Abstract
The class I myosin, Myo1b, is a calmodulin- and actin-associated molecular motor widely expressed in mammalian tissues. Analytical ultracentrifugation studies indicate that Myo1b purified from rat liver has a Stokes radius of 6.7 nm and a sedimentation coefficient, s20,w, of 7.0 S with a predicted molar mass of 213 kg/mol. These results indicate that Myo1b is monomeric and consists primarily of a splice variant having five associated calmodulins. Molecular modeling based on the analytical ultracentrifugation studies are supported by electron microscopy studies that depict Myo1b as a single-headed, tadpole-shaped molecule with outer dimensions of 27.9 × 4.0 nm. Above a certain Myo1b/actin ratio, Myo1b bundles actin filaments presumably by virtue of a second actin-binding site. These studies provide new information regarding the oligomeric state and morphology of Myo1b and support a model in which Myo1b cross-links actin through a cryptic actin-binding site.
INTRODUCTION
Multiple class I myosins exist in higher organisms, one of which is Myo1b. The gene for Myo1b, previously called MI130 (Coluccio and Conaty, 1993) or MYR1 (Ruppert et al., 1993), can code for up to 6 IQ motifs which form alpha helices that bind calmodulin (Ruppert et al., 1993). In the cell, Myo1b is alternatively spliced to yield forms with four, five, or six IQ domains (Ruppert et al., 1993). Unlike its closest relative, Myo1a (intestinal brush border myosin I or BBMI), whose expression is confined essentially to intestine (Hoshimaru and Nakanishi, 1987; Garcia et al., 1998), expression of Myo1b is ubiquitous (Ruppert et al., 1993; Sherr et al., 1993).
Localization studies in tissue culture cells place Myo1b at the plasma membrane and in punctae within the cell body (Ruppert et al., 1995). It is associated with several different purified rat liver membrane fractions including plasma membrane and microsomes (Ruppert et al., 1995; Balish et al., 1999; Raposo et al., 1999); moreover, some Myo1b is presumably associated with the cytoskeleton (Balish et al., 1999).
Myo1b supports the translocation of actin filaments in vitro; movement is regulated by the calcium ion concentration (Williams and Coluccio, 1994). The slow kinetics exhibited by Myo1b suggest that Myo1b has evolved to participate in cytoskeletal rearrangements or maintenance of cortical tension (Coluccio and Geeves, 1999; Geeves et al., 2000). A role for Myo1b in the endocytic pathway has been proposed (Raposo et al., 1999).
Purified Myo1b from rat liver contains one heavy chain of ∼130 kDa and multiple calmodulin light chains of 17 kDa (Coluccio, 1994). For this study analytical ultracentrifugation and electron microscopy (EM) were used to provide data about the oligomeric state and morphology of Myo1b. The centrifugation studies show that Myo1b is an elongated monomer. Electron micrographs are consistent with this and show what are probably the motor domain and lever arm regions of the molecule. The studies predict that Myo1b from rat liver is predominantly the five IQ variant. We also show here that Myo1b binds to and cross-links actin filaments. The degree of actin cross-linking exhibited by Myo1b depends on the molar ratio of Myo1b/actin. The studies provide important structural information on Myo1b and predict that Myo1b bundles actin filaments by means of a second actin-binding site.
EXPERIMENTAL PROCEDURES
Preparation of protein
Myo1b was purified by gel filtration, cation, and anion exchange chromatography after homogenization of rat livers in the presence of ATP as previously described (Coluccio and Conaty, 1993). Homogeneity was checked by SDS-polyacrylamide gel electrophoresis. The resulting protein was used within one or two days after purification or, for some of the EM studies, was frozen dropwise in liquid N2 and stored at −80°C until use. Rabbit skeletal muscle actin was prepared as described (Spudich and Watt, 1971) and subjected to gel filtration (MacLean-Fletcher and Pollard, 1980). Actin was frozen as F-actin in liquid N2, and then stored at −80°C until use.
Analytical ultracentrifugation
Myo1b at protein concentrations ranging from 0.1–0.5 mg/ml in 10 mM Tris, pH 8.1, 100 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 1 mM DTT was analyzed at 4°C in a Beckman Optima XL-I analytical ultracentrifuge. Data collection and analyses, including determination of diffusion coefficient and molecular weight from sedimentation velocity data, were performed as previously described using the software SEDANAL (Stafford, 1992, 1994; Stafford and Sherwood, 2004) and SEDFIT (Schuck, 2000). Velocity runs were carried out with interference optics at 0.15, 0.056, and 0.020 mg/ml and run at 50,000 RPM at 4°C. Sedimentation equilibrium runs were analyzed using the equilibrium fitting program NONSIM (Margossian and Stafford, 1982; Brenner et al., 1990) now incorporated into SEDANAL as a global fitter. Equilibrium runs were performed at three loading concentrations and run at 10,000, 14,000, and 20,000 rpm at 4°C. Loading concentrations were 0.14, 0.07, and 0.03 mg/ml. Attainment of equilibrium was determined using WinMatch. A wavelength of 230 nm was used. An extinction coefficient at 230 nm was calculated from the aromatic residue and peptide backbone contributions (n-π* transition) using values of 6400, 3689, and 80 a.u.-cm−1-L-mol−1 for tryptophan, tyrosine, and the peptide bond, respectively, in 0.1 M phosphate at pH 7.0 (cf. http://omlc.ogi.edu/spectra/PhotochemCAD/abs_html/) (Gratzer, 1967; Du et al., 1998). Values of the extinction coefficients of Myo1b, actin and calmodulin at 230 nm were calculated to be 2.10, 2.66, and 1.15 a.u.-cm−1-mL-mg−1, respectively. Global analysis of the nine data sets was carried out with SEDANAL with a conservation of mass constraint and fitting for the relative amounts of each component as global fitting parameters. Fitting was performed by minimizing the L-1 norm (i.e., the sum of the absolute values of the residuals). Error estimates of the fitted parameters were obtained by bootstrap analysis. The partial specific volume and hydration used in analyses of ultracentrifugation data were calculated from the mass averaged contributions of individual amino acids (Kuntz and Kauzmann, 1974; Perkins, 1986).
From the amino acid composition, a value of 0.734 cc/g was computed for the partial specific volume, and a value of 0.415 g H2O/g protein was computed for the hydration. The frictional ratio using the hydration correction, 0.862, was computed from:
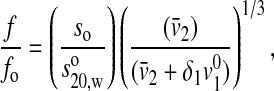 |
where f/fo is the frictional ratio due to shape alone, v2 is the partial specific volume, δ1 is the hydration in grams of water per gram of protein, 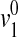 is the specific volume of pure water, and so is the sedimentation coefficient for the corresponding unhydrated sphere.
is the specific volume of pure water, and so is the sedimentation coefficient for the corresponding unhydrated sphere.
Molecular bead modeling
Myo1b structure was modeled with eight beads, one for the head domain (80 kDa), one for each of the five IQ domains with calmodulin bound (17.7 kDa each), and two for the tail domain (16.9 kDa each). The head and tail portions were bent away from the axis of the IQ domains as shown in Fig. 2. The sedimentation coefficient of the model was computed from the bead model using the program HYDRO (Garcia de la Torre et al., 1994). Other models with different degrees of bending of the head and tail sections were considered, but the model shown in Fig. 2 was the best representation of most of the images seen by EM.
FIGURE 2.

Bead model of Myo1b. A string of eight beads, one representing the motor domain, one for each of five calmodulin molecules, and two for the tail region (see text for details), was used to model the structure of Myo1b. The angles of both the motor and tail domains relative to the IQ region were selected based on the parameters obtained by analytical ultracentrifugation consistent with the EM images.
Electron microscopy
Myo1b in 10 mM MOPS, pH 7.0, 80 mM KCl, 1 mM EGTA, 1 mM MgCl2, and 0.2 mM ATP, was applied to UV-treated carbon films, negatively stained in 1% uranyl acetate (Walker et al., 1985), then viewed in a JEOL 1200ex electron microscope operating at 80 kV. Actin-Myo1b samples were applied to collodion- and carbon-coated copper grids for 30 s followed by application of 2% aqueous uranyl acetate for 45 s before viewing with a Philips 200 electron microscope.
Actin-binding assays
Myo1b and actin were incubated at various molar ratios for the times indicated in a total volume of 100 μl of 10 mM Tris, pH 8.0, 100 mM KCl, 1 mM MgCl2, 0.5 mM DTT, and 1 mM EGTA. Control samples included actin only and Myo1b only. After the incubation period, the samples were centrifuged in a TLA 100 rotor in a Beckman tabletop ultracentrifuge at 245,000 × g for 20 min or in a microfuge at low speed at 14,000 × g for 15 min to check for bundling (Glenney et al., 1981). To check for reversibility of binding, in some cases ATP was added to 10 mM for 10 min before high-speed centrifugation.
Supernatants were separated from pellets resulting from both the high- and low-speed spins. The pellets were resuspended in 1 M Tris base, then SDS-PAGE sample buffer and boiled. The supernatants were precipitated in 10% trichloroacetic acid before resuspending in 1 M Tris base and preparing for SDS-PAGE by the addition of sample buffer and boiling. After analysis by gel electrophoresis (Laemmli, 1970), the amount of Myo1b heavy chain, calmodulin, and/or actin in the supernatants and pellets was determined by densitometry using the program NIH Image 1.63.
RESULTS
Analytical ultracentrifugation
Equilibrium and velocity sedimentation analysis were performed on Myo1b to gain information concerning its size, shape, and possible association properties. The main question to be answered was whether Myo1b was monomeric or dimeric under these conditions. Sedimentation velocity analysis using time derivative analysis with SEDANAL and c(s) analysis with SEDFIT showed a single, unimodal boundary at 7.0 S (Fig. 1) with a small amount of other material sedimenting at 3.5, 4.9, and 8.3 S indicating the presence of small amounts of impurities. The precise location and amplitude of the smaller peaks in the c(s) analysis were dependent on the exact choice of data span and number of points used in the analysis and, therefore, no definite assignments could be made. Overlaying of normalized g(s) plots showed that there was no appreciable concentration dependence in this system ruling out any significant reversible self-association of Myo1b. Sedimentation equilibrium analysis with SEDANAL (a global fit to nine data sets at three concentrations and three speeds) showed the presence of three components with molar mass of 213 ± 5 kg/mol (Myo1b), 41.2 ± 7 kg/mol (actin), and 16.6 ± 5 kg/mol (calmodulin) present in amounts expressed as weight fractions of 0.71 ± 0.01, 0.11 ± 0.04, and 0.16 ± 0.03, respectively. The mean absolute residual was ± 0.009 a.u. Residuals were random and showed no systematic trends. The parameter error estimates were obtained by bootstrap analysis. The small amounts of an aggregate (8.3 S) suggested by the c(s) analysis were not seen in significant amounts in the analysis of the equilibrium data and indicated that no significant dimer was being formed under these conditions. Fits that included a dimer species (432 kg/mol) returned a mole fraction of dimer of only 0.03 ± 0.01. This unequivocally demonstrates that Myo1b is monomeric under these conditions and strongly implies the existence of two actin-binding sites on each molecule which are necessary to cause actin bundling. This value combined with the value obtained for the sedimentation coefficient gives a calculated value of 3.0 × 10−7cm2/s for the diffusion coefficient (Table 1). Since three splice forms of Myo1b may exist in these preparations (Ruppert et al., 1993), the sedimentation velocity data were processed with the program SEDFIT using c(s) analysis to look for these splice forms. SEDFIT showed a single sharp peak indicating the absence of any significant amounts of the other two splice forms. Modeling with SEDANAL showed that SEDFIT would have been able to detect and resolve the other two splice forms if present at 5–10% assuming an M2/3 dependence of s on M. Peaks corresponding to the other slice forms, had they been present, would have been expected at ∼6.6 S (M = 199 kg/mol) and ∼7.4 S (M = 232 kg/mol). The preparation runs as a single band on SDS-polyacrylamide gel electrophoresis (see Fig. 4). The combination of these data predicts a Stokes radius of 7.1 nm for the five IQ variant.
FIGURE 1.
Plots of the sedimentation velocity profiles of Myo1b determined by analytical ultracentrifugation. After dialysis against 100 mM KCl, 10 mM Tris, pH 8.1, 1 mM MgCl2, 1 mM EGTA, and 1 mM DTT, Myo1b at three different protein concentrations (0.15, 0.056, and 0.020 mg/ml) was centrifuged at 4°C and 50,000 rpm in a Beckman Optima XL-I analytical ultracentrifuge. Data were analyzed with the DCDT/SEDANAL package (left-hand y axis) and the c(s)/SEDFIT package (right-hand y axis 0.15 mg/ml). The overlapping curves are normalized g(s) curves showing that there is no significant concentration dependence of Myo1b. The narrow curve is a high resolution c(s) analysis of the data which demonstrated that the preparation contains mainly the 5-IQ splice form and does not contain detectable amounts of the other two splice forms. The exact position and amplitude of the smaller peaks in the c(s) analysis was dependent on the choice of data span and number of scans used. But they did reflect the presence of material seen in both the g(s) plots and in equilibrium sedimentation experiments. Global analysis of the equilibrium data (see the text) was consistent with the presence of small amounts of free calmodulin and actin in substantial agreement with the c(s) analysis.
TABLE 1.
Physical properties of Myo1b
| Molar mass | 213 kg/mol |
| Partial specific volume | 0.734 cc/g |
| Hydration | 0.415 g/g |
Sedimentation coefficient 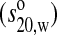
|
7.0 S |
| Stokes radius | 7.1 nm |
| Diffusion coefficient | 3.0 F |
| Frictional ratio (f/fo) (corrected for hydration) | 1.57 |
| Ellipsoid analysis | |
| Axial ratio (Perrin, prolate) | 10.5 |
| Length of prolate ellipsoid (hydrated) | 43.6 nm |
| Diameter of prolate ellipsoid | 4.2 nm |
FIGURE 4.
SDS-PAGE of actin-binding assays. Actin (2.4 μM) was incubated in buffer containing 1 mM EGTA with increasing amounts of Myo1b (0–0.48 μM) for 1 h at room temperature, then centrifuged to separate into supernatants (panels A and C) or pellets (panels B and D) at either low speed (panels A and B) or high speed (panels C and D). Pellets and supernatants were analyzed by SDS-PAGE. The position of the Myo1b heavy chain (Myo1b), actin (Actin), and calmodulin (Calm) are marked at right. Lane 1, 0.48 μM Myo1b; lane 2, 0.48 μM Myo1b, 2.4 μM F-actin (1:5); lane 3, 0.24 μM Myo1b; lane 4, 0.24 μM Myo1b, 2.4 μM F-actin (1:10); lane 5, 0.16 μM Myo1b; lane 6, 0.16 μM Myo1b, 2.4 μM F-actin (1:15); lane 7, 0.10 μM Myo1b; lane 8, 0.10 μM Myo1b, 2.4 μM F-actin (1:25); lane 9, 0.08 μM Myo1b; lane 10, 0.08 μM Myo1b, 2.4 μM F-actin (1:30); lane 11, 2.4 μM F-actin.
Bead modeling
Using the EM images as a guide, a bead model of Myo1b consisting of eight beads was used to model the frictional coefficient obtained from the sedimentation data (Fig. 2; Table 1). A single bead of mass 80 kDa was used to model the motor domain, five beads each of 20 kDa were used to model the IQ domains occupied by calmodulin, and two beads each of 16.5 kDa were used to model the tail domain. The contour length of the model was ∼30.5 nm. The longest chord of the bead model with the head positioned at a 45° angle to the rest of the molecule is 26.8 nm. The predicted sedimentation coefficient for this bead model was 7.3 S in good agreement with the measured value of 7.0 S. Other models (not shown) in which the head and tail sections assumed other angles were also simulated. A model in which both the head and the tail domain were bent 90° with respect to the IQ domain predicted a sedimentation coefficient of 7.8 S. Another model in which both the head and tail sections were straight with respect to the IQ domains gave a sedimentation coefficient of 7.2 S. Although this is not significantly different from the observed value or from the value for the straight model, the model in which the head and tail sections are bent 45° most closely represents both the EM and the sedimentation data, especially since the maximum chord length for the 45° model agreed with the value found by analysis of the EM pictures. The modeling was also consistent with the monomeric nature of Myo1b under the conditions of both the sedimentation experiments and EM.
Electron microscopy
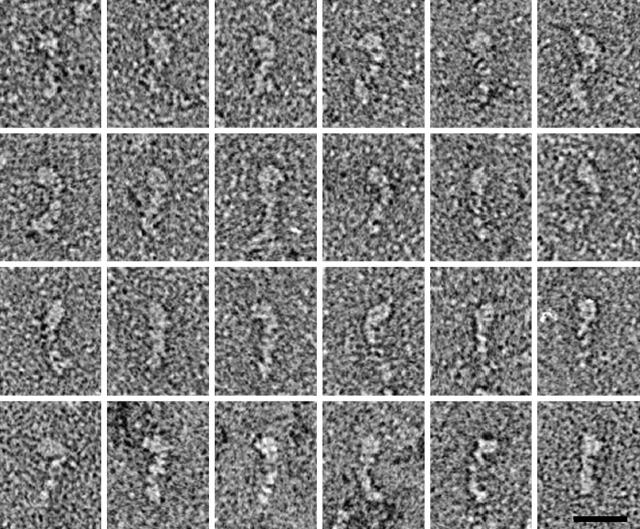
Electron microscopic images of negatively stained Myo1b preparations showed mostly tadpole-shaped objects whose shapes and size were consistent with monomeric Myo1b molecules. The molecules, shown in a montage in Fig. 3, have a bulbous region ∼8-nm long at one end which we interpret to be the motor domain. The remainder of the molecule is ∼20-nm long which is consistent with a lever arm consisting of five calmodulins. The motor domain was found at varying orientations with respect to the lever. Some of this apparent flexibility may result from passive bending of the molecule, for instance at a pliant region, similar to what has been identified in myosin II (Houdusse et al., 2000); however, the most sharply bent molecules (e.g., Fig. 3, bottom left) may result from active movements fuelled by the ATP in the buffer. The average length of the molecules determined analytically was 27.9 ± 0.5 SE. This agrees very well with the longest chord length computed from the bead model with head and tail bent at 45° for which a value of 27.0 nm was obtained. The values for the straight and 90° bent models were 19.0 and 30.4 nm, respectively.
FIGURE 3.
Montage of negatively-stained Myo1b molecules. The tadpole-shaped molecules are oriented with their motor domains at top. Variable bending between the motor domain and the rest of the molecule is indicative of flexibility at or near the pliant region. Bar, 20 nm.
Interaction of Myo1b with actin
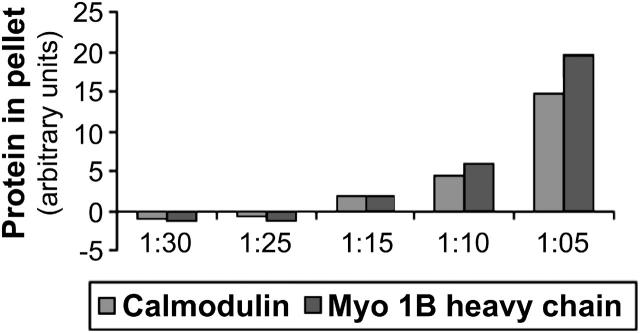
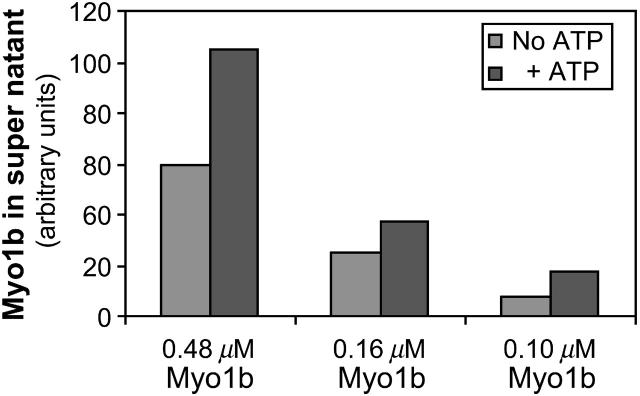
To examine the interaction of actin filaments with Myo1b, different molar ratios of Myo1b/actin (1:5; 1:10; 1:15; 1:25; and 1:30) and control samples containing either actin only or Myo1b only were incubated together, then centrifuged at high speed (Fig. 4, C and D). The supernatants (Fig. 4 C) and pellets (Fig. 4 D) were separated and analyzed by SDS-PAGE. A fraction of Myo1b heavy chain pelleted at high speed in the absence of F-actin (Fig. 4 D, lane 1). With increasing amounts of Myo1b, the amount of both Myo1b heavy chain and calmodulin found in the high-speed pellet with F-actin increased (Fig. 4 D and Fig. 5); however, not all the available Myo1b heavy chain associated with F-actin. In some cases, ATP was added to 10 mM for 10 min after a 1-h incubation, but before high-speed centrifugation; ∼50% more Myo1b appeared in the supernatants after ATP treatment indicating that association of Myo1b with actin is partially reversible (Fig. 6).
FIGURE 5.
Analyses of Myo1b-actin co-sedimentation assays. The relative amounts of Myo1b heavy chain and calmodulin in the pellets as determined using NIH Image 1.63 software are plotted.
FIGURE 6.
ATP reverses binding of Myo1b to actin. Myo1b at the concentrations indicated was incubated with actin at 2.4 μM for 1 h then treated with 10 mM ATP for 10 min before high-speed centrifugation. Roughly twice as much Myo1b remained in the supernatant after ATP treatment as compared to untreated samples.
When the Myo1b/actin mixtures were subjected to low-speed rather than high-speed centrifugation (Fig. 4, A and B), essentially all the actin, except for the critical concentration, pelleted in the presence of high molar ratios of Myo1b/actin (Fig. 4 B, lanes 2 and 4). These results are consistent with Myo1b bundling actin filaments and allowing sedimentation under conditions when most of the actin would normally remain in the supernatant (Fig. 4 A, lane 11, and Fig. 7).
FIGURE 7.
Low speed centrifugation of Myo1b-actin complexes. The relative amount of actin pelleting at low speed with various amounts of Myo1b is plotted. These results are indicative of higher-ordered actin structures being formed in the presence of Myo1b.
EM of negatively stained images was used to visualize the actin structures formed after incubation of Myo1b and actin. At Myo1b/actin ratio of 1:5, the actin filaments associated into tight bundles. Almost no single filaments were observed after incubation overnight in EGTA (Fig. 8 A). In some places (arrows and at higher magnification in Fig. 8 A′) the actin filaments resembled a ladder with Myo1b presumably as the rungs. The bundles formed under these conditions were stable to dilution to final concentrations as low as 1:40 even after an additional 20 h (data not shown). At lower ratios of Myo1b/actin such as 1:10 and 1:15, groups of filaments were more loosely associated and single filaments were frequently observed (Fig. 8 B). Although no obvious decoration of the actin filaments was apparent at Myo1b/actin ratios of 1:25, the Myo1b-actin filaments were considerably straighter (Fig. 8 C) than control actin filaments which were more bent under these buffer conditions (Fig. 8 D). Myo1b also associates with actin in buffer containing ∼60 μM free Ca2+. There were no obvious differences in the amount of Myo1b pelleting with actin or the appearance of the resulting actin structures as determined by EM (data not shown).
FIGURE 8.

Electron micrographs of actin filaments after incubation with Myo1b. Myo1b and actin at a ratio of 1:10 (panel A) 1:15 (panel B) and 1:25 (panel C) were incubated in 10 mM Tris, pH 7.8, 100 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 0.5 mm DTT overnight, applied to grids, and stained with uranyl acetate. A higher magnification of the area in panel A indicated by arrows is shown in panel A′ (bar, 0.2 μm). Control samples containing actin only are shown in panel D. Bundling of actin filaments was proportional to the Myo1b concentration. Bar (0.2 μm) in panel D applies to all panels except A′.
DISCUSSION
Using the complementary approaches of analytical ultracentrifugation and EM we have shown that Myo1b exists as a monomer with a distinct head and tail region. The analytical ultracentrifugation studies indicate that Myo1b preparations from rat liver have a molar mass of 213 kDa. This compares favorably to the size expected for a complex of one Myo1b heavy chain (Ruppert et al., 1993) together with five molecules of calmodulin (17 kDa). Northern blot analysis has previously shown that the 4 IQ and 5 IQ variants are predominantly expressed in rat liver (Ruppert et al., 1993). Apparently, the 5 IQ isoform is preferentially selected during the purification.
In EM images Myo1b is elongated in shape with one end wider than the other. This tadpole shape is consistent with other myosins, which contain a motor domain followed by a lever arm (neck) and tail regions. From comparisons with myosin II crystal structures, e.g., (Houdusse et al., 2000), the motor domain is expected to be ∼8-nm long and each IQ motif and its calmodulin can be expected to lengthen the lever arm by ∼4 nm. The measured overall length of Myo1b of 28 nm is therefore similar to what is predicted for the motor domain and a lever arm with five calmodulins. The ∼300 amino acids C-terminal to the lever arm are not resolved as a separate region in the images, but are therefore likely to be folded compactly. The incidence of images showing both sharply bent and relatively straight molecules may result from large conformational changes brought about during steady-state ATP hydrolysis. These shape changes are thought to form the basis of the stepping mechanism along actin filaments.
The EM studies provide the first look at single Myo1b molecules. The average length of the Myo1b molecule as calculated from the electron micrographs is 28 nm ± 0.5 SE; n = 24. This is consistent with the length of the longest cord of a Myo1b molecule whose head is at a 45° angle to the remainder of the molecule (26.8 nm) using a bead model based on the analytical ultracentrifugation results. The bent model is consistent with the results from EM and the sedimentation analyses; whereas the straight model although not ruled out by a conservative analysis of the sedimentation data is not consistent with the majority of the structures seen by EM. Like other myosins, it is likely that Myo1b is flexible so we consider this model for the average conformation.
Actin filaments incubated with low ratios of Myo1b in EGTA were noticeably straighter, but the micrographs did not have sufficient contrast to allow unambiguous identification of the individual Myo1b molecules. Myo1b associates with actin filaments orthogonally (Arthur, Lin, Coluccio, and Milligan, manuscript in preparation) as has been observed with cryoelectron microscopy of actin filaments after incubation with BBMI (Jontes et al., 1998). EM confirmed that Myo1b forms sedimentable structures. With increasing ratios of Myo1b/actin, the actin filaments form networks or bundles. Bundling of F-actin in vitro has previously been observed for Acanthamoeba myosin I (Fujisaki et al., 1985), brush border myosin I (Coluccio and Bretscher, 1987; Conzelman and Mooseker, 1987), and the liver myosin I isoforms (Coluccio and Conaty, 1993).
Actin cross-linking can be accounted for in one of two ways: through oligomerization of two molecules each with one actin-binding site or via a single molecule containing two (or more) actin-binding sites. The lower eukaryotic class I myosins bundle actin filaments through a second actin-binding site (Lynch et al., 1986) in their extended tail region (termed TH2 and TH3; Hammer, 1991); however, no such second actin-binding site is obvious from review of the Myo1b sequence making the mechanism of actin cross-linking by Myo1b unclear. Nevertheless, our studies suggest that a second actin-binding site exists on Myo1b to explain how monomeric Myo1b supports actin cross-linking. Additionally, a baculovirus-expressed truncated Myo1b fragment representing the motor domain and 1 IQ region does not cross-link actin filaments (Perreault-Micale et al., 2000). Efforts to express soluble tail regions to use to determine if they contain an actin-binding site have so far failed.
Cross-linking of actin by Myo1b suggests a role for Myo1b in the maintenance of actin-actin interactions in the cytoskeletal-rich regions of cells. One possibility is that Myo1b is responsible for the generation of tension by cross-linking actin filaments. Dictyostelium amoebae overexpressing Myo1b are deficient in their ability to form actin-rich protrusions and to become polarized providing evidence for the amoeboid myosin I's involvement in generating tension (Novak and Titus, 1997). Similar mechanisms might exist for myosin I in mammalian cells.
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM068080) and March of Dimes (501) to L.M.C., National Science Foundation (BIR-9513060) to W.F.S., and National Institutes of Health (AR40964) and BBSRC to J.A.T.
References
- Balish, M. F., E. F. Moeller 3rd, and L. M. Coluccio. 1999. Overlapping distribution of the 130- and 110-kDa myosin I isoforms on rat liver membranes. Arch. Biochem. Biophys. 370:285–293. [DOI] [PubMed] [Google Scholar]
- Brenner, S. L., A. Zlotnick, and W. F. Stafford. 1990. RecA protein self-assembly. II. Analytical equilibrium ultracentrifugation studies of the entropy-driven self-association of RecA. J. Mol. Biol. 216:949–964. [DOI] [PubMed] [Google Scholar]
- Coluccio, L. M. 1994. Differential calmodulin binding to three myosin-1 isoforms from liver. J. Cell Sci. 107:2279–2284. [DOI] [PubMed] [Google Scholar]
- Coluccio, L. M., and A. Bretscher. 1987. Calcium-regulated cooperative binding of the microvillar 110K-calmodulin complex to F-actin: formation of decorated filaments. J. Cell Biol. 105:325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluccio, L. M., and C. Conaty. 1993. Myosin-I in mammalian liver. Cell Motil. Cytoskeleton. 24:189–199. [DOI] [PubMed] [Google Scholar]
- Coluccio, L. M., and M. A. Geeves. 1999. Transient kinetic analysis of the 130-kDa myosin I (myr 1 gene product) from rat liver: a myosin I designed for maintenance of tension? J. Biol. Chem. 274:21575–21580. [DOI] [PubMed] [Google Scholar]
- Conzelman, K. A., and M. S. Mooseker. 1987. The 110-kD protein-calmodulin complex of the intestinal microvillus is an actin-activated MgATPase. J. Cell Biol. 105:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, H., R. A. Fuh, A. Corkan, and J. S. Lindsey. 1998. PhotochemCAD: a computer-aided design and research tool in photchemistry. Photochem. Photobiol. 68:141–142. [Google Scholar]
- Fujisaki, H., J. P. Albanesi, and E. D. Korn. 1985. Experimental evidence for the contractile activities of Acanthamoeba myosins IA and IB. J. Biol. Chem. 260:11183–11189. [PubMed] [Google Scholar]
- Garcia de la Torre, J., S. Navarro, M. C. Lopez Martinez, F. G. Diaz, and J. J. Lopez Cascales. 1994. HYDRO: a computer program for the prediction of hydrodynamic properties of macromolecules. Biophys. J. 67:530–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, J. A., A. G. Yee, P. G. Gillespie, and D. P. Corey. 1998. Localization of myosin-I-beta near both ends of tip links in frog saccular hair cells. J. Neurosci. 18:8637–8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeves, M. A., C. Perreault-Micale, and L. M. Coluccio. 2000. Kinetic analyses of a truncated mammalian myosin I suggest a novel isomerization event preceding nucleotide binding. J. Biol. Chem. 275:21624–21630. [DOI] [PubMed] [Google Scholar]
- Glenney, J. R., Jr., P. Kaulfus, P. Matsudaira, and K. Weber. 1981. F-actin binding and bundling properties of fimbrin, a major cytoskeletal protein of microvillus core filaments. J. Biol. Chem. 256:9283–9288. [PubMed] [Google Scholar]
- Gratzer, W. 1967. Ultraviolet Spectra of Polypeptides. Marcel Dekker, New York.
- Hammer, J. A. I. 1991. Novel myosins. Trends Cell Biol. 1:50–56. [DOI] [PubMed] [Google Scholar]
- Hoshimaru, M., and S. Nakanishi. 1987. Identification of a new type of mammalian myosin heavy chain by molecular cloning. Overlap of its mRNA with preprotachykinin B mRNA. J. Biol. Chem. 262:14625–14632. [PubMed] [Google Scholar]
- Houdusse, A., A. G. Szent-Gyorgyi, and C. Cohen. 2000. Three conformational states of scallop myosin S1. Proc. Natl. Acad. Sci. USA. 97:11238–11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jontes, J. D., E. M. Ostap, T. D. Pollard, and R. A. Milligan. 1998. Three-dimensional structure of Acanthamoeba castellanii myosin-IB (MIB) determined by cryoelectron microscopy of decorated actin filaments. J. Cell Biol. 141:155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz, I. D., Jr., and W. Kauzmann. 1974. Hydration of proteins and polypeptides. Adv. Protein Chem. 28:239–345. [DOI] [PubMed] [Google Scholar]
- Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685. [DOI] [PubMed] [Google Scholar]
- Lynch, T. J., J. P. Albanesi, E. D. Korn, E. A. Robinson, B. Bowers, and H. Fujisaki. 1986. ATPase activities and actin-binding properties of subfragments of Acanthamoeba myosin IA. J. Biol. Chem. 261:17156–17162. [PubMed] [Google Scholar]
- MacLean-Fletcher, S., and T. D. Pollard. 1980. Identification of a factor in conventional muscle actin preparations which inhibits actin filament self-association. Biochem. Biophys. Res. Commun. 96:18–27. [DOI] [PubMed] [Google Scholar]
- Margossian, S. S., and W. F. Stafford. 1982. Calcium-induced dimerization of troponin-C. J. Biol. Chem. 257:1160–1165. [PubMed] [Google Scholar]
- Novak, K. D., and M. A. Titus. 1997. Myosin I overexpression impairs cell migration. J. Cell Biol. 136:633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, S. J. 1986. Protein volumes and hydration effects. The calculations of partial specific volumes, neutron scattering matchpoints and 280-nm absorption coefficients for proteins and glycoproteins from amino acid sequences. Eur. J. Biochem. 157:169–180. [DOI] [PubMed] [Google Scholar]
- Perreault-Micale, C., A. D. Shushan, and L. M. Coluccio. 2000. Truncation of a mammalian myosin I results in loss of Ca2+ sensitive motility. J. Biol. Chem. 275:21618–21623. [DOI] [PubMed] [Google Scholar]
- Raposo, G., M. N. Cordonnier, D. Tenza, B. Menichi, A. Durrbach, D. Louvard, and E. Coudrier. 1999. Association of myosin I alpha with endosomes and lysosomes in mammalian cells. Mol. Biol. Cell. 10:1477–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert, C., J. Godel, R. T. Müller, R. Kroschewski, J. Reinhard, and M. Bähler. 1995. Localization of the rat myosin I molecules myr 1 and myr 2 and in vivo targeting of their tail domains. J. Cell Sci. 108:3775–3786. [DOI] [PubMed] [Google Scholar]
- Ruppert, C., R. Kroschewski, and M. Bähler. 1993. Identification, characterization and cloning of myr 1, a mammalian myosin-I. J. Cell Biol. 120:1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck, P. 2000. Size distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78:1606–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr, E. H., M. P. Joyce, and L. A. Greene. 1993. Mammalian myosin I alpha, I beta, and I gamma: new widely expressed genes of the myosin I family. J. Cell Biol. 120:1405–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich, J. A., and S. Watt. 1971. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246:4866–4871. [PubMed] [Google Scholar]
- Stafford, W. F. 1994. Boundary analysis in sedimentation velocity experiments. Methods Enzymol. 240:478–501. [DOI] [PubMed] [Google Scholar]
- Stafford, W. F., and P. J. Sherwood. 2004. Analysis of heterogeneous interacting systems by sedimentation velocity: curve fitting algorithms for estimation of sedimentation coefficients, equilibrium and kinetic constants. Biophys. Chem. 108:231–243. [DOI] [PubMed] [Google Scholar]
- Stafford, W. F. 1992. Boundary analysis in sedimentation transport experiments: a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal Biochem. 203:295–301. [DOI] [PubMed] [Google Scholar]
- Walker, M., P. Knight, and J. Trinick. 1985. Negative staining of myosin molecules. J. Mol. Biol. 184:535–542. [DOI] [PubMed] [Google Scholar]
- Williams, R., and L. M. Coluccio. 1994. Novel 130-kDa rat liver myosin-1 will translocate actin filaments. Cell Motil. Cytoskeleton. 27:41–48. [DOI] [PubMed] [Google Scholar]