinium chloride (des-amino ethidium) were synthesized as described previously (Firth et al., 1983). Ligand purities were determined by 1H NMR and the ligands stored in dry crystalline form at −20°C until used. Fresh solutions were prepared for each experiment by dissolving in BPES buffer. Solutions were filtered with 0.45-μm syringe filters (Millipore, Billerica, MA) before use in DNA binding experiments. Ligand concentrations were determined from visible absorbance maxima using previously determined molar extinction coefficients (ethidium bromide, ɛ478 = 5680 M−1 cm−1; 3-amino ethidium, ɛ437 = 4500 M−1 cm−1; 8-amino ethidium, ɛ430 = 3380 M−1 cm−1; and des-amino ethidium, ɛ377 = 4770 M−1 cm−1. For reference, see Firth et al., 1983). Calf thymus DNA was purchased from Sigma-Aldrich and prepared as described by Chaires et al. (1982). DNA solutions were stored at 4°C until used. The quality of the DNA was confirmed by ultraviolet and circular dichroism spectroscopy. DNA concentrations were determined spectrophotometrically using a molar extinction coefficient of ɛ260 = 13,200 cm−1 M−1 (bp) (Graves et al., 1981).
Determination of DNA binding constants
Binding isotherms were obtained using fluorescence spectroscopy based on the procedure of Qu and Chaires (2000). A series of DNA stock solutions were prepared spanning a concentration range of 1 nM to 1 mM (bp) at 0.25 log10 units. Hence, 25 data points defined a binding isotherm that covered six log10 concentration units. Nonlinear least-squares fitting of these data using ORIGIN v6.0 (Microcal, Northampton, MA) was employed to estimate values for the DNA binding constant and the fluorescence intensities of the free and bound ligands.
For an individual experiment, 3.0 mL of each DNA stock solution was pipetted into a separate vial, to which was added a small volume of a concentrated ligand solution (typically a few microliters). An appropriate concentration of ligand solution was selected to achieve a final ligand concentration of 3.33 μM with the addition of only a few microliters; in this way, dilution of the DNA solutions was negligible. The selected ligand concentration was the same for each ligand and was selected to be as low as possible while yielding an acceptable fluorescence signal. Use of low ligand concentrations were desired to obtain good estimates of the DNA binding constant while minimizing ligand-ligand stacking (Correia and Chaires, 1994).
Each vial was wrapped in aluminum foil before addition of ligand to prevent the possibility of any light-sensitive reactions. A fluorescence emission spectrum of each DNA-ligand solution was recorded on the Fluoromax-3 spectrofluorometer (Jobin Yvon Horiba, Edison, NJ) at 25°C with the data collection software DATAMAX v2.10. The excitation wavelength was set at the ligand-absorbance maximum and fluorescence emission was collected from 15 nm above the excitation wavelength up to 900 nm. It is important to set the lower limit of the emission scan >10-nm higher than the excitation wavelength to avoid a significant scattered-light component in the spectrum. Spectral measurements were typically performed with a bandwidth of 4 nm.
Primary spectra were imported into Microcal ORIGIN v6.0. In all cases, a shift in ligand emission maximum was observed with change in DNA concentration. For each complete binding isotherm the raw data were examined and an appropriate wavelength selected at which the change in fluorescence intensity with DNA concentration was maximized. Fluorescence intensity versus DNA concentration data were fit using the nonlinear least-squares method of Qu and Chaires (2000) by considering the equations
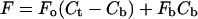 |
(1) |
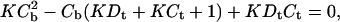 |
(2) |
where F is the observed fluorescence intensity at each DNA concentration, Fo is the fluorescence intensity of the free ligand, Fb is the fluorescence intensity of the bound ligand, Ct is the total ligand concentration, Cb is the bound ligand concentration, Dt is the total DNA concentration, and K is the DNA association constant.
A plot of F versus Dt at fixed Ct allows the determination of values for K, Fo, and Fb. Values were averaged from at least three, and as many as five, duplicate experiments for each ligand. Data were more easily visualized as a logarithmic abscissa plot.
Determination of quantum efficiencies
The quantum efficiency, Q, of a ligand is a measure of the amount of energy transferred from DNA to ligand upon binding and is evaluated from the ratio of the quantum efficiency of ligand bound to DNA (qb) to the quantum efficiency of free ligand (qf) using the equation (LePecq and Paoletti, 1967),
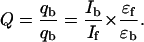 |
(3) |
The quantum efficiency for each of the ligands was estimated by determining the fluorescence intensities of bound (Ib) and free (If) ligand. These values were obtained during the construction of the fluorescence binding isotherm. It was then a matter of determining the molar extinction coefficients for each of the bound ligands (ɛb). Those for the free ligands (ɛf) were already known (Firth et al., 1983).
To determine the molar extinction coefficients of the bound ligands, small aliquots (5 or 10 μL) of a concentrated calf thymus DNA solution (∼7 mM (bp)) were titrated into a micromolar ligand solution. Using a Cary 4 UV-visible spectrophotometer (Varian, Walnut Creek, CA) at 25°C, the absorbance spectrum of the ligand solution was recorded in the absence of DNA and then after complete mixing of each serial titration. Cylindrical 10-cm pathlength quartz cells were selected for greater accuracy. For each ligand, a decrease in absorbance at the λmax was observed upon addition of increasing amounts of DNA, with each absorbance profile being a composite of the contribution of both free and bound ligands. Accounting for dilution, each observed absorbance was subtracted from the expected absorbance of a free ligand solution at an identical concentration. The free ligand absorbance was calculated from Beer's law, using the molar extinction coefficient for the free ligand. This subtraction gave the change in absorbance (ΔA) resulting from the increasing contribution of bound ligand. Values of ΔA were plotted against the inverse DNA concentration to obtain an exponential plot, the y intercept of which would yield the change in ligand absorbance as [DNA] approaches infinity, i.e., the change in ligand absorbance upon reaching a theoretically fully bound state. This value was obtained from a second-order polynomial fit of these data. The determination of ɛb was then derived from the equation
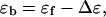 |
(4) |
where
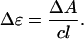 |
(5) |
In Eq. 5, c represents the total ligand concentration and is taken as the concentration of the starting (free ligand) solution, and l is the cell pathlength.
Determination of molar extinction coefficients of bound ligands were the average of three to four repeats. Quantum efficiencies were calculated from the determined fluorescence intensities of free and bound drug from each binding isotherm experiment, and were the average of between three and five determinations.
DNA binding energetics using isothermal titration calorimetry studies
Binding enthalpies were determined following the model independent excess-site method (Haq et al., 2001, 2000) using the CSC 4200 isothermal titration calorimeter (Calorimetry Sciences Corporation, Lindon, UT) at 25°C. All solutions were thoroughly degassed before use. The reference cell was filled to capacity with BPES buffer and the sample cell with calf thymus DNA (1 mM (bp); ∼1.3 mL; stir rate 297 rpm). Using the excess-site method, serial titration of ligand solution (1 mM; 20 × 10 μL; 400 s injection interval) into the sample cell, after thermal equilibration, yields identical titration peaks. Integration of these peaks (power, in microWatts, versus time, in seconds) using the software BINDWORKS v3.0 (Calorimetry Sciences Corporation), followed by normalization with respect to the number of moles of ligand, allowed a direct determination of the enthalpy of reaction. The observed enthalpies of reaction were corrected for the enthalpy of dilution of the ligand by performing separate experiments in which the ligand solution was titrated into a sample cell filled with BPES buffer. Subtraction of this enthalpy of dilution from the enthalpy of reaction for the titration of ligand into DNA yielded a value for the enthalpy of binding of the ligand to DNA. Ligand self-association was found to contribute a negligible heat effect (Ren et al., 2000). Enthalpies of ligand binding were averages of at least three repeats, and enthalpies of dilution were determined from averaging 3–5 experiments.
RESULTS
Fig. 2 shows a representative binding isotherm for ethidium bromide. Shown are fluorescence emission spectra after baseline subtraction for buffer for the 25 DNA concentrations ranging from 1 nM to 1 mM (bp) (Fig. 2 A). Fig. 2 B shows a plot of fluorescence intensity at λmax as a function of increasing DNA concentration; the inset is a plot in the form of log DNA concentration. The parameters K, Fo, and Fb were determined from the nonlinear least-squares fit of these data. The data represented in Fig. 2 were typical for the parent compound as well as the 3-amino and 8-amino ethidium analogs. For these three compounds the ligand emission maxima exhibited an increasing hyperchromic and hypsochromic (blue-shift) change in λmax with increasing DNA concentration. Conversely, the des-amino ethidium analog displayed hypochromicity upon DNA binding with no obvious shift in the wavelength of the emission maximum (Fig. 3).
FIGURE 2.
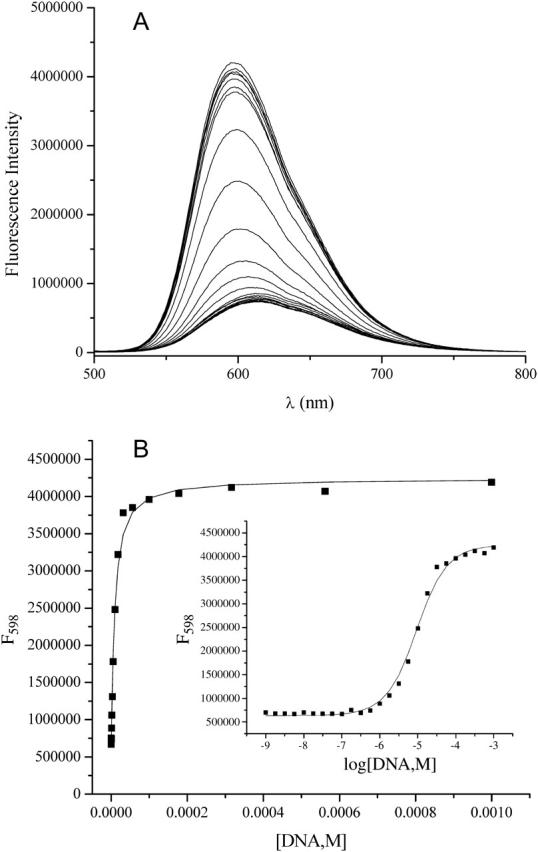
Determination of DNA binding constant for ethidium bromide using fluorescence spectroscopy. (A) Raw data in the form of fluorescence intensity versus wavelength. Note that the fluorescence profile of the ligand exhibits both hyperchromism and hypsochromism (blue shift) in the presence of increasing concentrations of calf thymus DNA. (B) Plot of fluorescence intensity at λmax as a function of increasing DNA concentration; the inset is a plot in the form of log DNA concentration. Binding parameters are determined from the nonlinear least-squares fitting routine described earlier.
FIGURE 3.
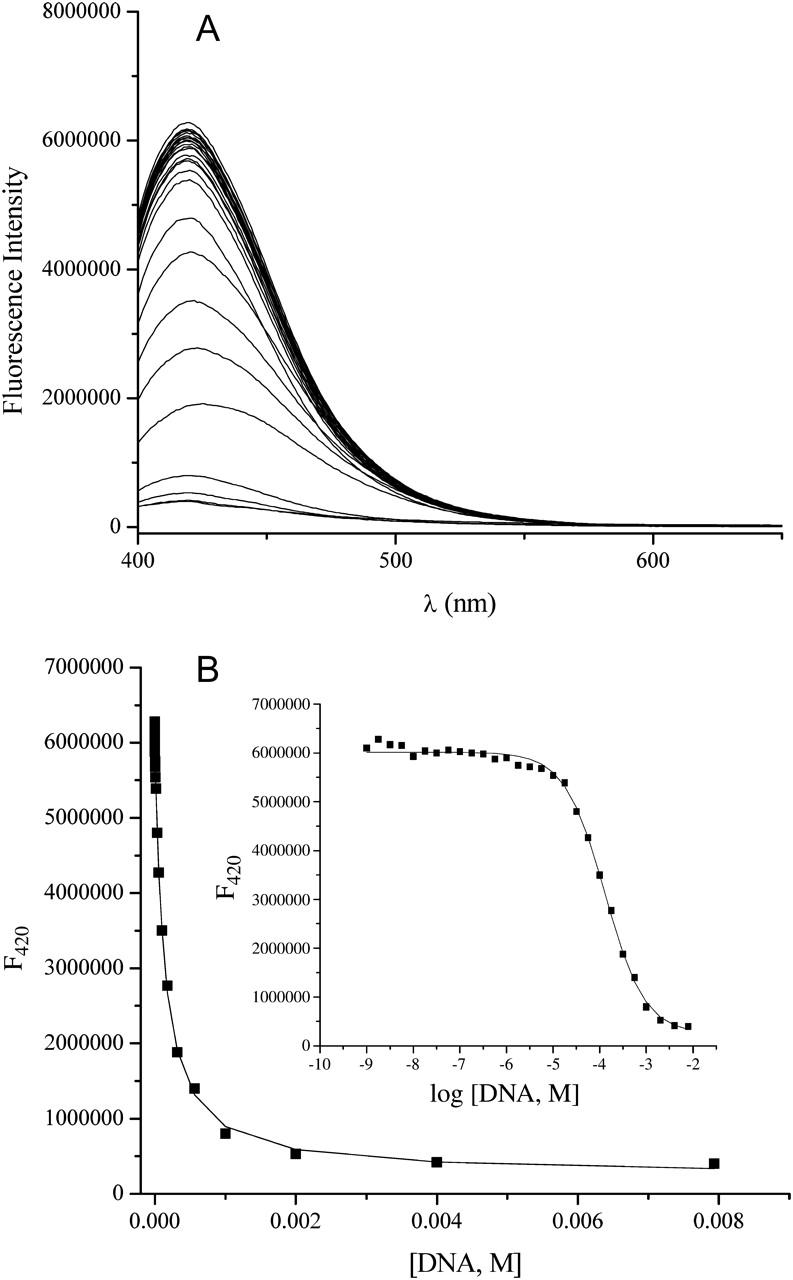
Determination of DNA binding constant for des-amino ethidium using fluorescence spectroscopy. (A) Raw data in the form of fluorescence intensity versus wavelength. Note that in contrast with Fig. 2, the fluorescence profile of the ligand exhibits hypochromism and no significant shift in wavelength maximum in the presence of increasing concentrations of calf thymus DNA. (B) Plot of fluorescence intensity at λmax as a function of increasing DNA concentration; the inset is a plot in the form of log DNA concentration. Binding parameters are determined from the nonlinear least-squares fitting routine described earlier.
Binding of ethidium and the ethidium analogs to DNA was found to be sensitive to both the number and the position of the 3- or 8-amino substituents. Ethidium bromide (3,8-diamino) exhibited the strongest binding affinity, i.e., (1.23 ± 0.07) × 105 M (bp)−1. In contrast, removal of one of the amino groups at either the 3- or 8-position resulted in a significant decrease in the binding affinity, i.e., 3-amino ethidium, (4.32 ± 0.54) × 104 M (bp)−1; and 8-amino ethidium, (5.83 ± 0.57) × 104 M (bp)−1. In the case of the absence of both amino groups (des-amino), the binding affinity decreased an additional order of magnitude to (7.56 ± 0.40) × 103 M (bp)−1 (Table 1). Although the binding constants for 3-amino and 8-amino ethidium were not identical, when considering the standard deviation of the experimental data they were of same order of magnitude and, hence, the binding of these analogs must be considered to exhibit only a very small difference in binding affinity.
TABLE 1.
DNA association constants and quantum yields determined at 25°C
| Ligand | K (M−1) | Q |
|---|---|---|
| Ethidium bromide | (1.23 ± 0.07) × 105 | 17.5 ± 0.3 |
| 3-Amino ethidium | (4.32 ± 0.54) × 104 | 13.3 ± 0.2 |
| 8-Amino ethidium | (5.83 ± 0.57) × 104 | 6.4 ± 0.2 |
| Des-amino ethidium | (7.56 ± 0.40) × 103 | 0.1 ± 0.0 |
Although the binding affinities for the 3-amino and 8-amino ethidium analogs are similar, the quantum efficiencies for these analogs were found to be markedly different. The value of Q determined for the parent ethidium was 17.5 ± 0.3. This can be reasonably compared with a reported value of 21 with solution conditions of 0.1 M NaCl and 0.1 M Tris-HCl (pH 7.5) and given the fact that the authors also report a dependence of DNA-bound ethidium bromide fluorescence on pH such that the observed fluorescence at pH 7.0 would be expected to be slightly lower than that at pH 7.5 (LePecq and Paoletti, 1967). A relative fluorescence enhancement of ∼15 has also been reported (Dedon, 2000). The quantum efficiency of 3-amino ethidium (13.3 ± 0.2) was much higher than that exhibited by 8-amino ethidium (6.4 ± 0.2), whereas des-amino ethidium displayed a quantum efficiency of <1 (0.1 ± 0.0), indicative of quenching of the fluorescence upon complex formation. These fluorescence data clearly indicate distinct differences in the DNA interaction geometries of the ethidium analogs that may be driven by the positions of the amino substituents.
Significant differences in the energetics of DNA complex formation due to the number and position of amino moieties on the phenanthridinium ring are also apparent from the calorimetry studies. Enthalpies of binding were determined using the model independent excess site method (Haq et al., 2000). Integration of the primary data and normalization for the number of moles of ligand added allowed the direct determination of the binding enthalpy. The enthalpy of binding of ligand to DNA was corrected for the enthalpy of dilution as discussed earlier. Significant variations in the binding enthalpies were evident for the ethidium compounds reflecting differences in the nature of the DNA binding interactions for each ligand (Fig. 4). The parent ethidium is characterized by a favorable binding enthalpy whereas the binding of des-amino is entropy-driven. The energetics of the 3- and 8-amino ethidium analogs lie in between these two extremes.
FIGURE 4.
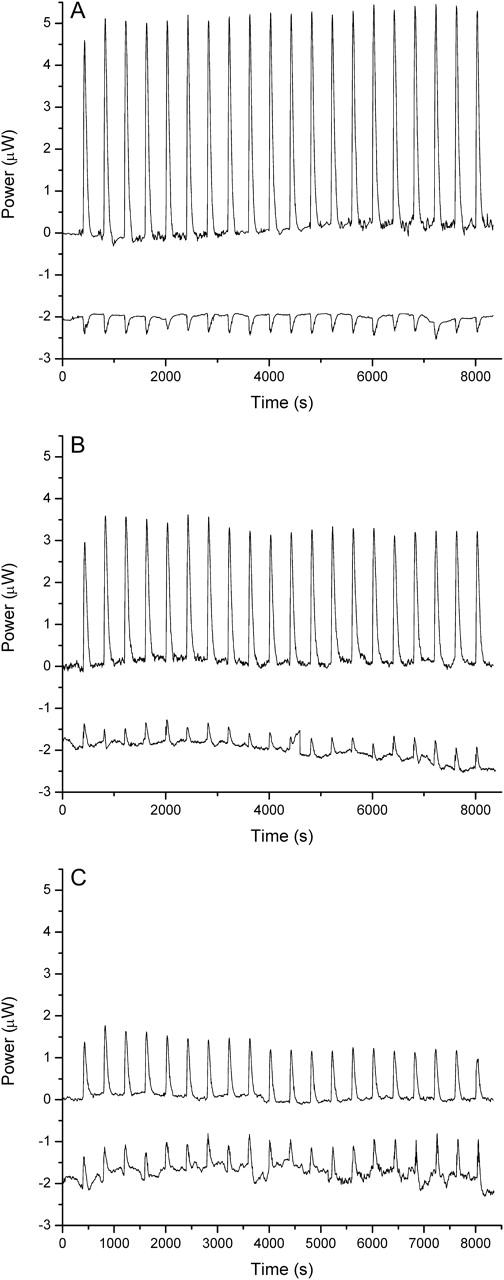
Representative energetic data from isothermal titration calorimetry studies. Raw ITC data for ethidium bromide (A), 3-amino ethidium (B), and des-amino ethidium (C) in the form of power (in microWatts) versus time (in seconds). The binding titration (top trace) represents a serial titration of ligand solution into a sample cell containing calf thymus DNA sufficiently concentrated to provide an excess of binding sites throughout the titration. The heats associated with ligand dilution are shown (bottom trace). Note that the CSC software represents exothermic events as positive peaks and endothermic events as negative peaks. These data were integrated and normalized for the number of moles of ligand, and appear in Table 2. Note that the raw binding titration data shown here represents heats from both ligand binding and dilution; the latter must be subtracted to obtain 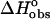 shown in Table 2.
shown in Table 2.
Use of a model independent excess site ITC method did not permit the determination of binding constants; however, these were determined from the fluorescence binding isotherms as discussed previously. Calculation of binding free energies were determined from the fluorescence binding constants using the equation 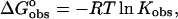 where R is the gas constant (1.987 cal K−1 mol−1) and T is the temperature in degrees Kelvin (298.15 K). Knowledge of the binding enthalpy from ITC allowed partitioning of the binding free energy into both enthalpic and entropic components:
where R is the gas constant (1.987 cal K−1 mol−1) and T is the temperature in degrees Kelvin (298.15 K). Knowledge of the binding enthalpy from ITC allowed partitioning of the binding free energy into both enthalpic and entropic components: 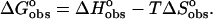
All ethidium compounds demonstrated favorable binding free energies. The parent ethidium exhibited the largest free energy of binding (−6.9 ± 0.1 kcal mol−1). Removal of one of the amino groups from either the 3- or 8-positions resulted in a 0.5-kcal mol−1 less-favorable binding free energy. Removal of both amino substituents resulted in a further decrease of 1 kcal mol−1 for des-amino ethidium.
The binding free energies reveal an enthalpy-entropy compensation commonly seen for ligand-DNA binding events. The binding of all compounds is characterized by a favorable enthalpic contribution which becomes increasingly less negative (less favorable) and is paralleled by an increasingly positive (favorable) entropic contribution (Table 2). The differences in binding entropies are likely to reflect changes in the ordered waters surrounding the ethidium chromophore resulting from the presence, absence, or position of the amino groups.
TABLE 2.
Thermodynamic characterization of the binding of ethidium bromide and its amino analogs to calf thymus DNA
| Ligand |
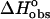 (kcal mol−1)* (kcal mol−1)*
|
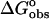 (kcal mol−1)† (kcal mol−1)†
|
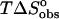 (kcal mol−1)‡ (kcal mol−1)‡
|
ΔHdiln (kcal mol−1)§ |
|---|---|---|---|---|
| Ethidium bromide | −8.7 ± 0.4 | −6.9 ± 0.1 | −1.8 ± 0.4 | 0.7 ± 0.1 |
| 3-Amino ethidium | −5.2 ± 0.6 | −6.3 ± 0.1 | 1.1 ± 0.5 | −0.6 ± 0.1 |
| 8-Amino ethidium | −4.5 ± 0.3 | −6.5 ± 0.1 | 2.0 ± 0.2 | −1.3 ± 0.2 |
| Des-amino ethidium | −0.8 ± 0.3 | −5.3 ± 0.1 | 4.5 ± 0.2 | −1.0 ± 0.2 |
Enthalpy of binding of the ligand to DNA. The ITC monitored titration of ligand into DNA yielded a direct determination of the enthalpy of reaction applying the model independent excess site method. To determine the enthalpy of binding a subtraction was made for the enthalpy of ligand dilution. (Also see footnote§.)
Determined from 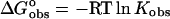 using association constants given in Table 1.
using association constants given in Table 1.
From 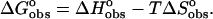
Heat of dilution of the ligand was determined from separate experiments in which the ligand solution was titrated into a sample cell filled with BPES buffer.
The heats of ligand dilution provide key insights into the interactions of the ligands within an aqueous environment. Ethidium bromide is shown to exhibit a positive (unfavorable) heat of dilution reflecting the disruption of favorable solvent interactions upon ligand dilution. All of the remaining ethidium analogs exhibit negative (favorable) heats of dilution suggesting differences in the nature of the solvation environment with the formation of more favorable solvent interactions upon dilution (Fig. 4).
DISCUSSION
The 3- and 8-amino groups of ethidium bromide play pivotal roles in directing the energetics of DNA complex formation and subsequent structural and fluorescent properties of the ligand-DNA complex. The aim of this study was to discern the influence of each amino group on both energetic contributions and concomitant fluorescent properties associated with complex formation. To this end, analogs of ethidium were synthesized with single amino groups at either the 3- or 8-positions or with neither of the amino groups on the phenanthridinium ring. Hence, these three ethidium analogs provided key ligand intermediates that could be used to dissect the distinct roles each of the amino substituents play in influencing DNA binding energetics and the fluorescent properties of the ligand-DNA complex. Moreover, the fluorescence properties of these analogs may provide insights regarding the environments of the amino groups within the DNA intercalation complex. The underlying reasons for the substantial enhancement in the fluorescence properties of intercalated ethidium bromide have not yet been fully described, and study of the ethidium analogs may serve to test the 1967 proposal of LePecq and Paoletti that the observed fluorescence enhancement of ethidium bromide upon binding to DNA under high salt conditions resulted from shielding of the ethidium amino groups within the intercalation site from quenching by aqueous solvent (LePecq and Paoletti, 1967).
Binding affinities were significantly influenced by the presence or absence of amino substituents on the phenanthridinium ring system. Ethidium bromide exhibited an association constant on the order of 105 M (bp)−1 with a decrease of one order of magnitude for the removal of a single amino substituent at either the 3- or 8-position. A further order-of-magnitude decrease in binding affinity was observed when both the 3- and 8-amino groups were removed. These observations translate into modest changes in binding free energy with an ∼0.5 kcal mol−1 decrease for the 3- and 8-amino ethidium analogs compared with the parent molecule and an additional 1-kcal mol−1 decrease in the binding free energy for the loss of both amino substituents. However, in the absence of both amino groups, a significant free energy of binding is observed, indicating that the stacking, hydrophobic, and van der Waals interactions of the phenanthridinium ring system within the intercalation site provide significant favorable contributions to the free energy of binding upon complex formation.
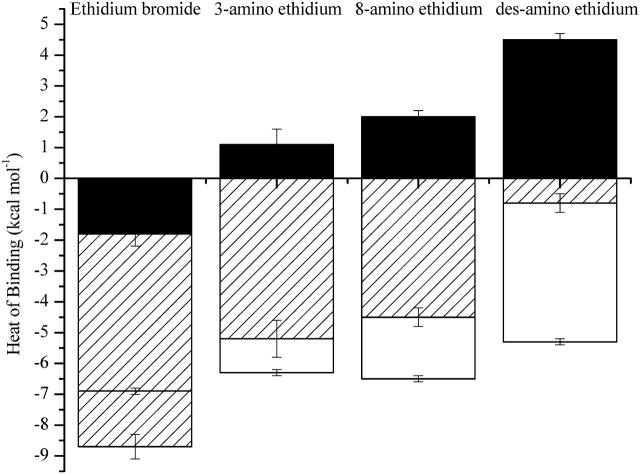
The modest differences observed in the free energies of complex formation underplay the considerable changes to both the enthalpic and entropic binding contributions upon removal of one or both amino substituents (Fig. 5). The DNA interaction of the parent ethidium is characterized by a very favorable binding enthalpy (−8.7 ± 0.4 kcal mol−1). However, removal of a single amino substituent from the phenanthridinium chromophore at either the 3- or 8-position results in an ∼4-kcal mol−1 decrease in favorable binding enthalpy. Furthermore, removal of both the 3- and 8-amino substituents results in an additional 4-kcal mol−1 decrease in favorable binding enthalpy to yield a near-zero enthalpy for the interaction of the des-amino ethidium analog with DNA. These large changes in binding enthalpy are indicative of the distinct role of the amino substituents in stabilizing bonding interactions between the ligand and DNA within the intercalation site. Furthermore, the presence of the amino substituents play a significant role in directing the thermodynamic mechanism of the binding interaction as the binding changes from an enthalpy- to an entropy-driven binding event with the removal of the amino substituents.
FIGURE 5.
Energetics of the interaction of ethidium bromide and its amino analogs with calf thymus DNA. (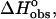 hatched bars;
hatched bars; 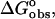 unfilled bars; and
unfilled bars; and 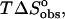 solid bars.)
solid bars.)
The significant entropy of binding (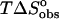 of +4.5 kcal mol−1) is the driving force behind the favorable interaction of des-amino ethidium with DNA. Such a large positive entropy change is likely to reflect the disruption of ordered water surrounding solvated des-amino ethidium molecules. In contrast, binding of the parent ethidium is enthalpically driven, with an unfavorable entropic contribution that may be a result of disrupted structured water because of the interactions of the amino groups with water at each end of the phenanthridinium chromophore. Upon first examination these observations may seem counterintuitive since the implication is that removal of the amino substituents appears to aid solvation. In fact, it is likely that the observations reflect subtle differences in the nature of the solvation environment of the ligand and DNA-ligand complex for each of the ethidium analogs. It is difficult to understand the energetic consequences completely, but some broad comments are appropriate. Consider that the ITC experiments reflect the titration of a concentrated ligand solution into either DNA (binding experiment) or buffer (dilution experiment), and that these observations reflect the dilution of the ligand solution into these media, and subsequent binding when the medium is DNA. Water forms a hydrogen-bonded solvation shell around polar solutes dispersing the solute throughout the solution; however, as water cannot bond with apolar substrates these are surrounded by a lattice of ordered water, a highly entropically costly process. For des-amino ethidium there is a significant entropic gain associated with the release of this structured water when disrupted and released back into the bulk medium as a result of its accommodation within the DNA intercalation site. However, the hydrogen-bonded solvation shells surrounding the polar amino groups on ethidium bromide will disrupt the ordered water lattice surrounding the phenanthridinium ring system. This will result in less entropic gain associated with DNA binding; and moreover, there will be a penalty for disruption of the favorable solvation shells associated with the amino groups. The binding entropies for the 3-amino and 8-amino ethidium analogs reflect solvation interactions intermediate between ethidium bromide and des-amino ethidium. A larger binding entropy is exhibited by the 8-amino analog where the presence of the 6-phenyl substituent lying perpendicular to the plane of the phenanthridinium chromophore results in anisotropic shielding of the amino group. Partial shielding of the 8-amino group from the formation of a hydrogen-bonded solvation shell would result in less disruption of the ordered water lattice surrounding the phenanthridinium ring. In contrast, the 3-amino ethidium analog has the 5-ethyl group in an equivalent position, which would confer considerably less shielding, resulting in the 3-amino group being more exposed to water facilitating hydrogen-bond formation and permitting less organization of the bulk water. The release of less structured water upon intercalation would be apparent as a less favorable entropy of binding.
of +4.5 kcal mol−1) is the driving force behind the favorable interaction of des-amino ethidium with DNA. Such a large positive entropy change is likely to reflect the disruption of ordered water surrounding solvated des-amino ethidium molecules. In contrast, binding of the parent ethidium is enthalpically driven, with an unfavorable entropic contribution that may be a result of disrupted structured water because of the interactions of the amino groups with water at each end of the phenanthridinium chromophore. Upon first examination these observations may seem counterintuitive since the implication is that removal of the amino substituents appears to aid solvation. In fact, it is likely that the observations reflect subtle differences in the nature of the solvation environment of the ligand and DNA-ligand complex for each of the ethidium analogs. It is difficult to understand the energetic consequences completely, but some broad comments are appropriate. Consider that the ITC experiments reflect the titration of a concentrated ligand solution into either DNA (binding experiment) or buffer (dilution experiment), and that these observations reflect the dilution of the ligand solution into these media, and subsequent binding when the medium is DNA. Water forms a hydrogen-bonded solvation shell around polar solutes dispersing the solute throughout the solution; however, as water cannot bond with apolar substrates these are surrounded by a lattice of ordered water, a highly entropically costly process. For des-amino ethidium there is a significant entropic gain associated with the release of this structured water when disrupted and released back into the bulk medium as a result of its accommodation within the DNA intercalation site. However, the hydrogen-bonded solvation shells surrounding the polar amino groups on ethidium bromide will disrupt the ordered water lattice surrounding the phenanthridinium ring system. This will result in less entropic gain associated with DNA binding; and moreover, there will be a penalty for disruption of the favorable solvation shells associated with the amino groups. The binding entropies for the 3-amino and 8-amino ethidium analogs reflect solvation interactions intermediate between ethidium bromide and des-amino ethidium. A larger binding entropy is exhibited by the 8-amino analog where the presence of the 6-phenyl substituent lying perpendicular to the plane of the phenanthridinium chromophore results in anisotropic shielding of the amino group. Partial shielding of the 8-amino group from the formation of a hydrogen-bonded solvation shell would result in less disruption of the ordered water lattice surrounding the phenanthridinium ring. In contrast, the 3-amino ethidium analog has the 5-ethyl group in an equivalent position, which would confer considerably less shielding, resulting in the 3-amino group being more exposed to water facilitating hydrogen-bond formation and permitting less organization of the bulk water. The release of less structured water upon intercalation would be apparent as a less favorable entropy of binding.
The nature of the solvation environment may be extended to consider the heats of ligand dilution. The des-amino ethidium analog exhibited a negative (favorable) heat of dilution reflecting the release of water molecules that had been associated with the structured clathrate-like solvation network within the concentrated ligand solution to participate in the hydrogen-bond network of bulk water. Heats of dilution for the 3- and 8-amino ethidium analogs were essentially similar to that for des-amino ethidium, suggesting that the energetic penalties associated with the disruption of the hydrogen-bonded solvation shells associated with the amino substituents has little energetic consequence for ligand dilution. However, the positive (unfavorable) heat of dilution observed for ethidium bromide suggests a more significant energetic penalty associated with the dilution of an ethidium analog with amino substituents in both the 3- and 8-positions. It is plausible that dilution may disrupt additional interactions that are maintained in a more concentrated ligand solution; such as, for example, intermolecular hydrogen-bonding between the amino groups.
Examination of the fluorescence quantum efficiencies supports the earlier conclusions regarding the differences between the interactions of the ethidium compounds with the solvation medium. As mentioned earlier, the quantum efficiency, Q, of a ligand is a measure of the amount of energy transferred from DNA to ligand upon binding and is evaluated as the ratio of the quantum efficiency of ligand bound to DNA to the quantum efficiency of free ligand. Therefore, a quantum efficiency of 1 would be expected for the circumstances where there is no transfer of energy from DNA to ligand, or where there is no difference in the relative energies of free and bound ligand. A value >1 indicates enhancement of the energy of the bound ligand, and a value <1 indicates quenching upon binding.
Des-amino ethidium displays hypochromicity of fluorescence emission upon binding with DNA, i.e., quenching, which is reflected in a Q-value of 0.1. All other analogs exhibit fluorescence enhancements in the presence of increasing concentrations of DNA and Q-values >1. The fluorescence enhancement of bound ligand compared with free ligand is indicative of greater retention of fluorescence energy by the bound ligand because of shielding within the DNA binding site from fluorescence quenching by solvent. Conversely, des-amino ethidium exhibits greater fluorescence when free in solution, which indicates that loss of fluorescence energy is more efficient when bound to DNA. These observations may suggest that a loss of fluorescence energy from the free ligand could occur more efficiently through interactions of the amino groups with water. In this way the fluorescence of des-amino ethidium would be more poorly quenched by solvent interactions with the chromophore when free in solution than by interactions of the chromophore within the DNA binding site. Conversely, the fluorescence of all ligands possessing amino substituents is more effectively quenched through interactions with the solvent than when the amino groups are shielded within the DNA molecule. The large disparity in quantum efficiencies between ethidium bromide (17.5 ± 0.3) and the 3-amino (13.3 ± 0.2) and 8-amino (6.4 ± 0.2) analogs suggests distinct differences in the environments of the 3- and 8-amino substituents within the DNA intercalation site, and may indicate differences in ligand binding orientation. Additional fluorescence studies are currently being completed to further understand these differences.
The studies presented here have focused on binding to calf thymus DNA. Competition dialysis studies in the Chaires laboratory have shown that ethidium bromide displays a preference for binding to RNA and DNA-RNA hybrid structures; the studies also showed that ethidium binds well to most normal duplex DNAs and the triplex structure poly(dA)-[poly(dT)]2 (Ren and Chaires, 1999). Application of competition dialysis studies to examine the binding preferences of the ethidium analogs may reveal important insights regarding the role of the amino groups in structural selectivity.
Acknowledgments
This work was supported by grant MCB-0334785 from the National Science Foundation.
References
- Chaires, J. B. 1998. Drug-DNA interactions. Curr. Opin. Struct. Biol. 8:314–320. [DOI] [PubMed] [Google Scholar]
- Chaires, J. B., N. Dattagupta, and D. M. Crothers. 1982. Studies on interaction of anthracycline antibiotics and deoxyribonucleic acid: equilibrium binding studies on the interaction of daunomycin with deoxyribonucleic acid. Biochemistry. 21:3933–3940. [DOI] [PubMed] [Google Scholar]
- Correia, J. J., and J. B. Chaires. 1994. Analysis of drug-DNA binding isotherms: a Monte Carlo approach. Methods Enzymol. 240:593–614. [DOI] [PubMed] [Google Scholar]
- Dedon, P. C. 2000. Determination of binding thermodynamics. In Current Protocols in Nucleic Acid Chemistry. E.W. Harkins, editor. John Wiley & Sons, Indianapolis, IN. [DOI] [PubMed]
- Firth, W. J., C. L. Watkins, D. E. Graves, and L. W. Yielding. 1983. Synthesis and characterization of ethidium analogs: emphasis on amino and azido substituents. J. Heterocycl. Chem. 20:759–765. [Google Scholar]
- Fuller, W., and M. J. Waring. 1964. A Molecular model for the interaction of ethidium bromide with deoxyribonucleic acid. Ber. Bunsen Ges. Phys. Chem. 68:805–808. [Google Scholar]
- Graves, D. E., C. L. Watkins, and L. W. Yielding. 1981. Ethidium bromide and its photoreactive analogues: spectroscopic analysis of deoxyribonucleic acid binding properties. Biochemistry. 20:1887–1892. [DOI] [PubMed] [Google Scholar]
- Haq, I., B. Z. Chowdhry, and T. C. Jenkins. 2001. Calorimetric techniques in the study of high-order DNA-drug interactions. Methods Enzymol. 340:109–149. [DOI] [PubMed] [Google Scholar]
- Haq, I., T. C. Jenkins, B. Z. Chowdhry, J. Ren, and J. B. Chaires. 2000. Parsing free energies of drug-DNA interactions. Methods Enzymol. 323:373–405. [DOI] [PubMed] [Google Scholar]
- Jain, S. C., C. C. Tsai, and H. M. Sobell. 1977. Visualization of drug-nucleic acid interactions at atomic resolution. II. Structure of an ethidium/dinucleoside monophosphate crystalline complex, ethidium:5-iodocytidylyl (3′-5′) guanosine. J. Mol. Biol. 114:317–331. [DOI] [PubMed] [Google Scholar]
- Laugaa, P., A. Delbarre, J.-B. Le Pecq, and B. P. Roques. 1983. Comparative binding of ethidium and three azido analogs to dinucleotides: affinity and intercalation geometry. Eur. J. Biochem. 134:163–173. [DOI] [PubMed] [Google Scholar]
- LePecq, J.-B., and C. Paoletti. 1967. A fluorescent complex between ethidium bromide and nucleic acids. Physical-chemical characterization. J. Mol. Biol. 27:87–106. [DOI] [PubMed] [Google Scholar]
- Qu, X., and J. B. Chaires. 2000. Analysis of drug-DNA binding data. Methods Enzymol. 321:353–369. [DOI] [PubMed] [Google Scholar]
- Ren, J., T. C. Jenkins, and J. B. Chaires. 2000. Energetics of DNA intercalation reactions. Biochemistry. 36:8439–8447. [DOI] [PubMed] [Google Scholar]
- Ren, J., and J. B. Chaires. 1999. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry. 38:16067–16075. [DOI] [PubMed] [Google Scholar]
- Tsai, C. C., S. C. Jain, and H. M. Sobell. 1977. Visualization of drug-nucleic acid interactions at atomic resolution. I. Structure of an ethidium/dinucleoside monophosphate crystalline complex, ethidium:5-iodouridylyl (3′-5′) adenosine. J. Mol. Biol. 114:301–315. [DOI] [PubMed] [Google Scholar]
- Waring, M. J. 1965. Complex formation between ethidium bromide and nucleic acids. J. Mol. Biol. 13:269–282. [DOI] [PubMed] [Google Scholar]