Abstract
Heme-copper oxidases are membrane proteins found in the respiratory chain of aerobic organisms. They are the terminal electron acceptors coupling the translocation of protons across the membrane with the reduction of oxygen to water. Because the catalytic process occurs in the heme cofactors positioned well inside the protein matrix, proton channels must exist. However, due to the high structural divergence among this kind of proteins, the proton channels previously described are not necessarily conserved. In this work we modeled the structure of the quinol oxidase from Acidianus ambivalens using comparative modeling techniques for identifying proton channels. Additionally, given the high importance that water molecules may have in this process, we have developed a methodology, within the context of comparative modeling, to identify high water probability zones and to deconvolute them into chains of ordered water molecules. From our results, and from the existent information from other proteins from the same superfamily, we were able to suggest three possible proton channels: one K-, one D-, and one Q-spatial homologous proton channels. This methodology can be applied to other systems where water molecules are important for their biological function.
INTRODUCTION
Heme-copper oxidases are the terminal oxidases of respiratory chains in eukaryotic and prokaryotic organisms. These transmembrane proteins pump protons across the membrane (Antonini et al., 1993; Capitanio et al., 1997; Pereira et al., 1999; Purschke et al., 1997; Verkhovskaya et al., 1997; Wikstrom, 2004) while catalyzing the reduction of oxygen to water (Babcock and Wikstrom, 1992) with the consumption of four electrons and four protons. The proton pumping mechanism contributes to the generation of an electrochemical gradient that is used by the organisms to synthesize ATP. The homology of heme-copper oxidases along different organisms is quite low. For example, whereas the mitochondrial cytochrome c oxidase is constituted of 13 different subunits, the oxidases from prokaryotic organisms have on average only four subunits, while retaining similar functional efficiency (Abramson et al., 2001; Pereira et al., 2001). Nevertheless, and despite these differences, subunits I and II are very similar among all organisms. Subunit I contains the binuclear center, composed of one high-spin heme and one copper atom (CuB), where the reduction of oxygen occurs. The electrons needed for oxygen reduction come from the low-spin heme found at close distance from this binuclear center (Namslauer et al., 2002; Verkhovsky et al., 2001). These electrons may arrive at this heme from different sources, depending on the type of oxidase. Although in subunit II of high-potential iron-sulfur protein (HiPIP) oxidases we find bimetallic copper centers acting as electron bridges between HiPIP and the low-spin heme (Pereira et al., 2002, 1999, 2001), in quinol oxidases the electrons are transferred directly to the low-spin heme by soluble molecules (like the caldariella quinone in the quinol oxidase of Acidianus ambivalens (Giuffrè et al., 1997; Tricone et al., 1989)), given that no copper center is found in subunit II.
Throughout the years, several hypotheses emerged concerning the mechanism responsible for the proton pumping. Musser and Chan (1998), trying to propose an evolutionary relationship between all heme-copper oxidases, suggested that the proton pumping mechanism would occur differently in cytochrome c oxidases and in quinol oxidases (Musser and Chan, 1998). However, only after taking into account amino acid analysis (Pereira et al., 2001), mutagenic (Aagaard et al., 2000; Backgren et al., 2000; Brändén et al., 2002; Gennis, 1998c; Hellwig et al., 2001; Karpefors et al., 1998a,b; Ma et al., 1999; Meunier, 2001; Tomson et al., 2003), and structural experiments (Abramson et al., 2001; Gennis, 1998a; Iwata et al., 1995; Michel et al., 1998; Ostermeier et al., 1997; Pereira et al., 1999; Souliname et al., 2000a,b; Svensson-Ek et al., 2002; Tsukihara et al., 1995, 1996; Yoshikawa et al., 1998) was it possible to suggest the existence of proton channels responsible not only for the proton pumping mechanism across the membrane, but also for the control of protons accessing the binuclear center for catalytic purposes (Brzezinski and Larsson, 2003; Ferguson-Miller and Babcock, 1996; Gennis, 1998b,c; Gomes et al., 2001; Michel, 1998; Mills and Ferguson-Miller, 1998; Mills et al., 2000; Pereira et al., 2002, 1999, 2001; Verkhovsky et al., 1999; Wikstrom, 1998, 2000; Wikstrom et al., 2000, 2003). The two main proton channels initially identified in heme-copper oxidases were the D- and the K-proton channels present in mitochondrial-like enzymes (type A oxidase; Pereira et al., 2001; Pereira and Teixeira, 2004). The D-proton channel can be divided into two main regions: one hydrophilic and one hydrophobic region (Iwata et al., 1995; Ostermeier et al., 1997). The first one is composed of a series of charged and polar residues going from the cytoplasmic surface of the protein to the key residue Glu-278 (Paracoccus denitrificans numbering), whereas the second one goes from the former residue to the binuclear center (Iwata et al., 1995; Ostermeier et al., 1997). However, the residues identified as important to this proton channel are not conserved in all heme-copper oxidases. For example, the cytochrome c oxidase aa3 from Thermus thermophilus (Mather et al., 1993) and the caa3 HiPIP/oxygen oxidoreductase from Rhodothermus marinus (Pereira et al., 1999), show all the conserved residues found in the P. denitrificans oxidase with the exception of Glu-278 (type A2 oxidase; Pereira et al., 2001). In these two proteins it was suggested that a motif consisting of one Tyr and one Ser would substitute Glu-278 from the P. denitrificans oxidase in delivering the protons to the binuclear center (Pereira et al., 1999; Santana et al., 2001). We have suggested before in the R. marinus oxidase (Pereira et al., 1999; Santana et al., 2001) that the Tyr residue of this motif could either be a protonable residue or a link in an ordered chain of water molecules. Our recent studies (Soares et al., 2004) give more support to the latter hypothesis, given that Tyr-262 (R. marinus numbering) is not proton active, whereas Glu-278 from P. denitrificans aa3 oxidase seems to be, even if it is mostly protonated in the states analyzed. Other oxidases (denominated as B-type oxidases; Pereira et al., 2001; Pereira and Teixeira, 2004), like the ba3 from T. thermophilus (Souliname et al., 2000a,b) and the quinol oxidase aa3 from A. ambivalens (Gilderson et al., 2001; Gomes et al., 2001), do not seem to show any conserved residues in this channel, although they are able to pump protons across the membrane. The other mentioned proton channel (K-channel) is much more conserved in all oxidases when compared to the D-channel (Pereira et al., 2001; Pereira and Teixeira, 2004). This channel is also composed of various charged and polar residues that are used to deliver protons directly to the binuclear center. In the aa3 oxidase from P. denitrificans this channel is composed of Lys-354, Thr-351, Ser-291, and Tyr-280 (covalently linked to His-276 coordinating the copper atom; MacMillan et al., 1999). Recent articles have also described a conserved glutamate residue found in subunit II in the majority of terminal oxidases (Glu-101 in Rhodobacter sphaeroides oxidase (Brändén et al., 2002; Tomson et al., 2003), Glu-89 in Escherichia coli ubiquinol oxidase (Ma et al., 1999) and Glu-78 in P. denitrificans cytochrome c oxidase (Kannt et al., 1998)) evidencing interesting features in the proton pumping mechanism through the K-channel. According to several authors (Brändén et al., 2002; Ma et al., 1999; Tomson et al., 2003), the region around this glutamate residue coincides with the beginning of the proton channel, therefore evidencing the high importance that this residue may have to the proton translocation process through the K-channel. Despite the mentioned high conservation found in the K-channel of different oxidases, the small differences in its composition (Pereira et al., 2001) do not seem to highly affect the proton pumping mechanism.
In this work we study the quinol oxidase aa3 from the thermoacidophilic archaeon A. ambivalens (Aa-aa3). This protein belongs to a very simple membrane-bound respiratory chain where the intermediate caldariella quinone (a quinone specific to this organism) transfers the electrons to the final membranar quinol oxidase (Aagaard et al., 1999; Das et al., 2004; Gomes et al., 2001). Previous studies have revealed unusual properties of of this protein, among other things, in what concerns the proton-pumping mechanism (Aagaard et al., 1999; Das et al., 1999; Gilderson et al., 2001; Giuffrè et al., 1997; Gomes et al., 2001; Hellwig et al., 2003; Purschke et al., 1997). To understand the structural reasons for the unusual properties of this protein, a three-dimensional (3D) model is needed. Because no experimental structure is available, we have modeled the 3D structure of Aa-aa3 with comparative modeling techniques. A previous theoretical model of subunit I of this protein had already been generated (Gomes et al., 2001). However, given that this model was based only on crystallographic structures with low sequence identity with Aa-aa3 (the only ones available at that time), the quality of the generated model could not be very good. Presently, crystallographic structures evidencing a higher sequence identity with Aa-aa3 are available, allowing, therefore, the design of a much more accurate model of this oxidase.
The translocation of protons through the proton channels found in heme-copper oxidases was previously suggested to occur by a Grotthus-type mechanism (Hofacker and Schulten, 1998; Pereira et al., 2001). According to this mechanism the proton transport is the result of a bond formation/breaking process, where the proton is transferred between chains of water molecules positioned in hydrophilic protein cavities (Luecke et al., 1998, 1999; Nagle and Morowitz, 1978; Nagle and Tristam-Nagle, 1983; Sham et al., 1999). Consequently, the identification of the positions of internal water molecules in the structure of the protein may be very useful to understand proton pumping mechanisms. Taking into account all these considerations we have modeled subunits I and II of Aa-aa3 with internal water molecules. From our results we were able to confirm the presence of a previously suggested K- and D-spatial homologous proton channel in subunit I of this protein (Gomes et al., 2001). Additionally, it was also possible to suggest another proton channel, named as Q-spatial homologous channel, due to its spatial similarity with the Q-channel found in the cytochrome c oxidase from T. thermophilus (Koutsoupakis et al., 2004; Pereira et al., 2001).
MATERIALS AND METHODS
Structure modeling
The three-dimensional structure of subunits I and II of Aa-aa3 were modeled based on the same two subunits from the ubiquinol oxidase (bo3) structure from E. coli (Ec-bo3) (PDB entry 1FFT; Abramson et al., 2000) and from the cytochrome c oxidase (ba3) structure from T. thermophilus (Tt-ba3) (PDB entry 1EHK; Souliname et al., 2000a). These two structures were chosen as templates given that these are the ones showing higher sequence homology with Aa-aa3 among all available terminal oxidase structures. For modeling subunit I of Aa-aa3 both template proteins were used, even if the corresponding subunit from Tt-ba3 showed a higher sequence homology (24% identity) when compared with the Ec-bo3 (19% identity). In the case of subunit II a more complex alignment had to be used. The transmembrane segment domain showed a much higher sequence homology with the corresponding domain of Tt-ba3 than with Ec-bo3. Therefore, for this region, only the Tt-ba3 sequence was used. In contrast, for the soluble domain the opposite was observed, and in this case only the Ec-bo3 structure was used. It should be mentioned in this section that the sequence of subunit II of Aa-aa3 used in the modeling protocol was the one coded by the doxC gene and not by the doxA gene (the doxA gene had been previously denominated by Purchke et al., 1997 as the one coding for the pseudosubunit II of Aa-aa3). This was done taking into account the fact that Kletzin et al. (2004) have proven that the doxA gene does not code for the pseudosubunit II of Aa-aa3 but rather for one subunit of the thiosulfate/quinone oxidoreductase.
Despite the fact the template structures used in this work did not show a high sequence identity with Aa-aa3, it should be mentioned that the main structural fold of subunits I and II in heme-copper oxidases are extremely similar (Pereira et al., 2001), revealing that the folds of these proteins are quite resistant to changes in the primary structure.
Modeller v6.1 (Sali and Blundell, 1993) was used to model Aa-aa3 by comparative modeling methods, using the sequence of the target protein and the structures of the templates. The templates were structurally aligned and the sequence of Aa-aa3 was aligned against this profile. Models were generated using this initial alignment and then used to derive new alignments to improve the final quality of the protein structure model. This quality is evaluated using two criteria: the observed restraint violations reported by Modeller and the conformational and stereochemistry of the generated models (performed by program Procheck; Laskowski et al., 1993). After this optimization, 300 different models of subunits I and II of the Aa-aa3 protein were generated, and the one evidencing the lowest value for the objective function from Modeller (Sali and Blundell, 1993) was chosen as the best model.
Modeling of water molecules
The identification of internal water molecules in terminal oxidases is of outmost interest to understand how this kind of proteins is able to pump protons across the membrane, and also to understand how the protons needed to catalyze the reduction of oxygen arrive at the binuclear center. However, the models of Aa-aa3 did not contain water molecules. To model them, we have used the program Dowser (Zhang and Hermans, 1996), which identifies water molecules by measuring their interaction energy at different places in the protein. A cutoff of −10 kcal/mol was used to insert a water molecule (Hofacker and Schulten, 1998; Zhang and Hermans, 1996). Dowser has already been used in the modeling of internal water molecules in other terminal oxidases (Hofacker and Schulten, 1998; Zheng et al., 2003). Our tests, for instance, with the Tt-ba3 protein, showed that this procedure was successful in predicting crystallographic water molecules.
For the problem at hand, other more sophisticated and probably more accurate procedures could be used to predict individual water positions more precisely, using, for instance, grand canonical Monte Carlo (Resat et al., 1997). However, these procedures are much more computationally demanding and, for this reason, not adequate for the size and the number of different structures we were to analyze (300 structures). Therefore, we followed a less computationally demanding procedure such as Dowser.
Water probability density function
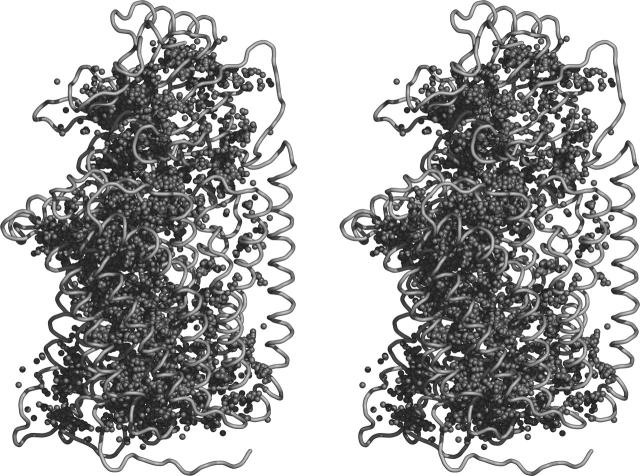
Models obtained by comparative modeling techniques using low-homology templates present considerable conformational variability. This variability is essentially present on exposed loops that have no correspondence on the templates and on the conformation of side chains. For the most variable zones (i.e., these exposed loops), no clustering of water molecules is observed. Therefore, depending on the model used, a different set of water molecules would be added. To circumvent this we have implemented a general protocol that can capture the average distribution of water molecules in the protein. This protocol generates a water probability density function (WPDF) and then searches this density function for the most probable regions to position water molecules. First, all 300 generated protein models (with water molecules added by Dowser) were fitted to the one with the lowest value of the objective function of Modeller. In Fig. 1 the best protein model is shown with all water molecules added to all generated models. As can be seen, water molecules are concentrated in specific places of the structure, evidencing a heterogeneous distribution inside the protein. The WPDF was then built based on all these water molecules. To define the function we started by embedding all water molecules inside a three-dimensional grid of 53 × 86 × 61 points, with a grid mesh of 1 Å (the dimensions of the grid were chosen so that all water molecules were within the grid). Then, the occupancy of each water molecule is split and assigned to the closest eight grid points using a trilinear weighting as done in Edmonds et al. (1984) and Klapper et al. (1986). In the end, this value is normalized considering the total number of conformations. See Fig. 2 a for more details about the graphic representation of the WPDF.
FIGURE 1.
Stereo representation of the best model of Aa-aa3 generated by Modeller (Sali and Blundell, 1993). In this figure all water molecules added to all generated models are represented by their oxygen atoms rendered as small spheres. Figure generated using Pymol (DeLano, 2003).
FIGURE 2.
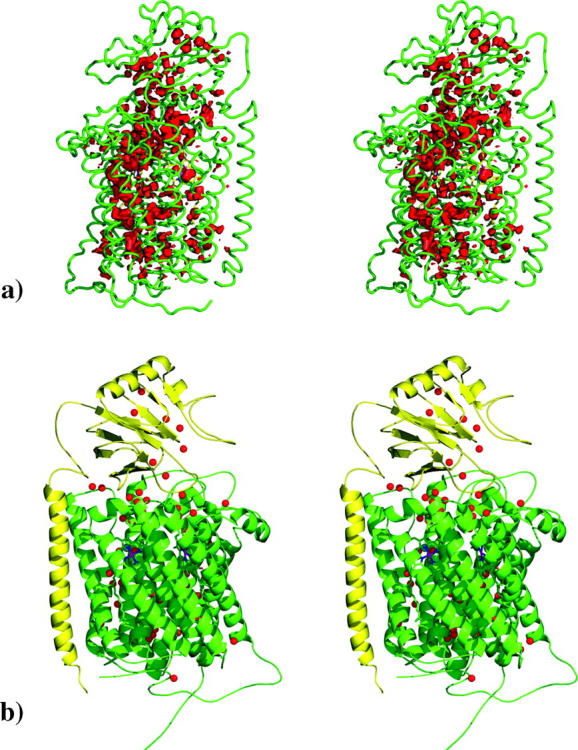
(a) Graphical representation of the WPDF in Aa-aa3. The protein model (subunit I in green and subunit II in yellow) is rendered as ribbon, and the WPDF is rendered as a red contour. Only the regions with WPDF >0.005 are shown. (b) Stereo view of the best model of Aa-aa3 in a cartoon representation, with internal water molecules represented as small red spheres. Subunit I is colored in green, and subunit II is colored in yellow. Heme cofactors are shown in dark blue. This figure was generated with the program Pymol (DeLano, 2003).
Modeling of water molecules as Gaussian functions
After computing the WPDF, the next step was to choose a method to model the water molecules back in the model, considering this average situation, instead of the individual structures. Therefore, we decided to mathematically model each water molecule as a Gaussian function
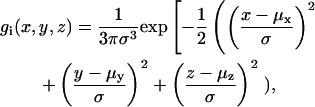 |
(1) |
where σ represents the spread of the water molecule across the grid, x, y, and z are the three-dimensional coordinates of a grid point and μx, μy, and μz are the coordinates of the considered water molecule. To compute the global water density generated by the water conformations we calculated the sum of all Gaussian functions
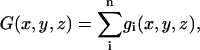 |
(2) |
which represents all the n water molecules being simulated. The σ parameter in Eq. 1, specifying the dimensions of a single water molecule over the grid, was assigned the value yielding the best overlap between the global Gaussian function of an ensemble of water molecules and the WPDF function. This best value was determined by performing Metropolis Monte Carlo simulations at constant temperature in the waters position space using different values for the σ parameter. Note that for these and for other calculations we used the grid containing the WPDF and we did not take into account any protein conformational information. This is because we are only interested in the water positions that are fully determined by the probability values present in the grid. An ensemble of 59 water molecules was used in all simulations, given that this corresponded to the average number of water molecules among all different models generated by Modeller. The initial spatial position of this ensemble of water molecules was randomly generated. In each simulation 6000 Monte Carlo steps (6000 spatial random movements for each water molecule) were performed. The constant temperature used in each simulation was chosen as the temperature where an acceptance/rejection Monte Carlo step ratio closest to unity could be obtained. With this, a good search of the conformational space was achieved. At each Monte Carlo step the overlap between the global Gaussian function of the ensemble of water molecules G(x,y,z) and the WPDF f(x,y,z) was computed as
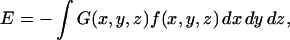 |
(3) |
which is taken as the energy function (in arbitrary units) to be used in the Monte Carlo simulations. Each time two water molecules of the chosen ensemble were within a distance <2.758 Å (the distance between two oxygen atoms from two water molecules establishing a hydrogen bond), an overlapping configurational penalty energy of 1000 arbitrary energy units was added to Eq. 3. This prevents clashes between water molecules.
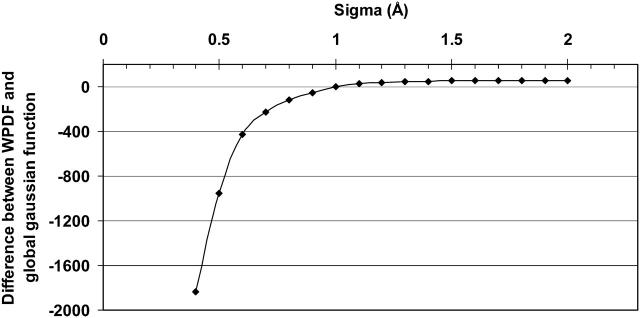
As can be seen in Fig. 3, the difference between the probabilities generated by the Monte Carlo simulations (Eq. 3) and the WPDF is close to zero when the σ value is set to 1.0 Å.
FIGURE 3.
Difference between the WPDF and the probability generated by Monte Carlo simulations (Eq. 2) using different σ values. The difference between the functions is close to zero at a σ value of 1.0 Å.
Locating best water positions
After developing a method to model the water molecules and after parameterizing the Gaussian function specifically for the system we are studying, we are in conditions to determine the most probable positions of the water molecules. To do that we performed Monte Carlo simulated annealing (MCSA), where the initial positions of the 59 water molecules were randomly generated, as previously described. Using the same energy function as before (Eq. 3), and with the σ parameter set to unity (Eq. 1), the MCSA simulation consisted on 24 consecutive cycles of Monte Carlo simulations, each one performed at a gradually lower temperature. The temperature used in the first temperature cycle was equal to 0.5 (arbitrary units). In each new temperature cycle, a temperature reduction factor of 0.7 was applied to the temperature used in the previous cycle. With this protocol, a good search of the conformational space was achieved, given that the first cycles present a high ratio of accepted/rejected Monte Carlo steps whereas in the last cycles this ratio is low. In each temperature cycle, 1000 Monte Carlo steps were performed. An initial maximum translation step of 5 Å is used in the first cycle, and this value is reduced in the following cycles using a factor 0.92. At the end of the 24 temperature cycles of the MCSA simulation, the 59 most probable positions for the water molecules in the protein were obtained.
After the determination of the most probable positions for the 59 water molecules, we have used again the program Modeller (Sali and Blundell, 1993), to generate 20 new models of Aa-aa3, using the same procedure as described before (structure modeling) but this time the 59 water molecules configurations previously determined were included in the comparative modeling procedure as any other residue. In this way the protein structure is modeled together with the water molecules. The model with the lowest value for the Modeller (Sali and Blundell, 1993) objective function was chosen as the best protein model (Fig. 2 b).
RESULTS AND DISCUSSION
In Fig. 2 b the best model of Aa-aa3 is presented. As can be seen in this figure, subunit I (colored in green) is constituted of 12 transmembrane helices connected by several different types of loops. Imbedded in the protein matrix we can find the heme a cofactor with its iron atom coordinated by His-70 and His-381. In close proximity to this cofactor we find the binuclear center composed of one heme a3 cofactor and one copper atom (CuB). The iron atom of the heme a3 is distally coordinated by His-379, whereas the CuB cofactor is coordinated by His-244, His-293, and His-294. Subunit II (colored in yellow in Fig. 2 b) is composed of one transmembrane helical segment, which is connected by a loop to one soluble domain, where no metallic cofactors are found. As can be seen in Fig. 2 b, several water molecules are positioned between the β-sheets. However, it is in subunit I that these molecules are found in larger concentration. As mention before, the positioning of water molecules in regions identified as possible proton channels can be quite revealing in what concerns the proton translocation process. Protons captured in the cytoplasm are translocated through the different proton channels and are directed to the binuclear center of the protein where the catalytic process occurs, or to the opposite side of the membrane, contributing to the chemiosmotic gradient.
Through a detailed analysis of the best Aa-aa3 model and by comparing it with the structure of other resolved oxidases (from Tt-ba3, Souliname et al., 2000a; and P. denitrificans, Iwata et al., 1995), we were able to suggest three different proton channels: one K-, one D-, and one Q-spatial homologous proton channel.
K-spatial homologous proton channel
As described before, Aa-aa3 is a B-type oxidase, not having a conserved K-proton channel as found in the A-type oxidases (like the P. denitrificans heme-copper oxidase) (Pereira et al., 2001; Pereira and Teixeira, 2004). Despite this, Aa-aa3 has other polar and charged residues that may be able to form proton channels directly to the binuclear center (Gomes et al., 2001). The K-pathway identified in the P. denitrificans oxidase goes from Lys-354, near the cytoplasm, to Tyr-280, close to the binuclear center (Table 1) (P. denitrificans numbering). As in other heme-copper oxidases (Buse et al., 1999; MacMillan et al., 1999; Tomson et al., 2002), Tyr-280 is covalently linked to His-276, which coordinates the copper atom of the binuclear center. As can be seen in Fig. 4 and in Table 1, in the model of Aa-aa3 we can also find a tyrosine residue — Tyr-248 (A. ambivalens numbering) in a homologous position. In our model, from Tyr-248 toward the cytoplasmatic side, other residues with polar side chains can also be found: Tyr-283, Ser-319, Ser-279, Thr-322, Tyr-259, Asn-325, and Ser-272 (A. ambivalens numbering) (Fig. 4 and Table 1). Despite the fact that the only putative protonatable residues in this set are Tyr-283 and Tyr-259, it is possible that the polar side chains of other residues can be used in the stabilization of water molecules as previously described (Hofacker and Schulten, 1998). This suggestion becomes stronger when we analyze Fig. 4. As can be seen, between the polar residues previously described we were able to identify six different water molecules that may be involved in the proton translocation mechanism through this channel. These water molecules are apparently stabilized by the polar side chains of the residues constituting this channel and by the hydrogen bridges formed among them.
TABLE 1.
Putative 3D equivalent K-channel residues in different heme-copper oxidases
| A. ambivalens | T. thermophilus | P. denitrificans |
|---|---|---|
| Tyr-248* | Tyr-237* | Tyr-280* |
| Ser-319* | Ser-309* | Thr-351* |
| Tyr-283* | Phe-272 | Ile-315 |
| Val-255 | Tyr-244* | – |
| Thr-322* | Thr-312* | Lys-354* |
| Tyr-259* | Tyr-248* | Ser-291* |
| Asn-325* | Thr-315 | Ser-357 |
| Thr-329* | Ser-319 | Thr-361 |
| Ser-272* | Ser-261 | – |
| Val-15 (II) | Glu-15 (II)* | Glu-78 (II)* |
| Glu-8 (II)* | Lys-9 (II) | Thr-72 (II) |
Polar residues likely to be involved in the proton translocation are marked with an asterisk.
FIGURE 4.
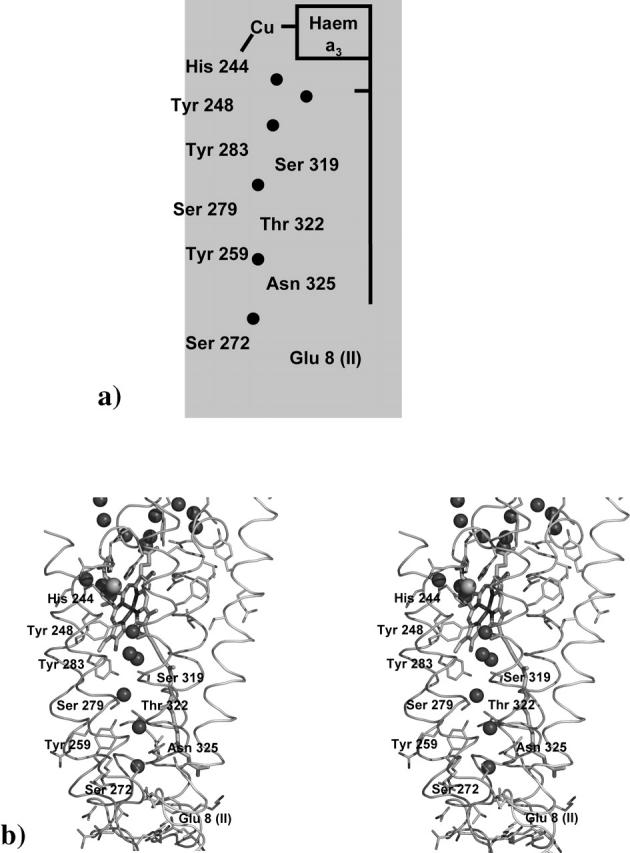
Proposed K-spatial homologous channel found in Aa-aa3. In panel a, a scheme of the proposed channel is presented, whereas in panel b the same region in the best generated model is shown. Water molecules (small spheres) and possible important residues found in this channel are identified. Panel b was prepared using the program Pymol (DeLano, 2003).
Comparing the spatial position of the residue side chains in the Aa-aa3 model with the same regions on the structure of Tt-ba3 (another B-type oxidase) and the cytochrome c oxidase from P. denitrificans (an A-type oxidase) we can see that Aa-aa3 is much more similar (in what concerns the residues composing it) to the former than to the latter. However, despite this similarity, the K-homologous channels in these two proteins are different (see Table 1).
As described for other heme-copper oxidases (Kannt et al., 1998; Ma et al., 1999; Soares et al., 2004; Tomson et al., 2003), it is possible to find in subunit II a glutamate residue that has been described as part of the K-channel (Glu-78 in P. denitrificans, Glu-71 in R. marinus, Glu-101 R. sphaeroides, Glu-89 in E. coli). Tomson et al. (2003) and Brändén et al. (2002) have described that the mutation of this residue led to the blocking of the proton conduction through the K-channel in R. sphaeroides. Similar results have been reported for the bo3 oxidase in E. coli (Ma et al., 1999). In Aa-aa3 (Fig. 4 and Table 1) we have found Glu-8(II) in a similar spatial position as those found in other heme-copper oxidases (see Fig. 4). FTIR experiments performed in Aa-aa3 (Hellwig et al., 2003) showed the existence of at least four acid residues in this protein where changes in its side-chain reorganization or protonation state were evident. Glu-8 from subunit II of Aa-aa3 is one strong candidate for this proton active behavior. In our recent studies (Soares et al., 2004) in the P. denitrificans and in the R. marinus oxidases it was possible to identify Glu-78 and Glu-71, respectively, as two homologous residues found in subunit II of both proteins with a proton active behavior, even if at a low degree. In conclusion, due to its structural position and to the previously described FTIR experiments we can suggest that Glu-8 in subunit II in Aa-aa3 is also a member of this K-spatial homologous channel.
D-spatial homologous proton channel
As previously described by other authors (Gomes et al., 2001; Pereira et al., 2002, 2001; Pereira and Teixeira, 2004), B-type oxidases do not have a conserved D-channel. In T. thermophilus cytochrome c oxidase (Souliname et al., 2000a,b), a channel has been identified that is spatially equivalent to the D-channel found in P. denitrificans oxidase (Iwata et al., 1995), which is composed of several polar residues (Table 2). When we compare the residues within the zone of the D-channel in the T. thermophilus and in the A. ambivalens oxidases, we can see that, although different, both proteins present several polar residues that can constitute the basis for proton channels. As can be seen in Fig. 5, the beginning of the D-channel in Aa-aa3 is close to Lys-93 and Lys-15. Above the position of these two residues we find one water molecule stabilized by the polar side chains of Tyr-89 and Thr-19. Another water molecule can also be found in close proximity (∼2.7 Å), which seems to be stabilized not only by the previous water molecule but also by the side chain of Asn-106. Above this second water molecule we have also identified another one stabilized by the side chain of Ser-174. Flanking this residue we found four different water molecules. Two of them are on the left of Ser-174 (closer to the membrane face of the protein), whereas the other two are buried deeper inside the protein. The first two water molecules seem to be stabilized by Thr-170, Asn-113, and two protonatable residues: Glu-80 and Asp-167. The side chains of these two residues are turned to each other, having a water molecule hydrogen bonded to both of them. In our model of Aa-aa3 we have not found the conserved and proton active Glu-278 from P. denitrificans (P. denitrificans numbering) as also have been previously described (Das et al., 1999; Gilderson et al., 2001; Gomes et al., 2001). However, it has been suggested (Gomes et al., 2001; Hellwig et al., 2003) that Glu-80 (A. ambivalens numbering) may act as a substitute for the Glu-278 from the P. denitrificans oxidase and other canonical heme-copper oxidases. A double mutation in R. sphaeroides oxidase tried to mimic Aa-aa3 (Aagaard et al., 2000), where Glu-286 was substituted by an alanine, whereas Ile-122 (R. sphaeroides numbering) was substituted by a glutamate, showing that in these conditions the protein was capable of proton pumping. This result and the results obtained in FTIR experiments (Hellwig et al., 2003) showed that the spatial arrangement observed in this region of Aa-aa3, although not similar with other arrangements observed in canonical heme-copper oxidases, allows the full functionality of the protein.
TABLE 2.
Putative 3D equivalent D-channel residues in different heme-copper oxidases
| A. ambivalens | T. thermophilus | P. denitrificans |
|---|---|---|
| Val-246 | Ile-235 | Glu-278* |
| Asp-167* | Phe-152 | Ser-189 |
| Glu-80* | Gln-82* | Ile-104 |
| Asn-113* | Ala-113 | Tyr-138 |
| Tyr-171* | Thr-156* | Ser-193* |
| Thr-170* | Ser-155* | Ser-192 |
| Phe-84 | Gln-86* | Gly-109 |
| Phe-109 | Ser-109* | Ser-134* |
| Val-22 | Phe-24 | Tyr-35* |
| Thr-19* | Thr-21* | Gly-32 |
| Asn-106* | Met-106 | Asn-131* |
| Tyr-89* | Tyr-91* | Tyr-114* |
| Ile-177 | Ile-162 | Asn-199* |
| Lys-93* | Arg-95 | Leu-118 |
| Lys-15* | Glu-17* | Asp-30 |
Polar residues likely to be involved in the proton translocation are marked with an asterisk.
FIGURE 5.
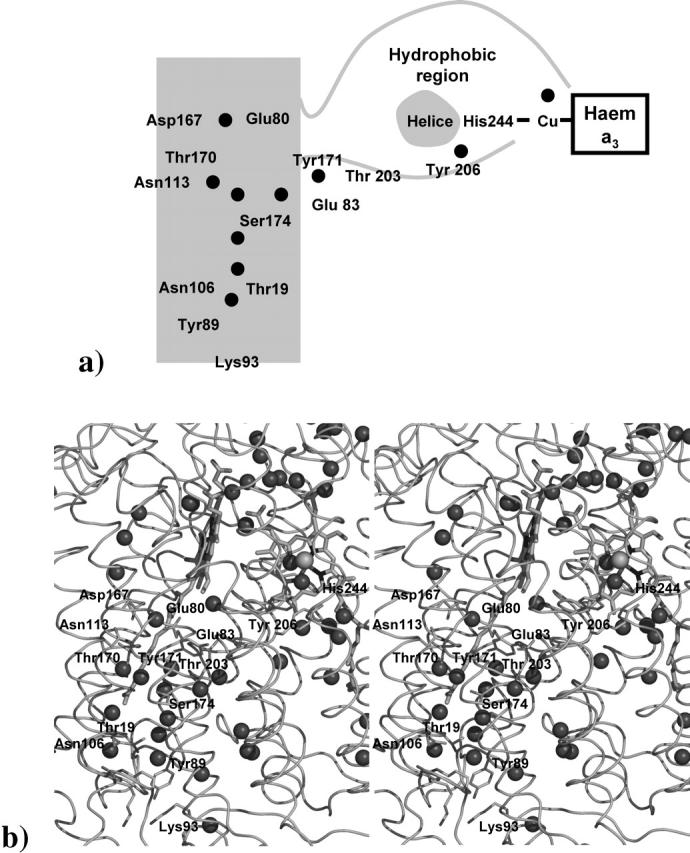
D-spatial homologous channel in Aa-aa3. In panel a, a scheme of the channel is presented, whereas in panel b a zoom in this region of Aa-aa3 is shown. Water molecules found in the structure (represented as small spheres) and possible important residues found in this channel are identified. Panel b was prepared using the program Pymol (DeLano, 2003).
The other two water molecules positioned in a more interior region of the protein near Ser-174 seem to be stabilized by three different residues: Tyr-171, Thr-203, and Glu-83. These three residues are structurally positioned in what seems to be an alternative path to translocate the protons through the D-channel to the catalytic center (Fig. 5). A path of several polar residues from this region to the catalytic center (ending near His-244, the residue coordinating the copper atom and covalently bound to the already described Tyr-248) can be found, suggesting a possible alternative way of delivering protons to the catalytic site of the protein (see Fig. 5). This alternative path can also be a way of connecting the D-channel and the K-channel in a process previously suggested (Wikstrom et al., 2003; Zheng et al., 2003).
Between the copper atom and the iron of the heme cofactor we can also find another water molecule. Its position here may suggest the place of a possible first escape path to where the water molecules are redirected after their generation in the catalytic process.
Q-spatial homologous proton channel
In Tt-ba3, a third proton channel was previously described that seems to have no similarity with any of the previously described proton channels existent in canonical oxidases (Koutsoupakis et al., 2004; Pereira et al., 2001). This channel, named as Q-channel, is situated between hemes b and a3 and was proposed to function as a proton-transfer mechanism or as a proton-pumping system that originates from the D-pathway (Pereira et al., 2001). In the Aa-aa3 model, it seems that this pathway is not present because no water molecules or conserved residues were found in similar positions as the ones observed in Tt-ba3. However, it is possible to observe in our model that several water molecules and residues with polar side chains are found in what seems to be a possible proton channel. This channel is positioned right behind the low-spin heme (heme a) and seems to start near Arg-406. Right above this residue we can find a chain of four water molecules stabilized by the side chains of several polar residues that may lead protons through the protein matrix to the catalytic site or to the other side of the membrane. This chain starts in one water molecule stabilized by Thr-404 and is followed by another one stabilized by two other residues: Ser-410 and Tyr-397. In an upper position we can find a third water molecule stabilized by Thr-481 and His-417. The last water molecule in this channel is stabilized by Tyr-421, which is closer to His-381 (coordinating the iron atom of heme a) in this channel. From this point, the Q-spatial homologous proton channel seems to be divided in two: in one possible path, protons flowing through the channel can go to the opposite side of the membrane, whereas through the other possible path protons may be translocated to the region close to the heme a3 propionates where the catalytic process occurs. No experimental evidences are available to support these suggestions. However, we propose that the Q-spatial homologous proton channel may be an alternative way of leading the protons to the catalytic center or to the opposite site of the membrane.
CONCLUDING REMARKS
The Aa-aa3 is a B-type oxidase with very particular characteristics in what concerns its putative proton channels. To understand in detail the structural factors coupled to this mechanism, we have used theoretical tools in the construction of a three-dimensional model of Aa-aa3 with explicit internal water molecules. In our model we have identified three possible proton channels: the K-, the D-, and the Q-spatial homologous proton channel. The K-channel in Aa-aa3, although the spatial equivalent to the one found in other heme-copper oxidases of the A-type, does not show any conservation in what concerns the residues composing it. Despite this, the K-spatial homologous proton channel is composed of other residues with polar side chains that can lead the protons directly to the catalytic center. Several water molecules, stabilized by the side chains of these residues, were also found in a spatial distribution that may allow its full participation in the proton pumping mechanism through this channel. The D-spatial homologous proton channel suggested from our model is also quite different when compared with the D-channels identified in all other heme-copper oxidases with available structures. For example, we could find in this channel neither the glutamate residues characteristic of the type-A1 oxidases, nor the Tyr-Ser motif from the type-A2 oxidases. However, we were able to identify Glu-80 (A. ambivalens numbering) as a possible key residue of the D-spatial homologous channel. Due to its spatial position, this residue may act, as previously suggested (Gomes et al., 2001), as a possible substitute for the Glu-278 from P. denitrificans in the proton translocation mechanism through the D-channel. The positioning of several water molecules in this channel was very helpful in the identification of other residues with polar side chains that may be important in the proton translocation process through this channel. The third proton channel suggested from the analysis of Aa-aa3 was the Q-spatial homologous proton channel. As its name describes, this channel is positioned in the protein in a similar region (although not entirely) as the suggested Q-channel found in Tt-ba3. According to our model this channel is positioned right behind the low-spin heme of Aa-aa3, and is constituted of a chain of residues with polar side chains and several water molecules in a distribution that may allow the redirection of protons to the catalytic center.
With our model we were able to understand in more detail the structural features of Aa-aa3 that may be responsible for the proton-pumping capability previously described. The methodology developed and applied here can be very useful for the study of other systems where structural experimental information is not available.
FIGURE 6.
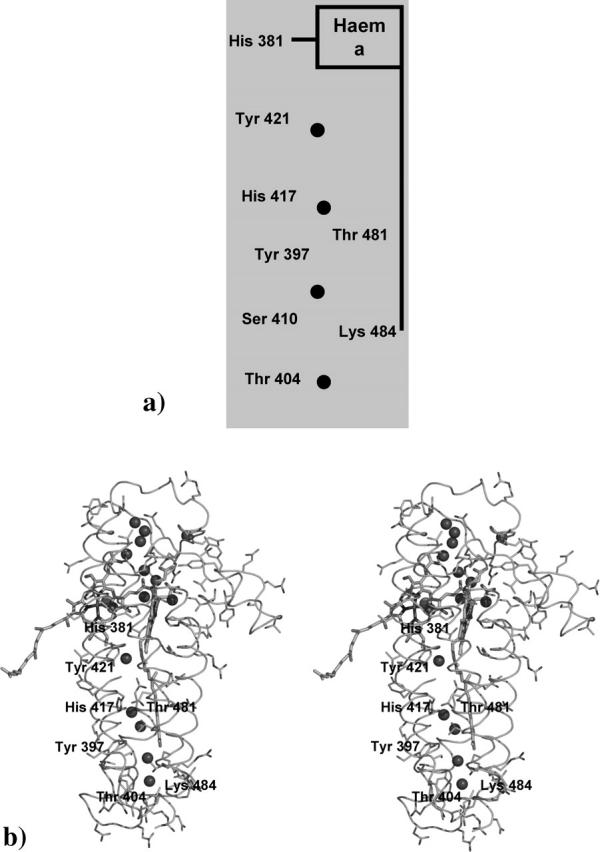
Q-spatial homologous channel in Aa-aa3. In panel a, a scheme of the channel is presented, whereas in panel b a zoom in this region of Aa-aa3 is shown. Water molecules found in the structure (represented as small spheres) and possible important residues found in this channel are identified. Panel b was prepared using the program Pymol (DeLano, 2003).
Acknowledgments
We thank Professor Miguel Teixeira, Dr. Cláudio M. Gomes, and Tiago Bandeiras for fruitful discussions.
This work was supported by grant POCTI/BME/32789/99 and by fellowship SFRH/BD/10622/2002 from Fundação para a Ciência e a Tecnologia, Portugal.
References
- Aagaard, A., G. Gilderson, C. M. Gomes, M. Teixeira, and P. Brzezinski. 1999. Dynamics of the binuclear center of the quinol oxidase from Acidianus ambivalens. Biochemistry. 38:10032–10041. [DOI] [PubMed] [Google Scholar]
- Aagaard, A., G. Gilderson, D. A. Mills, S. Ferguson-Miller, and P. Brzezinski. 2000. Redesign of the proton-pumping machinery of the cytochrome c oxidase: proton pumping does not require glu(I-286). Biochemistry. 39:15847–15850. [DOI] [PubMed] [Google Scholar]
- Abramson, J., S. Riistama, G. Larsson, A. Jasaitis, M. Svensson-Ek, L. Laakkonen, A. Puustinnen, S. Iwata, and M. Wilkstrom. 2000. The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. Nat. Struct. Biol. 7:910–917. [DOI] [PubMed] [Google Scholar]
- Abramson, J., M. Svensson-Ek, B. Byrne, and S. Iwata. 2001. Structure of cytochrome c oxidase: a comparison of the bacterial and mitochondrial enzymes. Biochim. Biophys. Acta. 1544:1–9. [DOI] [PubMed] [Google Scholar]
- Antonini, G., F. Malatesta, P. Sarti, and M. Brunori. 1993. Proton pumping by cytochrome c oxidase as studied by time-resolved stopped-flow spectrophotometry. Proc. Natl. Acad. Sci. USA. 90:5949–5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock, G. T., and M. Wikstrom. 1992. Oxygen activation and the conservation of energy in cell respiration. Nature. 356:301–309. [DOI] [PubMed] [Google Scholar]
- Backgren, C., G. Hummer, M. Wikstrom, and A. Puustinen. 2000. Proton translocation by cytochrome c oxidase can take place without the conserved glutamic acid in subunit I. Biochemistry. 39:7863–7867. [DOI] [PubMed] [Google Scholar]
- Brändén, M., F. Tomson, R. B. Gennis, and P. Brzezinski. 2002. The entry point of the K-proton-transfer pathway in cytochrome c oxidase. Biochemistry. 41:10794–10798. [DOI] [PubMed] [Google Scholar]
- Brzezinski, P., and G. Larsson. 2003. Redox-driven proton pumping by heme-copper oxidases. Biochim. Biophys. Acta. 1605:1–13. [DOI] [PubMed] [Google Scholar]
- Buse, G., T. Souliname, M. Dewor, H. E. Meyer, and M. Blüggel. 1999. Evidence for a copper-coordinated histidine-tyrosine cross-link in the active site of cytochrome oxidase. Protein Sci. 8:985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capitanio, N., G. Capitanio, E. De Nitto, and S. Papa. 1997. Vectorial nature of redox Bohr effects in bovine heart cytochrome c oxidase. FEBS Lett. 414:414–418. [DOI] [PubMed] [Google Scholar]
- Das, T. K., C. M. Gomes, T. M. Bandeiras, M. M. Pereira, M. Teixeira, and D. L. Rousseau. 2004. Active site structure of the aa3 quinol oxidase of Acidianus ambivalens. Biochim. Biophys. Acta. 1655:306–320. [DOI] [PubMed] [Google Scholar]
- Das, T., C. M. Gomes, M. Teixeira, and D. Rousseau. 1999. Redox-linked transient deprotonation at the binuclear site in the aa3-type quinol oxidase from Acidianus ambivalens: implications for proton translocation. Proc. Natl. Acad. Sci. USA. 96:9591–9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano, W. L. 2003. The Pymol Molecular Graphics System: Version 0.90. DeLano Scientific, San Carlos, CA.
- Edmonds, D., N. Rogers, and J. Sternberg. 1984. Regular representation of irregular charge distributions. Application to the electrostatic potentials of globular proteins. Mol. Phys. 52:1487–1494. [Google Scholar]
- Ferguson-Miller, S., and G. T. Babcock. 1996. Heme/copper terminal oxidases. Chem. Rev. 96:2889–2907. [DOI] [PubMed] [Google Scholar]
- Gennis, R. B. 1998a. Cytochrome c oxidase: one enzyme, two mechanisms? Science. 280:1712–1713. [DOI] [PubMed] [Google Scholar]
- Gennis, R. B. 1998b. How does cytochrome oxidase pump protons? Proc. Natl. Acad. Sci. USA. 95:12747–12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennis, R. B. 1998c. Multiple proton-conducting pathways in cytochrome oxidase and a proposed role for the active-site tyrosine. Biochim. Biophys. Acta. 1365:241–248. [Google Scholar]
- Gilderson, G., A. Aagaard, C. M. Gomes, P. Adelroth, M. Teixeira, and P. Brzezinski. 2001. Kinetics of electron and proton transfer during O2 reduction in cytochrome aa3 from A. ambivalens: an enzyme lacking Glu(I-286). Biochim. Biophys. Acta. 1503:261–270. [DOI] [PubMed] [Google Scholar]
- Giuffrè, A., C. M. Gomes, G. Antonini, E. D'Itri, M. Teixeira, and M. Brunori. 1997. Functional properties of the quinol oxidase from Acidianus ambivalens and the possible catalytic of its electron donor. Eur. J. Biochem. 250:383–388. [DOI] [PubMed] [Google Scholar]
- Gomes, C. M., C. Backgren, M. Teixeira, A. Puustinnen, M. Verkhovskaya, M. Wikstrom, and M. Verkhovsky. 2001. Heme-copper oxidases with modified D- and K-pathways are yet efficient proton pumps. FEBS Lett. 497:159–164. [DOI] [PubMed] [Google Scholar]
- Hellwig, P., B. Barquera, and R. Gennis. 2001. Direct evidence for the protonation of aspartate-75, proposed to be at a quinol binding site, upon reduction of cytochrome bo3 from E. coli. Biochemistry. 40:1077–1082. [DOI] [PubMed] [Google Scholar]
- Hellwig, P., C. M. Gomes, and M. Teixeira. 2003. FTIR spectroscopic characterization of the cytochrome aa3 from Acidianus ambivalens: evidence for the involvement of acidic residues in redox coupled proton translocation. Biochemistry. 42:6179–6184. [DOI] [PubMed] [Google Scholar]
- Hofacker, I., and K. Schulten. 1998. Oxygen and proton pathways in cytochrome c oxidase. Proteins. 30:100–107. [PubMed] [Google Scholar]
- Iwata, S., F. Osterberg, B. Ludwig, and H. Michel. 1995. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 376:660–669. [DOI] [PubMed] [Google Scholar]
- Kannt, A., R. D. Lancaster, and H. Michel. 1998. The coupling of electron transfer and proton translocation: electrostatic calculations on Paracoccus denitricans cytochrome c oxidase. Biophys. J. 74:708–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpefors, M., P. Ädelroth, A. Aagaard, H. Sigurdson, M. S. Ek, and P. Brzezinski. 1998a. Electron-proton interactions in terminal oxidases. Biochim. Biophys. Acta. 1365:159–169. [DOI] [PubMed] [Google Scholar]
- Karpefors, M., P. Ädelroth, Z. Yuejun, S. Ferguson-Miller, and P. Brzezinski. 1998b. Proton uptake controls electron transfer in cytochrome c oxidase. Proc. Natl. Acad. Sci. USA. 95:13606–13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapper, I., R. Hagstrom, R. Fine, K. Sharp, and B. Honig. 1986. Focusing of electric fields in the active site of Cu-Zn superoxide dismutase: effects of ionic strength and amino-acid modification. Proteins. 1:47–59. [DOI] [PubMed] [Google Scholar]
- Kletzin, A., T. Urich, F. Muller, T. M. Bandeiras, and C. M. Gomes. 2004. Dissimilatory oxidation and reduction of elemental sulfur in thermophilic archaea. J. Bioenerg. Biomembr. 36:77–91. [DOI] [PubMed] [Google Scholar]
- Koutsoupakis, C., T. Soulimane, and C. Varotsis. 2004. Probing the Q-proton pathway of ba3-cytochrome c oxidase by time-resolved Fourier transform infrared spectroscopy. Biophys. J. 86:2438–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291. [Google Scholar]
- Luecke, H., H. T. Richter, and J. K. Lanyi. 1998. Proton transfer pathways in bacteriorhodopsin at 2.3 Å resolution. Science. 280:1934–1937. [DOI] [PubMed] [Google Scholar]
- Luecke, H., B. Schobert, H. T. Richter, J. P. Cartailler, and J. K. Lanyi. 1999. Structural changes in bacteriorhodopsin at 2 Å resolution. Science. 286:255–260. [DOI] [PubMed] [Google Scholar]
- Ma, J., P. H. Tsatsos, D. Zaslavsky, B. Barquera, J. W. Thomas, A. Katsonouri, A. Puustinnen, M. Wikstrom, P. Brzezinski, J. O. Alben, and R. B. Gennis. 1999. Glutamate-89 in subunit II of cytochrome bo3 from Escherichia coli is required for the function of the heme-copper oxidase. Biochemistry. 38:15150–15156. [DOI] [PubMed] [Google Scholar]
- MacMillan, F., A. Kannt, J. Behr, T. Prisner, and H. Michel. 1999. Direct evidence for a tyrosine radical in the reaction of cytochrome c oxidase with hydrogen peroxide. Biochemistry. 38:9180–9184. [DOI] [PubMed] [Google Scholar]
- Mather, M. W., P. Springer, S. Hensel, G. Buse, and J. A. Fee. 1993. Cytochrome oxidase genes from Thermus thermophilus. J. Biol. Chem. 15:5395–5408. [PubMed] [Google Scholar]
- Meunier, B. 2001. Site-directed mutations in the mitochondrially encoded subunits I and III of yeast cytochrome oxidase. Biochem. J. 354:407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, H. 1998. The mechanism of proton pumping by cytochrome c oxidase. Proc. Natl. Acad. Sci. USA. 95:12819–12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, H., J. Behr, A. Harrenga, and A. Kannt. 1998. Cytochrome c oxidase: structure and spectroscopy. Annu. Rev. Biophys. Biomol. Struct. 27:329–356. [DOI] [PubMed] [Google Scholar]
- Mills, D. A., and S. Ferguson-Miller. 1998. Proton uptake and release in cytochrome c oxidase: separate pathways in time and space. Biochim. Biophys. Acta. 1365:46–52. [DOI] [PubMed] [Google Scholar]
- Mills, D. A., L. Florens, C. Hiser, J. Qian, and S. Ferguson-Miller. 2000. Where is “outside” in cytochrome c oxidase and how and when do protons get there? Biochim. Biophys. Acta. 1458:180–187. [DOI] [PubMed] [Google Scholar]
- Musser, S. M., and S. I. Chan. 1998. Evolution of the cytochrome c oxidase proton pump. J. Mol. Evol. 46:508–520. [DOI] [PubMed] [Google Scholar]
- Nagle, J. F., and H. J. Morowitz. 1978. Molecular mechanisms for proton transport in membranes. Proc. Natl. Acad. Sci. USA. 75:298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle, J. F., and S. Tristam-Nagle. 1983. Hydrogen bonded chain mechanisms for proton conduction and proton pumping. J. Membr. Biol. 74:1–14. [DOI] [PubMed] [Google Scholar]
- Namslauer, A., M. Brändén, and P. Brzezinski. 2002. The rate of internal heme-heme electron transfer in cytochrome c oxidase. Biochemistry. 41:10369–10374. [DOI] [PubMed] [Google Scholar]
- Ostermeier, C., A. Harrenda, U. Ermler, and H. Michel. 1997. Structure at 2.7 Å resolution of the Paracoccus denitrificans two-subunit cytochrome c oxidase complexed with an antibody FV fragment. Proc. Natl. Acad. Sci. USA. 94:10547–10553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, M., C. M. Gomes, and M. Teixeira. 2002. Plasticity of proton pathways in haem-copper oxygen reductases. FEBS Lett. 522:14–18. [DOI] [PubMed] [Google Scholar]
- Pereira, M., M. Santana, C. M. Soares, J. Mendes, J. Carita, A. Fernandes, M. Saraste, M. A. Carrondo, and M. Teixeira. 1999. The caa3 terminal oxidase of the thermohalophilic bacterium Rhodothermus marinus: a HiPIP:oxygen oxidoreductase lacking the key glutamate of the D-channel. Biochim. Biophys. Acta. 1413:1–13. [DOI] [PubMed] [Google Scholar]
- Pereira, M., M. Santana, and M. Teixeira. 2001. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim. Biophys. Acta. 1505:185–208. [DOI] [PubMed] [Google Scholar]
- Pereira, M. M., and M. Teixeira. 2004. Proton pathways, ligand binding and dynamics of the catalytic site in haem-copper oxygen reductases: a comparison between the three families. Biochim. Biophys. Acta. 1655:340–346. [DOI] [PubMed] [Google Scholar]
- Purschke, W., C. Schimitd, A. Petersen, and G. Schafer. 1997. The terminal quinol oxidase of the hyperthermophilic archaeon Acidianus ambivalens exhibits a novel subunit structure and gene organization. J. Bacteriol. 179:1344–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resat, H., T. J. Marrone, and J. A. McCammon. 1997. Enzyme-inhibitor association thermodynamics: explicit and continuum solvent studies. Biophys. J. 72:522–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali, A., and T. Blundell. 1993. Comparative protein modelling by satisfaction of spacial restraints. J. Mol. Biol. 234:779–815. [DOI] [PubMed] [Google Scholar]
- Santana, M., M. Pereira, N. Elias, C. M. Soares, and M. Teixeira. 2001. Gene cluster of Rhodothermus marinus high-potential iron-sulfur protein: oxygen oxidoreductase, a caa3-type oxidase belonging to the superfamily of heme-copper oxidases. J. Bacteriol. 183:687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham, Y., I. Muegge, and A. Warshel. 1999. Simulating proton translocations in proteins: probing proton transfer in the Rhodobacter sphaeroides reaction center. Proteins. 36:484–500. [PubMed] [Google Scholar]
- Soares, C. M., A. M. Baptista, M. M. Pereira, and M. Teixeira. 2004. Investigation of protonatable residues in Rhodothermus marinus caa3 haem-copper oxygen reductase: comparison with Paracoccus denitrificans aa3 haem-copper oxygen reductase. J. Biol. Inorg. Chem. 9:124–134. [DOI] [PubMed] [Google Scholar]
- Souliname, T., G. Buse, G. Bourenkov, H. Bartunik, R. Huber, and M. Than. 2000a. Structure and mechanism of the aberrant ba3-cytochrome c oxidase from Thermus thermophilus. EMBO J. 19:1766–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souliname, T., M. E. Than, M. Dewor, R. Huber, and G. Buse. 2000b. Primary structure of a novel subunit in ba3-cytochrome oxidase from Thermus Thermophilus. Protein Sci. 9:2068–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson-Ek, M., J. Abramson, G. Larsson, S. Tornroth, P. Brzezinski, and S. Iwata. 2002. The x-ray crystal structures of wild-type and eq(I-286) mutant cytochrome c oxidases from Rhodobacter Sphaeroides. J. Mol. Biol. 321:329–339. [DOI] [PubMed] [Google Scholar]
- Tomson, F., J. A. Bailey, R. B. Gennis, C. J. Unkefer, Z. Li, L. A. Silks, R. A. Martinez, R. J. Donohoe, R. B. Dyer, and W. H. Woodruff. 2002. Direct infrared detection of the covalently ring linked His-Tyr structure in the active site of the heme-copper oxidases. Biochemistry. 41:14383–14390. [DOI] [PubMed] [Google Scholar]
- Tomson, F. L., J. E. Morgan, G. Gu, B. Barquera, T. V. Vygodina, and R. B. Gennis. 2003. Substitutions for glutamate 101 in subunit II of cytochrome c oxidase from Rhodobacter sphaeroides result in blocking the proton-conducting K-channel. Biochemistry. 42:1711–1717. [DOI] [PubMed] [Google Scholar]
- Tricone, A., V. Lanzotti, B. Nicolaus, W. Zillig, M. De Rosa, and A. Gambacorta. 1989. Comparative lipid composition of aerobically and anaerobically grown Desulfurolobus ambivalens, a autotrophic thermophilic archaeobacterium. J. Gen. Microbiol. 135:2751–2757. [Google Scholar]
- Tsukihara, T., H. Aoyama, E. Yamashita, T. Tomizaki, H. Yamaguchi, K. Shinzawa-Itoth, R. Nakashima, R. Yaono, and S. Yoshikawa. 1995. Structures of metal sites of oxidised bovine heart cytochrome c oxidase at 2.8 Å. Science. 269:1069–1074. [DOI] [PubMed] [Google Scholar]
- Tsukihara, T., H. Aoyama, E. Yamashita, T. Tomizaki, H. Yamaguchi, K. Shinzawa-Itoth, R. Nakashima, R. Yaono, and S. Yoshikawa. 1996. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science. 272:1136–1144. [DOI] [PubMed] [Google Scholar]
- Verkhovskaya, M., A. Garcìa-Horsman, A. Puustinnen, J. L. Rigaud, J. Morgan, M. Verkhovsky, and M. Wikstrom. 1997. Glutamic acid 286 in subunit I of cytochrome bo3 is involved in proton translocation. Proc. Natl. Acad. Sci. USA. 94:10128–10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhovsky, M., A. Jasaitis, M. Verkhovskaya, J. Morgan, and M. Wilkstrom. 1999. Proton translocation by cytochrome c oxidase. Nature. 400:480–483. [DOI] [PubMed] [Google Scholar]
- Verkhovsky, M., A. Jasaitis, and M. Wikstrom. 2001. Ultrafast haem-haem electron transfer in cytochrome c oxidase. Biochim. Biophys. Acta. 1506:143–146. [DOI] [PubMed] [Google Scholar]
- Wikstrom, M. 1998. Proton translocation by the respiratory haem-copper oxidases. Biochim. Biophys. Acta. 1365:185–192. [DOI] [PubMed] [Google Scholar]
- Wikstrom, M. 2000. Proton translocation by cytochrome c oxidase: a rejoinder to recent criticism. Biochemistry. 39:3515–3519. [DOI] [PubMed] [Google Scholar]
- Wikstrom, M. 2004. Cytochrome c oxidase: 25 years of the elusive proton pump. Biochim. Biophys. Acta. 1655:241–247. [DOI] [PubMed] [Google Scholar]
- Wikstrom, M., A. Jasaitis, C. Backgren, A. Puustinnen, and M. Verkhovsky. 2000. The role of the D- and K-pathways of proton transfer in the function of the haem-copper oxidases. Biochim. Biophys. Acta. 1459:514–520. [DOI] [PubMed] [Google Scholar]
- Wikstrom, M., M. Verkhovsky, and G. Hummer. 2003. Water-gated mechanism of proton translocation by cytochrome c oxidase. Biochim. Biophys. Acta. 1604:61–65. [DOI] [PubMed] [Google Scholar]
- Yoshikawa, S., K. Shinzawa-Itoth, R. Nakashima, R. Yaono, E. Yamashita, N. Inoue, M. Yao, M. J. Fei, C. P. Libeu, T. Mizushima, H. Yamaguchi, T. Tomizaki, and T. Tsukihara. 1998. Redox-coupled crystal structural changes in bovine heart cytochrome c oxidase. Science. 280:1723–1729. [DOI] [PubMed] [Google Scholar]
- Zhang, L., and J. Hermans. 1996. Hydrophilicity of cavities in proteins. Proteins. 24:433–438. [DOI] [PubMed] [Google Scholar]
- Zheng, X., E. S. Medvedev, J. Swanson, and A. A. Stuchebrukhov. 2003. Computer simulation of water in cytochrome c oxidase. Biochim. Biophys. Acta. 1557:99–107. [DOI] [PubMed] [Google Scholar]