Abstract
Mechanical properties of single double-stranded DNA (dsDNA) in the presence of different binding ligands were analyzed in optical-tweezers experiments with subpiconewton force resolution. The binding of ligands to DNA changes the overall mechanic response of the dsDNA molecule. This fundamental property can be used for discrimination and identification of different binding modes and, furthermore, may be relevant for various processes like nucleosome packing or applications like cancer therapy. We compared the effects of the minor groove binder distamycin-A, a major groove binding α-helical peptide, the intercalators ethidium bromide, YO-1, and daunomycin as well as the bisintercalator YOYO-1 on λ-DNA. Binding of molecules to the minor and major groove of dsDNA induces distinct changes in the molecular elasticity compared to the free dsDNA detectable as a shift of the overstretching transition to higher forces. Intercalating molecules affect the molecular mechanics by a complete disappearance of the B-S transition and an associated increase in molecular contour length. Significant force hysteresis effects occurring during stretching/relaxation cycles with velocities >10 nm/s for YOYO-1 and >1000 nm/s for daunomycin. These indicate structural changes in the timescale of minutes for the YOYO-DNA and of seconds for the daunomycin-DNA complexes, respectively.
INTRODUCTION
The interaction of ligands with double-stranded DNA is fundamental for many intracellular processes. Especially proteins that bind to specific DNA target sequences control a variety of processes such as regulation, transcription, and translation. Small binding ligands with reduced or no sequence specificity are often able to interfere with those processes because they are capable of changing mechanical properties of the DNA strands and are, therefore, frequently used in cancer therapy (Hurley, 2002). Because of the complex double-helical structure of DNA, different binding modes are possible. Besides covalent binding there are several classes of specific or unspecific noncovalent binding modes: intercalation between basepairs (Reha et al., 2002), bisintercalation (Krishnamoorthy et al., 2002), minor groove binding (Reddy et al., 2001), major groove binding (Niidome et al., 1996; Eckel et al., 2003), a combination of those (Larsson et al., 1994), and binding via nonclassical modes (Lipscomb et al., 1996).
Intercalation is characterized by noncovalent stacking between adjacent basepairs via interaction with π-orbitals of these basepairs (Graves and Velea, 2000) and often combined with hydrogen bonding (Reha et al., 2002). Intercalation extends and frequently partially unwinds the DNA double strands, having large impact on the structure of the nucleosome (McMurray et al., 1991). Furthermore, side groups of intercalating parts of few ligands also influence the binding process and accordingly can cause sequence selective behavior.
Selective binding to the narrow minor groove of AT-rich sequences by van der Waals interaction, formation of hydrogen bonds, and electrostatic interaction is characteristic for minor groove binders (Reddy et al., 2001). Electrostatic interaction is characteristic for major groove binders (especially helical peptide ligands) as well (Eckel et al., 2003). Minor groove binding drugs, for instance, can interfere with the specific binding of regulatory proteins by changing the local bending of DNA (Zimmer and Wähnert, 1986), or disrupt the nucleosome in a selective way (Fitzgerald and Anderson, 1999).
Detailed information about the structural aspects of binding are given by x-ray diffraction (Coste et al., 1999) and NMR spectroscopy (Gelasco and Lippard, 1998). Additionally, procedures to detect binding properties by investigating contour lengths of ligand-DNA complexes by means of scanning force microscopy (SFM) techniques (Coury et al., 1996) have been introduced by placing those complexes onto a treated surface accessible for SFM survey.
Over the last 15 years different ultrasensitive techniques have been developed that allow measurements of inter- and intramolecular forces at the single-molecule level. Most common techniques are based on atomic force microscopy (AFM) (Binnig et al., 1986) and optical tweezers (Ashkin, 1970, 1997; Ashkin et al., 1986; Svoboda and Block, 1994). Recent works cover AFM force spectroscopy of single DNA molecules (Rief et al., 1999; Clausen-Schaumann et al., 2000) as well as of ligand-DNA complexes (Krautbauer et al., 2000; Anselmetti et al., 2000; Krautbauer et al., 2002a; Eckel et al., 2003), demonstrating their significant implications while investigating mechanical properties of ligand-complexed DNA observable in force-extension measurements.
Optical tweezers systems with their superior force sensitivity compared to AFM were utilized for measurements of elastic responses of immobilized single- and double-stranded DNA molecules (Smith et al., 1996; Wuite et al., 2000; Williams et al., 2001; Wenner et al., 2002; Sischka et al., 2003), whereas optical fiber setups were used for probing the molecular extension of a ligand complexed double-stranded DNA (Cluzel et al., 1996).
Most recently, optical tweezers experiments yield and reveal changes in the mechanical and elastic properties of double-stranded DNA molecules in the presence of binding ligands (Bennink et al., 1999; Husale et al., 2002; Sischka et al., 2003; Tessmer et al., 2003).
In this work a set of DNA binding agents was investigated, including a multitude of binding modes such as the minor groove binder distamycin-A and the supposed major groove binding α-helical peptide Ac-(Leu-Ala-Arg-Leu)3-NH-linker, intercalators ethidium bromide, YO-1, and daunomycin, and the bisintercalator YOYO-1. Distinct and characteristic changes within the mechanical response of DNA up to forces of 100 pN were identified and attributed to the corresponding binding mechanisms.
MATERIALS AND METHODS
Our single-beam optical-tweezers instrumentation was described recently (Sischka et al., 2003). Briefly, an infrared laser (1064 nm) combined with a commercial inverse microscope achieves maximum trapping forces of 150 pN at a laser power output up to 900 mW. The stability of the optical-tweezers system is based on dedicated optical and flow-system components allowing calibrated and precise force-extension measurements with a force resolution of 0.4 pN at 600 mW during a broad variety of experiments.
For all experiments we took streptavidin-coated polystyrene microspheres (Spherotech, Libertyville, IL) with a diameter of 3.18 μm, which we used in a diluted suspension of 5 × 10−4 % w/v. λ-DNA was biochemically modified (Sischka et al., 2003) to ensure tethering to the beads at the beginning of each force measurement.
Beads, λ-DNA, and binding ligands were dissolved in 10 mM Tris buffer (Sigma, Traufkirchen, Germany) (pH 8.0) containing 150 mM NaCl (Sigma). The concentration of λ-DNA was 15 pM whereas the binding ligands distamycin-A (Sigma), Ac-(Leu-Ala-Arg-Leu)3-NH-linker, ethidium bromide (Merck, Darmstadt, Germany), YO-1 (Molecular Probes, Eugene, OR), daunomycin (Sigma), and YOYO-1 (Molecular Probes) were used at a total concentration of 1 μM, respectively. All experiments were performed at 20°C.
RESULTS AND DISCUSSION
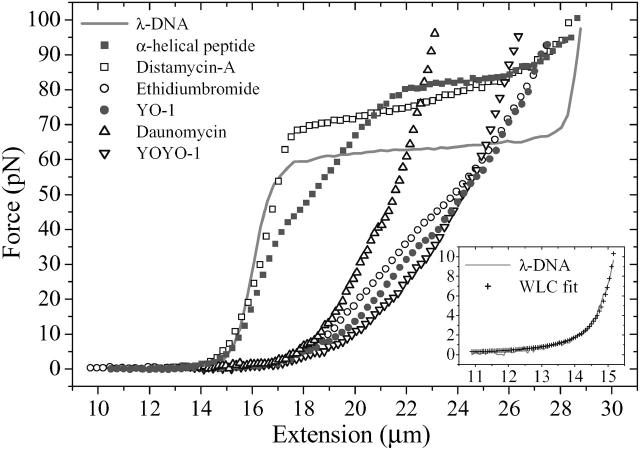
In Fig. 1, the mechanical response to an external force of free λ-DNA and λ-DNA complexed with distamycin-A (minor groove binder), the α-helical peptide Ac-(Leu-Ala-Arg-Leu)3-NH-linker (major groove binder), ethidium bromide, YO-1, daunomycin (intercalators), and YOYO-1 (bisintercalator) are presented. During these force measurements, a trap stiffness of 88 pN/μm combined with a molecular loading rate of 8.8 pN/s was established. Molecular extensions were converted from piezo stage movements using the given trap stiffness and the actual measured forces. To quantify the elastic properties of all measured curves in the medium- and low-force regime, we determined the molecule length at a constant external force of 40 pN as well as the contour and persistence length with the extended worm-like chain model (WLC) (Marko and Siggia, 1995; Bouchiat et al., 1999) in the lower force regime with an upper force limit of 10 pN (Table 1).
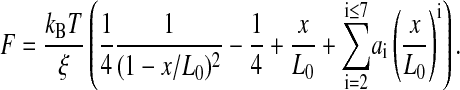 |
(1) |
FIGURE 1.
Single λ-phage DNA molecule and λ-DNA molecule complexed with minor groove binder (distamycin-A), major groove binder (α-helical peptide Ac-(Leu-Ala-Arg-Leu)3-NH-linker), intercalators (daunomycin, YO-1, ethidium bromide), and bisintercalator (YOYO-1), respectively, exhibit different elasticity curves indicating individual mechanical properties (force fingerprints). Total concentration of each binding ligand was 1 μM, and stretching velocity was 100 nm/s. (Inset) Worm-like-chain model fit on a free λ-DNA in the low-force regime up to 10 pN.
TABLE 1.
Molecular parameters for DNA-ligand complexes
| Complex | Binding mode | Molecule length at 40 pN | WLC contour length | WLC persistence length | Overstretching transition |
|---|---|---|---|---|---|
| Free dsDNA | – | 16.4 μm | 16.0 μm | 40.0 nm | 62–65 pN at 18–27 μm |
| Distamycin-A | Minor groove | 16.7 μm | 16.3 μm | 26.7 nm | 70–85 pN at 18–27 μm |
| α-Helical peptide | Major groove | 17.1 μm | 16.5 μm | 29.4 nm | 80–85 pN at 22–27 μm; crossover at 17–22 μm |
| Ethidium bromide | Intercalating | 22.5 μm | 20.4 μm | 20.7 nm | No transition |
| YO-1 | Intercalating | 23.2 μm | 19.8 μm | 29.2 nm | No transition |
| Daunomycin | Intercalating | 20.9 μm | 19.8 μm | 28.1 nm | No transition |
| YOYO-1 | Bisintercalating | 23.5 μm | 21.8 μm | 11.8 nm | No transition |
Molecular parameters extracted from worm-like-chain model fit of experimental data of free double-stranded λ-DNA and dsDNA complexed with distamycin-A, α-helical peptide Ac-(Leu-Ala-Arg-Leu)3-NH-linker, ethidium bromide, YO-1, daunomycin, and YOYO-1. The concentration of each binding ligand was set to 1 μM, respectively.
F denotes the applied force, ξ the persistence length, x the end-to-end distance, L0 the DNA contour length, and ai numerical coefficients (Bouchiat et al., 1999), respectively.
In the following, we discuss the experimental findings of the measured elasticity curves of Fig. 1.
Free dsDNA
The elastic response of a single λ-DNA molecule under an external force shows a distinct plateau, which was first attributed to a structural change from the dsDNA B-form to the overstretched S-form (Cluzel et al., 1996). Based on data obtained by experiments with different ionic strength, temperature, and pH conditions, Wenner et al. (2002) proposed a model where the overstretching plateau was attributed to a force-induced melting process where at the end of the overstretching process short helical domains of the DNA hold large melted strands together (Williams et al., 2001, 2002; Wenner et al., 2002). In the following we term this plateau “overstretching transition”. Further elongation results in a strongly increasing elastic response corresponding to a nonequilibrium melting process (Rief et al., 1999).
In our experiments we observed the overstretching transition at 64 pN up to an extension of 28 μm (170% of dsDNA contour length), which is in good agreement with the results of other groups under similar conditions such as temperature, ionic strength, and pH value (Cluzel et al., 1996; Williams et al., 2001, 2002). The extended worm-like chain model yielded a contour length of 16.0 μm and a persistence length of 40 nm (Table 1) consistent with previous studies (Husale et al., 2002; Wenner et al., 2002). On the basis of the molecular length at a force of 40 pN, we found a value of 16.4 μm, resulting in a λ-DNA (48,502 basepairs) basepair distance of 0.338 nm/basepair, in excellent agreement with previous studies (Husale et al., 2002; Tessmer et al., 2003).
Minor groove binders
The minor groove binder distamycin-A has only a small effect on the molecular length of the λ-DNA; at an extension force of 40 pN we observe a slightly increased value of 16.7 μm and a WLC contour length of 16.3 μm. In contrast to the results for the free dsDNA, the overstretching transition is shifted to higher force values (from 64 pN to 70–85 pN), and a drastic change in the persistence length from 40.0 to 26.7 nm can be observed. Noncovalent binding of distamycin-A to the minor groove of dsDNA is characterized by a combination of electrostatic, van der Waals, and bifurcated hydrogen bondings with a strong preference for AT-rich regions (Coll et al., 1987), which stabilize the double strands and resist the force-induced melting. AFM force spectroscopy studies with the minor groove binder netropsin and λ-DNA exhibit a comparable increase in the overstretching transition (Krautbauer et al., 2002a). Due to the distamycin-A concentration of 1 μM in our experiments a 1:1 binding motif is expected to be dominant with a high binding constant of 107–108 M−1 and a preference of binding to AT-rich regions (Pelton and Wemmer, 1989; Bielawski et al., 2001). Previous experiments with poly(dG-dC) ds-DNA and distamycin-A resulted in a slight lowering of the plateau value of the overstretching transition (Eckel et al., 2003). This phenomenon in combination with the observation of a distinct decreased binding affinity for GC-rich regions (Kassociation = 2 × 105 M−1; Bielawski et al., 2001) is indicative for different binding modes for AT and GC.
Solid-state NMR studies show that distamycin-A in the 1:1 motif effects a significant narrowing of the minor groove from 9.4 to 7.0 Å (Olsen et al., 2003). We observe a decreased persistence length, corresponding to an increased bending flexibility. For netropsin, where structural data result in a widening of the minor groove, an increased persistence length is described (Tessmer et al., 2003). This is an indication for a direct dependence of the persistence length of dsDNA complexed with minor groove binder and structural changes of the groove.
Major groove binders
The elastic response curve of λ-DNA complexed with the α-helical peptide Ac-(Leu-Ala-Arg-Leu)3-NH-linker, which binds in the major groove (Niidome et al., 1996; Eckel et al., 2003), is characterized by an intersected transition (between 17 and 22 μm) between the elastic stretching of B-DNA at low forces and the less pronounced overstretching transition (22–27 μm) at 80–85 pN. Similar to distamycin-A, the force extension curve exhibits a merging of the overstretching transition into the nonequilibrium melting transition at extensions beyond 28 μm. The molecule length at 40 pN and the WLC contour length is slightly increased to 17.1 and 16.5 μm, respectively, and a reduced persistence length of 29.4 nm was calculated. This observation can be associated with an electrostatic binding along with a compensation of the negatively charged DNA backbone by the guanidino groups of the peptide (Niidome et al., 1996), which neutralizes the intrinsic charge and extends the flexibility of the complexed dsDNA.
Recent investigations (Eckel et al., 2003) did not reveal the intersected transition within the complex of Ac-(Leu-Ala-Arg-Leu)3-NH-linker and poly(dG-dC) dsDNA, so we implicate our results to a different binding behavior of Ac-(Leu-Ala-Arg-Leu)3-NH-linker between GC-rich and AT-rich regions.
Recently, λ-DNA complexed with SYBR-Green I (Molecular Probes) has been investigated using a dual-beam optical-tweezers setup (Husale et al., 2002). Because SYBR-Green I is a major groove binder, the elasticity curve exhibits similarities to our results, such as a decreased persistence length, a faint and tilted overstretching plateau around 1.5 fractional extensions (f.e.) of B-DNA, and a characteristic intersected transition in the range between 1.1 and 1.35 f.e. (18–22 μm).
Intercalators
The effect of three different DNA monointercalating agents ethidium bromide, YO-1, and daunomycin, and the bisintercalating agent YOYO-1 was investigated. For all intercalators it was found that the plateau attributed to the overstretching transition completely disappeared, the molecule length at 40 pN and the WLC contour length increased, and the persistence length was considerably reduced compared to free dsDNA (see Table 1). In contrast to groove binding, intercalation is additionally stabilized by ionic interaction between a positively charged group (a protonated imino group in ethidium bromide and YO-1, and a protonated amino group in daunomycin and YO-1) of the intercalator and the negatively charged phosphate DNA backbone. This unspecific electrostatic binding of the intercalators reduces the net charge and extends the flexibility of the DNA, which explains the decrease of the persistence length.
Ethidium bromide and YO-1
The binding of ethidium bromide to dsDNA is structurally characterized by an increase of the basepair distance by 0.34 nm/per molecule (Coury et al., 1996). The contour length 20.4 μm indicates that on average every fourth intercalation site has been occupied by an ethidium bromide molecule, a result that was also found by Husale et al. (2002). The corresponding persistence length is reduced to 20.7 nm, which is in excellent agreement with recent results (Tessmer et al., 2003; Husale et al., 2002). The intercalator YO-1, which has been investigated by AFM techniques (Eckel et al., 2003), is characterized by a smaller reduction of the persistence length to 29.2 nm, whereas the contour length (19.8 μm) is almost equal to that of ethidium bromide. Force-extension curves of dsDNA complexed with ethidium bromide or YO-1 exhibit no hysteresis effects for stretching and relaxing velocities between 100 and 8000 nm/s.
Daunomycin
λ-DNA complexed with daunomycin (also known as cerubidine or daunorubicin in medical chemotherapy; Fig. 2 A), recently investigated with AFM force spectroscopy (Eckel et al., 2003), exhibits an increase of the contour length to 19.8 μm and a decrease of the persistence length to 28.1 nm. Daunomycin, like other anthracyclines, is stabilized during intercalation by its electron-deficient anthraquinone part, with hydrogen bonds and electrostatic interaction additionally enhancing the binding stability between the minor groove of the dsDNA and the amino sugar part of daunomycin (Wang et al., 1987). This explains the large resistance of daunomycin complexed dsDNA against an external force, indicated by a steep rise within the force-extension curve that yields a molecular length of 20.9 μm at 40 pN.
FIGURE 2.
(A) Force-extension curves of a single λ-phage DNA molecule and in the presence of DNA intercalator daunomycin (1 μM) obtained at a stretching velocity of 100 nm/s. (B) At higher velocities, daunomycin intercalated DNA reveals distinct hysteresis effects during extension relaxation cycles.
During extension/relaxation at cycle velocities beyond 1000 nm/s, we identified distinct hysteresis effects (Fig. 2 B). To our knowledge this is the first observation of nonequilibrium processes for monointercalating substances. This hysteresis finding is highly reproducible and can therefore not be attributed to melting hysteresis effects as reported (Krautbauer et al., 2002b).
YOYO-1
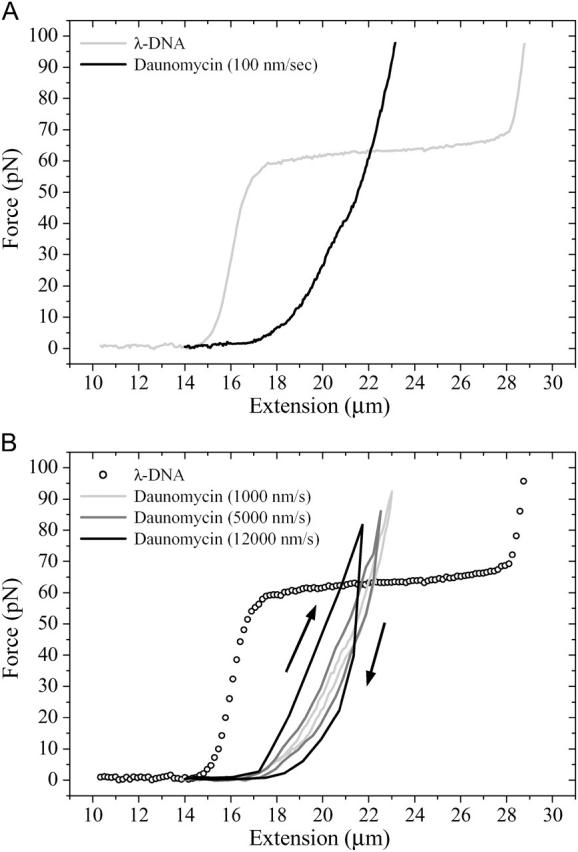
Similar to daunomycin, dsDNA complexed with the bisintercalating agent YOYO-1 exhibits distinct force hysteresis effects and is characterized by an increase of the contour length to 21.8 μm and a strong decrease of the persistence length to 11.8 nm (Fig. 3 A). These results have been obtained during stretching experiments down to velocities of 10 nm/s and at YOYO-1 concentrations of 1 μM. Similar to other intercalators the increase of the contour length can be explained by intercalating between adjacent basepairs. However, YOYO-1 stacks two aromatic ring systems (connected by an aliphatic diamine “backbone”) into two intercalation sites causing a “clamp-like” binding motif. We relate the strong decrease of the persistence length to two protonated amino and two protonated imino groups at one YOYO-1 molecule that reduce the intrinsic charge of the DNA backbone and strongly increase its flexibility.
FIGURE 3.
(A) YOYO-1 (1 μM) bisintercalated λ-phage DNA molecule reveals hysteresis effects during extension/relaxation loop even at low stretching and relaxing velocities. (B) At increased velocities, elasticity curves are shifted to lower extension values. (C) Hysteresis effects at different maximum forces during stretching/relaxation loops of YOYO-1 bisintercalated λ-phage DNA.
The force hysteresis was found to depend on the cycle velocity and the applied maximum force (Fig. 3, B and C) and is consistent with results of Bennink et al. (1999). All force hysteresis effects were found to be highly reproducible during stretching and relaxation and can, therefore, not be attributed to a melting hysteresis effect. The observed hysteresis is accompanied by a shift of the elasticity curve to smaller extension values with increased experimental velocities.
Retention force decay and hysteresis effects of daunomycin and YOYO-1
To investigate the hysteresis phenomenon in more detail we carried out the following experiment: while monitoring the force, a λ-DNA molecule in the presence of 1 μM daunomycin or YOYO-1 was rapidly overstretched (12,000 nm/s, 5000 nm/s, and 2000 nm/s) to different maximum forces. After stopping the extension, we observed an exponential decay of the retention force to a lower stable value (Fig. 4 A). During the force decay, the trapped bead is retreated toward the center of the optical trap, causing an elongation of the stretched DNA given by the trap stiffness divided by the force difference. In these experiments we observed elongations of <300 nm.
FIGURE 4.
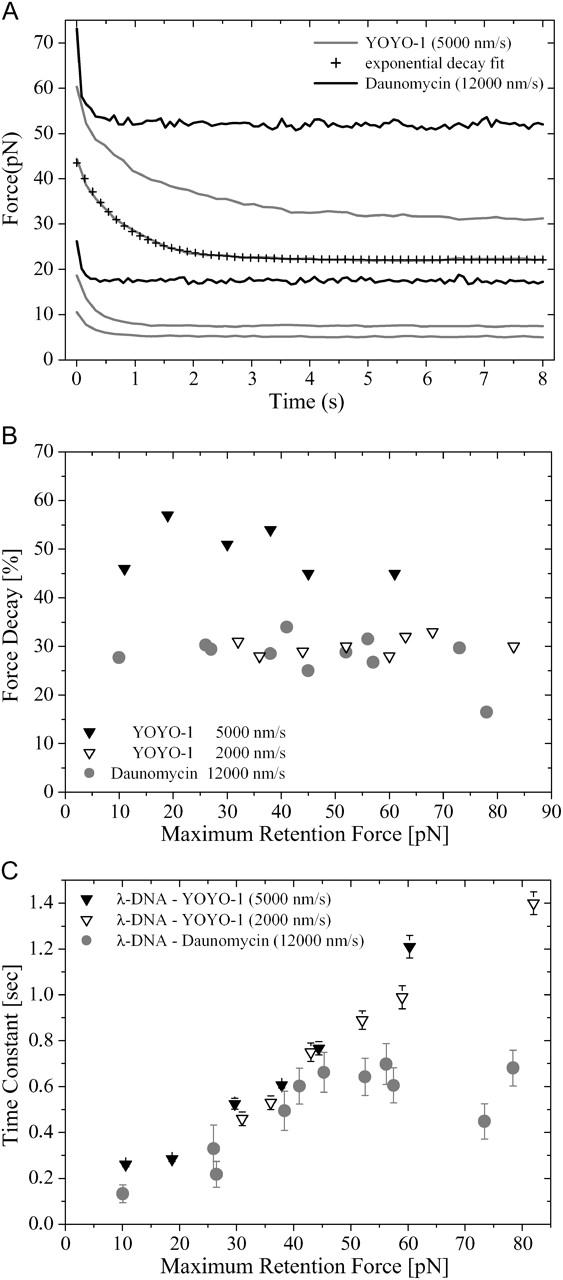
(A) Fast stretching of YOYO-1 bisintercalated and daunomycin intercalated λ-phage DNA with a velocity of 5000 nm/s and 12,000 nm/s, respectively, to a maximum force and immediate stopping unveils an exponential decay of the retention force with time. (B) Percentage of exponential decay of the retention force as a function of maximum retention force for YOYO-1 and daunomycin. (C) Relaxation time as a function of maximum retention force for YOYO-1 and daunomycin. Due to high-extension velocity for daunomcin, short-time data acquisition implicates less data averaging, which effects more statistical noise and larger error bars.
The relative force decays increase with the stretching velocities and are independent from the maximum retention force (Fig. 4 B). For dsDNA complexed with YOYO-1 the time constants derived from exponential fits show an almost linear dependence from the maximum retention forces and were independent from stretching velocities (Fig. 4 C). Values from 0.26 s for a maximal retention force of 10 pN to 1.21 s for 83 pN were observed. For daunomycin the linear dependence was only found for retention forces <45 pN, the values range from 0.13 s at 10 pN to 0.68 s at 45 pN. For higher forces the decay times are constant, which indicates changes in the molecular extension process.
The observed hysteresis and retention force decays suggest that the dsDNA strands complexed with these two intercalators are not in equilibrium at the given pulling speed. Rief et al. introduce a model based on a coupled two-level system for biopolymer extensibility, where segments of the molecule undergo conformational changes (Rief et al., 1998). Under an external force the length of each segment changes based on thermodynamics. With this model the hysteresis effects and the retention force can be explained. For YOYO-1 the process of segment elongation is remarkably slow. Pulling velocities of 10 nm/s with a molecular extension of a few microns result in a timescale of minutes, whereas for daunomycin the transition from equilibrium to the nonequilibrium state is in the timescale of seconds.
This elongation of the segments can in principle be attributed to force-induced changes in the molecular structure or to intercalation of additional molecules into the stretched DNA. Because of the high association constant of YOYO-1 (1012 M−1; Larsson et al., 1994) and even for the lower constant of daunomycin (105 M−1; Coury et al., 1996), both intercalators may associate to dsDNA by electrostatic interaction in a fast (for our experiments, undetectable) timescale, but the intercalation itself (especially those of both aromatic ring systems of YOYO-1 into dsDNA) takes place on a much slower timescale, as can be inferred from the calculated time constants.
CONCLUSIONS
Mechanical properties of λ-DNA (dsDNA) complexed with different binding ligands were analyzed in single-molecule optical-tweezers experiments. The differences between binding modes, such as minor groove binding, major groove binding, and (bis)intercalation could be distinguished by analyzing the mechanical response of a single dsDNA molecule to an applied external force. Different binding properties of the minor and major groove binder for AT- and GC-rich regions could be identified upon comparing our measurement with recently published AFM results. The persistence length of DNA complexed with minor groove binders might be related to changes in the width of the minor groove: narrowing results in a decreased persistence length, a topic to be investigated in more detail in the future.
Force hysteresis effects during stretching/relaxation cycles and retention force decays were found for the bisintercalator YOYO-1 and for the intercalator daunomycin, which we relate to a slow force-induced elongation of DNA segments. This can be attributed to a structural change or the intercalation of additional molecules to the stretched dsDNA. The observation that the other binding ligands investigated (i.e., distamycin-A, the α-helical peptide Ac-(Leu-Ala-Arg-Leu)3-NH-linker, ethidium bromide, and YO-1) lack force hysteresis still remains to be explained. It could be, for these ligands, that the applied force induces no additional time-dependent change in molecular conformation, but it is also possible that the said effect takes place on a much faster timescale than our experiment (milliseconds, or even faster). In further experiments with very high stretching velocities this question will be addressed. Experiments like our retention force-decay measurements with the force-clamp technique should be able to give quantitative data to describe this fundamental slow structural transition process.
For daunomycin we found in the decay time analysis an unexpected transition from a linear to a constant dependence on the maximal retention force at 45 pN. This gives an interesting hint to a change in the elongation process from a low- to a high-force regime, which will be addressed in further experiments.
Acknowledgments
We thank Martin Hegner and Wilfried Grange for support during the setup of the optical-tweezers system and Peter Reimann for helpful discussions.
We gratefully acknowledge funding from the collaborative research project SFB 613 from the Deutsche Forschungsgemeinschaft (DFG).
References
- Anselmetti, D., J. Fritz, B. Smith, and X. Fernandez-Busquets. 2000. Single molecule DNA biophysics with atomic force microscopy. Single Mol. 1:17–23. [Google Scholar]
- Ashkin, A. 1970. Acceleration and trapping of particles by radiation pressure. Phys. Rev. Lett. 24:156–159. [Google Scholar]
- Ashkin, A., J. M. Dziedzic, J. E. Bjorkholm, and S. Chu. 1986. Observation of a single-beam gradient force optical trap for dielectic particles. Opt. Lett. 11:288–290. [DOI] [PubMed] [Google Scholar]
- Ashkin, A. 1997. Optical trapping and manipulating of neutral particles using lasers. Proc. Natl. Acad. Sci. USA. 94:4853–4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennink, M. J., O. D. Schärer, R. Kanaar, K. Sakata-Sogawa, J. M. Schins, J. S. Kanger, B. G. de Grooth, and J. Greve. 1999. Single-molecule manipulation of double-stranded DNA using optical tweezers: interaction studies of DNA with RecA and YOYO-1. Cytometry. 36:200–208. [DOI] [PubMed] [Google Scholar]
- Bielawski, K., S. Wolczynski, and A. Bielawska. 2001. DNA-binding properties and cytotoxity of extended aromatic bisamidines in breast cancer mcf-7 cells. Pol. J. Pharmacol. 53:143–147. [PubMed] [Google Scholar]
- Binnig, G., C. F. Quate, and C. Gerber. 1986. Atomic force microscope. Phys. Rev. Lett. 56:930–933. [DOI] [PubMed] [Google Scholar]
- Bouchiat, C., M. D. Wang, J. F. Allemand, T. Strick, S. M. Block, and V. Croquette. 1999. Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophys. J. 76:409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen-Schaumann, H., M. Rief, C. Tolksdorf, and H. E. Gaub. 2000. Mechanical stability of single DNA molecules. Biophys. J. 78:1997–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluzel, P., A. Lebrun, C. Heller, R. Lavery, J. L. Viovy, D. Chatenay, and F. Caron. 1996. DNA: an extensible molecule. Science. 271:792–794. [DOI] [PubMed] [Google Scholar]
- Coll, M., C. A. Frederick, A. H.-J. Wang, and A. Rich. 1987. A bifurcated hydrogen-bonded conformation in the d(AT) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc. Natl. Acad. Sci. USA. 84:8385–8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste, F., J. M. Malinge, L. Serre, W. Shephard, M. Roth, M. Leng, and C. Zelwer. 1999. Crystal structure of a double-stranded DNA containing a cisplatin interstrand cross-link at 1.63 A resolution: hydration at the platinated site. Nucleic Acids Res. 27:1837–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coury, J. E., L. McFail-Isom, L. D. Williams, and L. A. Bottomley. 1996. A novel assay for drug-DNA binding mode, affinity, and exclusion number: scanning force microscopy. Proc. Natl. Acad. Sci. USA. 93:12283–12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel, R., R. Ros, A. Ros, S. D. Wilking, N. Sewald, and D. Anselmetti. 2003. Identification of binding mechanisms in single molecule-DNA complexes. Biophys. J. 85:1968–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald, D. J., and J. N. Anderson. 1999. Selective nucleosome disruption by drugs that bind in the minor groove of DNA. J. Biol. Chem. 274:27128–27138. [DOI] [PubMed] [Google Scholar]
- Gelasco, A., and S. J. Lippard. 1998. NMR solution structure of a DNA dodecamer duplex containing a cis-diammineplatinum(II) d(GpG) intrastrand cross-link, the major adduct of the anticancer drug cisplatin. Biochemistry. 37:9230–9239. [DOI] [PubMed] [Google Scholar]
- Graves, D. E., and L. M. Velea. 2000. Intercalative Binding of Small Molecules to Nucleic Acids. Curr. Org. Chem. 4:915–929. [Google Scholar]
- Hurley, L. H. 2002. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2:188–200. [DOI] [PubMed] [Google Scholar]
- Husale, S., W. Grange, and M. Hegner. 2002. DNA mechanics affected by small DNA interacting ligands. Single Mol. 3:91–96. [Google Scholar]
- Krautbauer, R., H. Clausen-Schaumann, and H. E. Gaub. 2000. Cisplatin changes the mechanics of single DNA molecules. Angew. Chem. Int. Ed. 39:3912–3915 [DOI] [PubMed] [Google Scholar]
- Krautbauer, R., S. Fischerländer, S. Allen, and H. E. Gaub. 2002a. Mechanical fingerprints of DNA drug complexes. Single Mol. 3:97–103. [Google Scholar]
- Krautbauer, R., L. H. Pope, T. E. Schrader, S. Allen, and H. E. Gaub. 2002b. Discriminating small molecule DNA binding modes by single molecule force spectroscopy. FEBS Lett. 510:154–158. [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy, G., G. Duportail, and Y. Mely. 2002. Structure and dynamics of condensed DNA probed by 1,1′-(4,4,8,8-tetramethyl-4,8-diazaundecamethylene)bis[4-[[3-methylbenz-1,3-oxazol-2-yl]methylidine]-1,4-dihydroquinolinium] tetraiodide fluorescence. Biochemistry. 41:15277–15287. [DOI] [PubMed] [Google Scholar]
- Larsson, A., C. Carlsson, M. Jonsson, and B. Albinsson. 1994. Characterization of the binding of the fluorescent dyes YO and YOYO to DNA by polarized light spectroscopy. J. Am. Chem. Soc. 116:8459–8465. [Google Scholar]
- Lipscomb, L. A., F. X. Zhou, S. R. Presnell, R. J. Woo, M. E. Peek, R. R. Plaskon, and L. D. Williams. 1996. Structure of a DNA-porphyrin complex. Biochemistry. 35:2818–2823. [DOI] [PubMed] [Google Scholar]
- Marko, J. F., and E. D. Siggia. 1995. Stretching DNA. Macromolecules. 28:8759–8770. [Google Scholar]
- McMurray, C., E. W. Small, and K. E. van Holde. 1991. Binding of ethidium to the nucleosome core particle. 2. Internal and external binding modes. Biochemistry. 30:5644–5652. [DOI] [PubMed] [Google Scholar]
- Niidome, T., N. Ohmori, A. Ichinose, A. Wada, H. Mihara, T. Hirayama, and H. Aoyagi. 1996. Binding of cationic alpha-helical peptides to plasmid DNA and their gene-transfer abilities into cells. J. Biol. Chem. 272:15307–15312. [DOI] [PubMed] [Google Scholar]
- Olsen, G. L., E. A. Louie, G. P. Drobny, and S. Th. Sigurdsson. 2003. Determination of DNA minor groove width in distamycin-DNA complexes by solid-state NMR. Nucleic Acids Res. 31:5084–5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelton, J. G., and D. E. Wemmer. 1989. Structure characterization of a 2:1 distamycin A d(CGCAAATTGGC) complex by two-dimensional NMR. Proc. Natl. Acad. Sci. USA. 86:5723–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, B. S. P., S. K. Sharma, and J. W. Lown. 2001. Recent developments in sequence selective minor groove DNA effectors. Curr. Med. Chem. 8:475–508. [DOI] [PubMed] [Google Scholar]
- Reha, D., M. Kabelac, F. Ryjacek, J. Sponer, J. E. Sponer, M. Elstner, S. Suhai, and P. Hobza. 2002. Intercalators. 1. Nature of stacking interactions between intercalators (ethidium, daunomycin, ellipticine, and 4′,6-diaminide-2-phenylindole) and DNA base pairs. Ab initio quantum chemical, density functional theory, and empirical potential study. J. Am. Chem. Soc. 124:3366–3376. [DOI] [PubMed] [Google Scholar]
- Rief, M., J. M. Fernandez, and H. E. Gaub. 1998. Elastically coupled two-level systems as a model for biopolymer extensibility. Phys. Rev. Lett. 81:4764–4767. [Google Scholar]
- Rief, M., H. Clausen-Schauman, and H. E. Gaub. 1999. Sequence-dependent mechanics of single DNA molecules. Nat. Struct. Biol. 6:346–349. [DOI] [PubMed] [Google Scholar]
- Sischka, A., R. Eckel, K. Tönsing, R. Ros, and D. Anselmetti. 2003. Compact, microscope based optical tweezers system for molecular manipulation. Rev. Sci. Instrum. 74:4827–4831. [Google Scholar]
- Smith, S. B., Y. Cui, and C. Bustamante. 1996. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 271:795–799. [DOI] [PubMed] [Google Scholar]
- Svoboda, K., and S. M. Block. 1994. Biological applications of optical forces. Annu. Rev. Biophys. Biomol. Struct. 23:247–285. [DOI] [PubMed] [Google Scholar]
- Tessmer, I., C. G. Baumann, G. M. Skinner, J. E. Molloy, J. G. Hoggett, S. J. B. Tendler, and S. Allen. 2003. Mode of drug binding to DNA determined by optical tweezers force spectroscopy. J. Mod. Optics. 50:1627–1636. [Google Scholar]
- Wang, A. H., G. Ughetto, G. J. Quigley, and A. Rich. 1987. Interactions between an anthracycline antibiotic and DNA: molecular structure of daunomycin complexed to d(CpGpTpApCpG) at 1.2-A resolution. Biochemistry. 26:1152–1163. [DOI] [PubMed] [Google Scholar]
- Wenner, J. R., M. C. Williams, I. Rouzina, and V. A. Bloomfield. 2002. Salt dependence of the elasticity and overstretching transition of single DNA molecules. Biophys. J. 82:3160–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, M. C., J. R. Wenner, I. Rouzina, and V. A. Bloomfield. 2001. Effect of pH on the overstretching transition of double-stranded DNA: evidence for force-induced DNA melting. Biophys. J. 80:874–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, M. C., I. Rouzina, and V. A. Bloomfield. 2002. Thermodynamics of DNA interactions from single molecule stretching experiments. Acc. Chem. Res. 35:159–166. [DOI] [PubMed] [Google Scholar]
- Wuite, G. J. L., R. J. Davenport, A. Rappaport, and C. Bustamante. 2000. An integrated laser trap/flow control video microscope for the study of single biomolecules. Biophys. J. 79:1155–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer, Ch., and U. Wähnert. 1986. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog. Biophys. Mol. Biol. 47:31–112. [DOI] [PubMed] [Google Scholar]