Abstract
Mechanical properties of living cells are important for cell shape, motility, and cellular responses to biochemical and biophysical signals. Although these properties are predominantly determined by the cytoskeleton, relatively little is known about the mechanical organization of cells at a subcellular level. We have studied the cell cortex of bovine pulmonary artery endothelial cells (BPAECs) using atomic force microscopy (AFM) and confocal fluorescence microscopy (CFM). We show that the contrast in AFM imaging of these cells derives in large part from differences in local mechanical properties, and AFM images of BPAEC reveal the local micromechanical architecture of their apical cortex at ∼125 nm resolution. Mechanically the cortex in these cells is organized as a polygonal mesh at two length scales: a coarse mesh with mesh element areas ∼0.5–10 μm2, and a finer mesh with areas <0.5 μm2. These meshes appear to be intertwined, which may have interesting implications for the mechanical properties of the cell. Correlated AFM-CFM experiments and pharmacological treatments reveal that actin and vimentin are components of the coarse mesh, but microtubules are not mechanical components of the BPAEC apical cortex.
INTRODUCTION
Cell mechanics are fundamental to cell shape, motility, division, tissue organization, and other biologically important properties and processes (Alberts et al., 1994; Howard, 2001; Boal, 2002). The mechanical properties of many cell types have been studied extensively, and living cells are known to be complex and heterogeneous visco-elastic structures (Elson, 1988; Evans, 1989; Pollard et al., 2000). Although the molecular underpinnings for cell mechanics are not fully understood, it is clear that cytoskeleton plays a central role. Actin, microtubules, and intermediate filaments are structural components of the cytoskeleton with unique mechanical properties, and rheological measurements of purified cytoskeletal preparations show that these have visco-elastic properties similar to whole cells (Janmey et al., 1991, 1994; Gittes et al., 1993; Kojima et al., 1994; Kurachi et al., 1995). Pharmacological disruption of actin causes a significant change in cell mechanics (Rotsch and Radmacher, 2000; Yamada et al., 2000; Wakatsuki et al., 2001); likewise, genetic disruption of cytoskeletal proteins has significant mechanical consequences (Janssen et al., 1996; Weber et al., 1999). As a result of this close relationship between cytoskeleton and mechanics, cytoskeletal organization is sometimes taken to reflect mechanical organization (e.g., Heidemann et al., 2000; Ingber et al., 2000). However, there is little direct information about the spatial micromechanical organization of living cells at a subcellular level.
The cytoskeleton is partitioned to carry out specific functions in a cell; one organizational unit is the cortical cytoskeleton (CS), which comprises the cytoskeleton connected to and in close proximity to the plasma membrane and is often treated separately in mechanical models of cells (Evans, 1989; Yeung and Evans, 1989; Dong et al., 1991; Karcher et al., 2003). Freeze fracture deep etch electron microscopy provides highly detailed views of the CS and reveals a complex filamentous network (Satcher et al., 1997; Heuser, 2000). Immunofluorescence microscopy of CS frequently shows a diffuse submembrane label (Tsukita and Yonemura, 1999; Flanagan et al., 2001). Where details of the cytoskeleton organization can be seen, it is difficult to unambiguously identify the cortical component, unless cells are very thin and thus have little non-CS (Lazarides, 1975; Galbraith et al., 1998).
Vascular endothelial cells are an interesting system for studying functionally important aspects of cytoskeleton and mechanics (Dudek and Garcia, 2001; Helmke and Davies, 2002; Lee and Gotlieb, 2002; Ogunrinade et al., 2002). These cells are found in a mechanically active environment and are required to withstand shear stress, blood pressure, and changes in pressure due to breathing cycles. The cortex of these cells faces the blood stream and is important for responding to external forces, and transmitting force, as well as controlling cell shape. In this study, we used atomic force microscopy (AFM), confocal fluorescence microscopy (CFM), and anti-cytoskeletal drugs to characterize the micromechanical architecture of the cortex in bovine pulmonary artery endothelial cells (BPAECs). We show that the cortex in these cells is mechanically organized as a polygonal mesh on two levels: a coarse mesh with dimensions on the order of several micrometers and a fine overlapping mesh with dimensions on the order of hundreds of nanometers. These meshes appear to be intertwined and are in part composed of actin and vimentin.
MATERIALS AND METHODS
Cell culture
BPAECs, Eagle's minimum essential medium, and fetal bovine serum were from American Type Culture Collection (Manassas, VA). BPAECs were maintained on petri dishes or gelatin coated glass coverslips in Eagle's minimum essential medium supplemented with 20% fetal bovine serum at 5% CO2 and 37°C. The cells were fed every 2–3 days and passaged when confluent. Passages 17–22 were used.
AFM imaging
AFM imaging was performed with a Multimode or Bioscope AFM equipped with large area scanners (>100 μm × 100 μm), with a Nanoscope IIIa controller (Digital Instruments, Santa Barbara, CA). The Bioscope was mounted on an Olympus inverted optical microscope. For imaging live cells in solution, unsharpened (radius of curvature ∼50 nm) silicon nitride cantilevers with nominal force constants of 0.01 or 0.03 Newtons/meter were used (Nanoprobes, Digital Instruments). Ambient tapping of fixed and dried cells was performed with single crystal silicon cantilevers (model TESP; Digital Instruments). Live cell imaging was performed in fluid contact mode at room temperature and atmospheric CO2. The imaging buffer was phosphate buffered saline (Invitrogen, Carlsbad, CA) supplemented with 1.2 mM CaCl2, 1.2 mM MgCl2, 5 mM Hepes, and 1 g/L glucose. For pharmacological treatments of BPAEC cortex, stock solutions of cytochalasin B (1 mg/ml in dimethylsulfoxide (DMSO)) and nocodazole (4 mg/ml in DMSO) were prepared. The final concentrations of drugs were determined after estimating the amount of imaging buffer present in the fluid holder before drug addition. Since DMSO can increase the temperature upon mixing with aqueous buffers, first 50–100 μl imaging buffer was smoothly pipetted out of the fluid holder channels and it was mixed with 1–5 μl of stock drug solution. After reaching room temperature, this solution was added to the fluid holder and mixed gently to ensure rapid drug delivery to cells. Imaging parameters were empirically optimized to produce clear images with minimal distortion or damage to the cells. Typically, scan rates were 60–120 μm/s, resulting in image acquisition times of 4–16 min depending on the scan size. BPAECs could be imaged for up to 4 h, during which time the cells remained adherent and high quality images could be collected. With extended imaging the fenestrae between cells began to expand, exposing the substrate. We interpret this as an indicator of cell deterioration in response to the AFM imaging. Further imaging resulted in loss of cells from the surface.
Local mechanical measurements
AFM force curves over confluent BPAEC monolayers were collected using the same cantilevers as for imaging at rates of ∼10 μm/s. The relative trigger on the microscope was set to 50 nm or less to prevent inadvertent damage to the cell. The force curves were used to determine the elastic modulus as described previously (Radmacher et al., 1996; Costa and Yin, 1999). Force calculations use nominal values for cantilever stiffness, and hence should be considered accurate to within a factor of two.
Confocal immunofluorescence microscopy
For correlated AFM-CFM experiments, cells cultured on gelatin (Sigma, St. Louis, MO) coated CELLocate glass coverslips (Brinkmann, Westbury, NY) were imaged by AFM and immediately fixed in 3.7% paraformaldehyde for 10 min. The fixative was added to the cells <1 min after completing the AFM imaging. Cells were then permeabilized with 0.2% Triton-X-100 in PBS for 5 min and blocked with 1% BSA in PBS for 30–60 min. The cells were then treated with Alexa Fluor 488 phalloidin (Molecular Probes, Eugene, OR), monoclonal anti-β-tubulin-Cy3 antibody (∼1:50–1:100) or monoclonal anti-vimentin-Cy3 antibody (∼1:50–1:100) (Sigma) in 1% BSA in PBS for 1 h. Finally, cells were washed with PBS, and mounted on slides with ProLong media (Molecular Probes). An UltraView Confocal microscope (PerkinElmer, Wellesley, MA) was used to collect immunofluorescent images. Areas to examine were determined by using bright field light micrographs collected during AFM imaging on the Bioscope in conjunction with the locator grid.
Scanning electron microscopy
Cells grown on coverslips were fixed with 3% glutaraldehyde, 0.1 M Hepes, 2 mM CaCl2 pH 7.2, for 30 min at room temperature. Cells were then rinsed twice using 0.1 M Hepes with 3 mM CaCl2 for 10 min, followed by a rinse in 0.1 M sodium cacodylate with 3 mM CaCl2 for 10 min, and post fixed in reduced osmium (1% OsO4, 0.8% K4FeCn6, 0.1 M sodium cacodylate, 3 mM CaCl2) for 1 h on ice. After a brief water rinse, cells were en-bloc stained in 2% uranyl acetate (filtered) for 1 h in the dark. Samples were dehydrated using a series of ethanol incubations and then treated with 100% hexamethyldisilazane (Polysciences, Warrington, PA) for 10 min. Coverslips were then placed on Whatman No. 1 filter paper (cell side up) and allowed to dry. Finally, coverslips were attached to carbon sticky tabs affixed to scanning EM stubs and evaporated with 1 nm of chromium in a Denton DV-502A high vacuum evaporator operating at 50 mA and 2 × 10−7 torr. Cells were viewed and digitized on a Leo 1530 Field Emission Scanning EM operating at 1 kV.
Image display and data analysis
AFM data were analyzed with Image SXM and SuperMapper, a custom software suite developed with Interactive Data Language (Research Systems, Boulder, CO). The deflection images were processed to optimize brightness and to enhance contrast. Immunofluorescence data were optimized for brightness and contrast using Adobe Photoshop. Correlated areas were determined visually by overlaying AFM deflection images on CFM images, and manually varying the transparency of the AFM imaged. Mesh element areas were estimated by ((A1 × A2)/2).
RESULTS
AFM images reveal the micromechanical organization of the cortex
BPAECs grow in confluent monolayers, and AFM images show a general morphology that is consistent with that seen by light and electron microscopy. Cells are typically tens of micrometers in diameter, and heights range from a few hundred nanometers at the periphery to ∼4 μm toward the center (Fig. 1, A–D). In addition, these cells display a complex highly organized filamentous network. The AFM is a sensitive surface probe; however, when soft materials such as living cells are probed with an AFM tip, local mechanical differences result in differential surface deformations and contribute to contrast in the images. Scanning EM and AFM images of fixed endothelial cells show a relatively smooth surface, with none of the filamentous features seen in AFM images of living cells (Fig. 1, C and D) (Chazov et al., 1981; Schaeffer et al., 1993). Hence the contrast in the AFM images of living BPAECs derives in substantial part from local mechanical properties; in particular the filamentous structures are mechanical features from the cortical regions of these cells.
FIGURE 1.
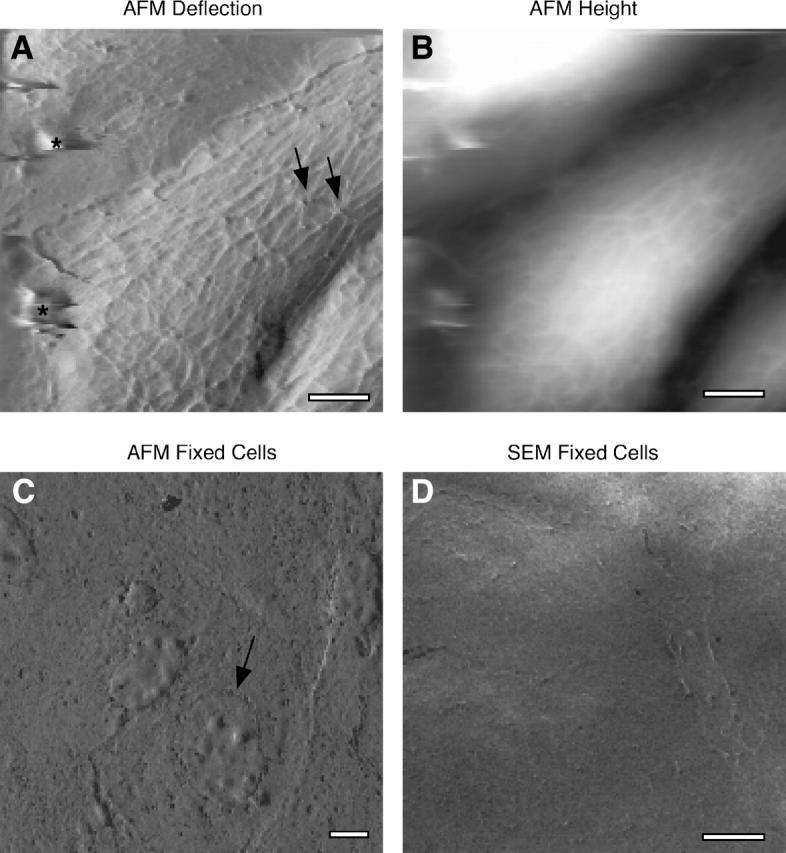
Mechanically based contrast in AFM images of BPAECs. (A) AFM deflection image. (B) Corresponding AFM height image of BPAECs imaged in a physiological saline. Generally the applied force was on the order of 1 nN, the scan rate was 60 μm/s, and the image acquisition time was between 4–16 min. Overall cell morphology is similar to that seen by light microscopy or scanning EM; however, significant additional detail is seen. The deflection images show an intricate mesh of filaments; other features seen in the images include focal points with 5–8 converging filaments (arrows). The smeared features (asterisk) are imaging artifacts. (C) AFM image of fixed BPAECs shows no filamentous structures; however, the cell boundaries and nuclei (arrow) are visible. The latter is likely visible because the membrane collapses over the nucleus during dehydration. (D) Likewise, scanning EM of BPAEC shows cell boundaries but no filamentous mesh. Thus, contrast for the fine features seen in A is attributed to differences in local mechanical properties. Z-range of the gray scale in B is 0–4 μm. Scale bars are 5 μm.
Architecture of the endothelial cortex
The filamentous network at the cell cortex is organized as a complex polygonal mesh. Visually the mesh appears to be structured on two length scales, one on the order of a few hundred nanometers and the other on the order of a few micrometers (Fig. 2). The larger mesh is composed of prominent and hence relatively stiff features, with a wide range of forms. The fine mesh is composed of thin filaments with weak contrast, and typically visible inside the large mesh elements. The fine mesh elements are more uniform in appearance, and have areas ∼0.05–0.5 μm2. In places the fine filaments run on top of the thicker filaments, and are thus closer to the plasma membrane. Other thin filaments appear to run underneath the thick filaments, although because of the weak contrast from the thin filaments the possibility that all thin filaments run on top of the thick filaments cannot be strictly excluded. Thus, the cell cortex might be described as a coarse mesh intertwined with a fine one, or possibly a fine mesh layer over a coarse mesh.
FIGURE 2.
Cortical mesh appears to be organized at two predominant length scales. (A–D) High magnification deflection AFM images of living BPAECs in a physiological saline. The filamentous mesh appears to be organized on two length scales, with coarse mesh (arrowheads) and fine mesh filaments (arrows). The coarse mesh forms larger elements and is easier to identify, whereas the fine mesh elements have much smaller dimensions. Further, contrast in the fine mesh is weak, making it difficult to completely trace. In places, the fine mesh runs over the coarse mesh and is thus more proximal to the membrane. In other places, the mesh may run under the coarse mesh, or is not visible due to the weak contrast. Thus, the two meshes are likely intertwined, although it is possible that the fine mesh is layered over the coarse mesh. Lateral resolution in these images is ∼125 nm. Scale bars are 1 μm.
Closer examination of the coarse mesh reveals that the thick filaments course across the monolayer surface, and in places give the appearance of being continuous with neighboring cells. Thick filaments often converge at “star-like” focal points, where, on average, 3–4 filamentous structures radiate from a single point; these are distributed over the entire apical surface. Such star-like focal points have previously been observed by electron microscopy, and it has been suggested that they are vertices of actin geodomes (Lazarides, 1975; Heuser and Kirschner, 1980). Less frequently there were also focal points with 5–8 converging filaments. To characterize the mesh elements we measured the long axis (A1), short axis (A2), and asymmetry (MA1/A2) in 67 mesh elements from nine different cells (Fig. 3). Individual elements were found to be relatively asymmetric with A1 = 4.4 ± 2.3 μm (mean ± SD), A2 = 1.6 ± 1 μm, and MA1/A2 = 2.8 ± 1.3. In elongated cells the major axis of mesh elements tends to align with the major axis of the cells. Examination of the internal branch angles in these mesh elements reveals a bimodal distribution with peaks at 53° and 103° (n = 66). These discrete distributions suggest that the branches do not form from random crossings of filaments. Proteins such as filamin, spectrin, and Arp2/3 can organize actin into networks with characteristic angles (Hartwig and Shevlin, 1986; Hansen et al., 1997; Pollard and Borisy, 2003). We note that scanning did not significantly perturb the lateral organization of the filaments. This is apparent from the shape of features in the image, which show little dependence on scan direction. Further, comparison of trace and retrace images showed little or no scan direction related orientation of the filaments (data not shown).
FIGURE 3.
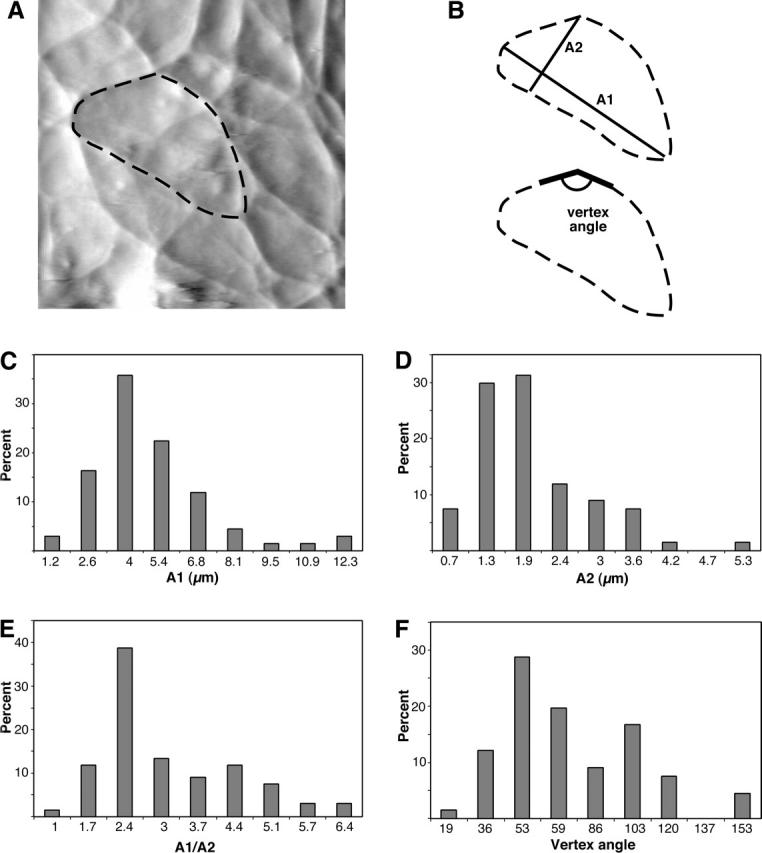
Coarse mesh elements are asymmetric and micrometer scale. Shapes of the coarse mesh elements were examined by hand tracing mesh boundaries (n = 67). To characterize the mesh, we described individual mesh elements by their longest axis (A1), longest axis perpendicular to A1 (A2), and the ratio of the two axes (MA1/A2). (A) Example of traced coarse mesh element in a BPAEC deflection image and (B) definitions of parameters examined. Histograms of measured dimensions show that (C) the long axis of the cells is on average 4.4 μm, (D) the short axis is on average 1.6 μm, and (E) the average aspect ratio is 2.8. (F) Internal angles of the coarse mesh elements show a bimodal distribution with peaks at 53° and 103° (n = 66).
Analysis of AFM deflection images of BPAECs reveals that the best resolution obtained is ∼125 nm along the fast scan axis, as determined from the spectral composition of the data (data not shown) (Joy, 2002). Thus, applying the Nyquist criterion, the resolution of 512 pixel/line images larger than 32 μm is limited by sampling frequency, whereas images smaller than 32 μm are limited by other aspects of the image acquisition. This resolution assessment agrees well with visual inspection of images, where the point-to-point resolution is determined from features such as spacing between individual filaments and their widths.
Cortex mechanics
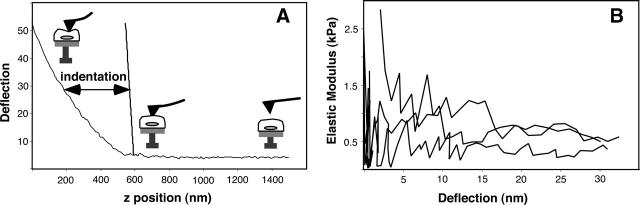
The AFM imaging presented above provides a detailed picture of the lateral mechanical organization of the cortex, but does not address variations as a function of distance beneath the plasma membrane. AFM force measurements can be used to quantitate local mechanical properties, and are also sensitive to changes in mechanical properties as a function of indentation depth (Radmacher et al., 1995; Costa and Yin, 1999). Micromechanical measurements on BPAECs using the AFM show that the average elastic modulus is in the range of 0.2–2 kPa, in agreement with previous studies on endothelial cells (Satcher and Dewey, 1996; Hochmuth, 2000; Mathur et al., 2000, 2001) (Fig. 4). Force curves collected at different positions on individual cells show that the cell body appears to be two- to threefold softer than the cell periphery, though it is known that the hard surface under a soft sample affects force measurements more as the sample thickness decreases (Costa and Yin, 1999). Thus, mechanical measurements on the cell body, where the cell reaches its maximum height, will have the least impact from the underlying hard surface (Charras et al., 2001; Dimitriadis et al., 2002). For force curves collected near the center of the cell, the measured modulus appears to be independent of indentation depth for indentations up to ∼1 μm (i.e., up to 25% strain). Furthermore, these force measurements show that at a force of 1 nanoNewton (nN) (typical imaging force), the upper indentation depth is ∼600 nm (at a scan rate of ∼10 μm/s). It should be noted that at small deflections these measurements are very noisy, and it is possible that some nonuniform behavior is buried in this portion of the data.
FIGURE 4.
Mechanical definition of the BPAEC cortex. (A) AFM force curves on BPAECs show an indentation of ∼600 nm at the imaging force, which set an upper limit for the thickness of the cortex. However, cell viscosity will contribute significantly, and the effective cortical region is likely to be on the order of a couple hundred nanometers. (B) Plots of elastic modulus as a function of deflection, corresponding to indentations up to 1 μm, show little variation as a function of indentation.
Actin is a major contributor to the cortical micromechanical architecture
To identify the molecular components that underlie the mechanical architecture visualized by AFM we performed correlated AFM-CFM and pharmacological treatments. For correlated AFM-CFM experiments, bright field light micrographs were collected during AFM imaging of living cells grown on locator coverslips. Cells were fixed immediately (within ∼1 min) after AFM imaging and stained for actin, vimentin, or microtubules. The locator grid then allowed fluorescence imaging of the same area where the AFM image was collected. CFM images show that actin filaments are present throughout the cell, and comparison of AFM images with fluorescence images show a direct correspondence for a number of filamentous and polygonal features (Fig. 5). In particular, comparing the more basal CFM slices with the AFM image, there are features in the thin periphery where the CFM and AFM images show the same features. However, toward the center of the cell, the CFM shows a number of strongly staining stress fibers that are not seen in the AFM image. In the more cortical slices the CFM staining is relatively weak and shows scattered spots of F-actin and few filamentous structures, in agreement with previous observations (Galbraith et al., 1998). Here a direct correlation is more difficult, but again the CFM appears to show several features similar to those seen in the AFM image. A double staining for actin and vimentin again shows a number of features in the AFM image that can be correlated with actin staining (Fig. 6). Vimentin filaments are more prevalent in the central regions of the cell than in the cell periphery. Generally the vimentin-based structures have a curly pattern of staining with filaments crossing each other multiple times, and in a few instances there are features that can be correlated to the AFM images. Staining for microtubules did not produce features that could be readily identified in the AFM images (data not shown). Thus, of the features that can be accounted for in the AFM images, actin is the most significant component, whereas vimentin is less prevalent and microtubules are not detected.
FIGURE 5.
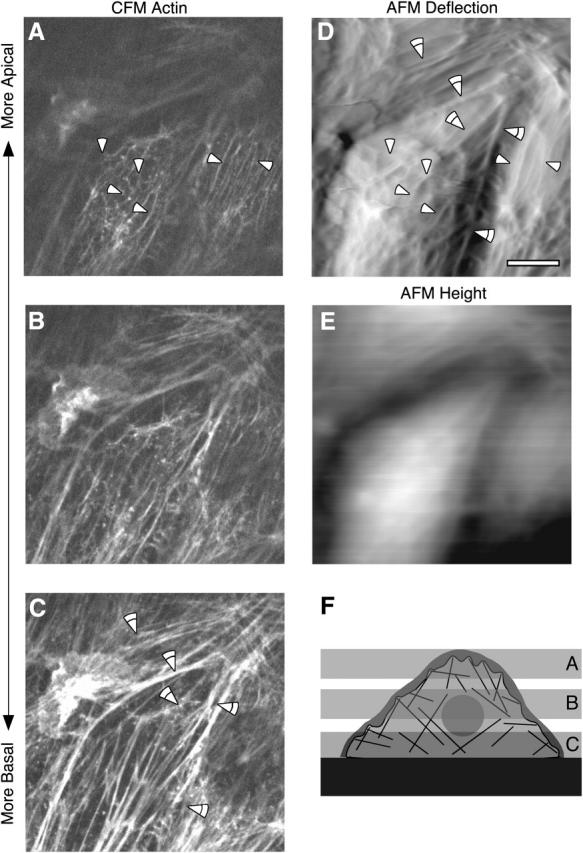
Correlated AFM and CFM of cells stained for actin. An AFM image of living BPAECs was collected and the cells were immediately fixed and processed for immunofluorescence. (A–C) Confocal microscopy planes of fixed cells. (D) AFM deflection image of living BPAEC in solution. (E) Corresponding height image. Cells were stained for actin using Alexa-phalloidin. The CFM images show that actin filaments are present throughout the cell. Note that the basal stress fibers are not seen in the AFM images. However, a careful comparison of AFM images with fluorescence images shows a direct correspondence for a number of filamentous and polygonal features. Correlated features are marked with arrowheads. (F) Schematic illustrating the spatial relationships between the AFM and confocal images. The horizontal gray planes represent optical confocal slices, whereas the dark gray area represents the cell cortex imaged by the AFM probe. Scale bar is 10 μm. Z-range in the height image is 0–4 μm.
FIGURE 6.
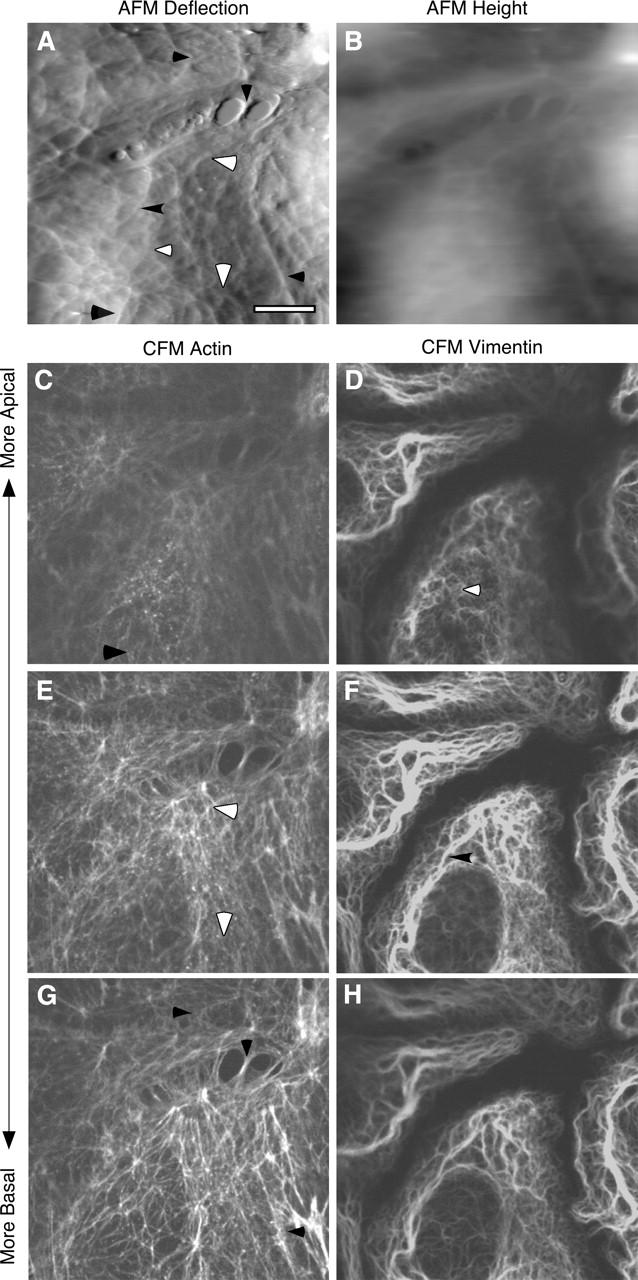
Correlated AFM and CFM of cells double stained for actin and vimentin. AFM image on living BPAECs was collected and the cells were immediately fixed and processed for immunofluorescence. (A) The deflection image shows details of the mesh-like cortical organization, whereas (B) the height image better reflects the overall cell shape. Confocal planes of cells stained for actin using Alexa-phalloidin (C, E, and G) and vimentin using Cy3-anti-vimentin antibody (D, F, and H). Correlated features are marked with arrowheads. Scale bar is 10 μm. Z-range in the height image is 0–4 μm.
To gain further insight into the molecular identity of the mechanical features seen in the AFM images we examined the effect of the actin disrupting drug cytochalasin B and microtubule disrupting drug nocodazole on the appearance of BPAECs. High concentration (50 μM) of cytochalasin B causes dramatic effects within 5 min: the filamentous network disappears, the cell surface becomes smooth, cells lose their mechanical strength, shape, and finally structural integrity (Fig. 7). During AFM imaging experiments we make sure that the probe applies the minimum force that gives good contrast. However, when cells lose their mechanical strength due to disruption of the cytoskeleton/mechanical elements, they cannot withstand even the small forces, and the probe ends up scraping cells from the surface. When a lower concentration (1 μM) of cytochalasin B is used, the cell maintains integrity for a significantly longer time. In this case the filamentous network disappears more gradually and the cells take on a somewhat granular appearance. In stark contrast to cytochalasin, treatment with nocodazole did not produce a loss of any features; instead there was an increase in the density of filaments (Fig. 8). Here it should be noted that nocodazole is known to promote actin polymerization (Ballestrem et al., 2000). These cells appear to reorganize the filamentous features with time, and the morphology of the cells appears to reflect an increase in tension in the plane of the monolayer. In conjunction with this apparent change in tension the cell monolayer also initially becomes smoother with smaller variations in height, although height variations are eventually restored (Fig. 9).
FIGURE 7.
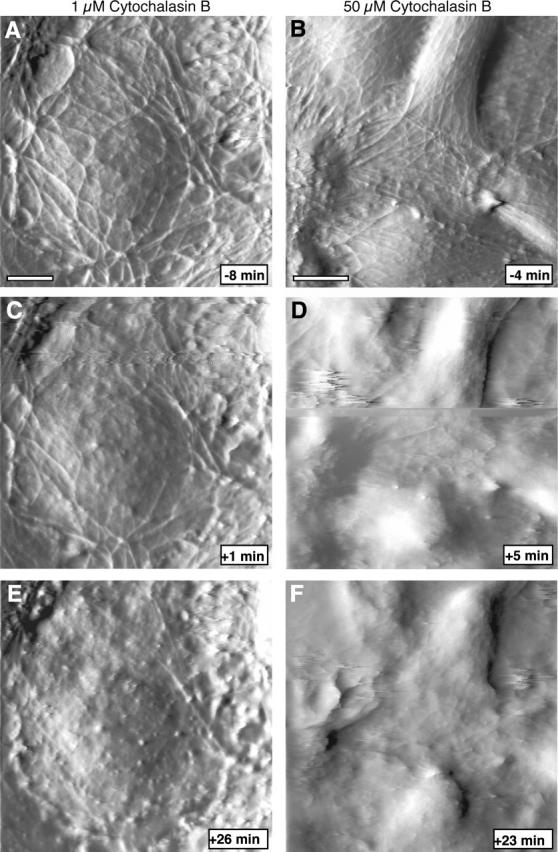
Treatment with cytochalasin B abolishes most filamentous features. Time series of AFM deflection images collected before and after exposure of cells to cyotochalasin. (A–C) Cells were treated with 1 μM cytochalasin B at time 0 min. Scale bar is 5 μm. (D–F) Cells were treated with 50 μM cytochalasin B at time 0 min. Scale bar is 10 μm. Low concentration of cytochalasin B results in gradual loss of filamentous structures, whereas high concentration abolishes all features within 5 min.
FIGURE 8.
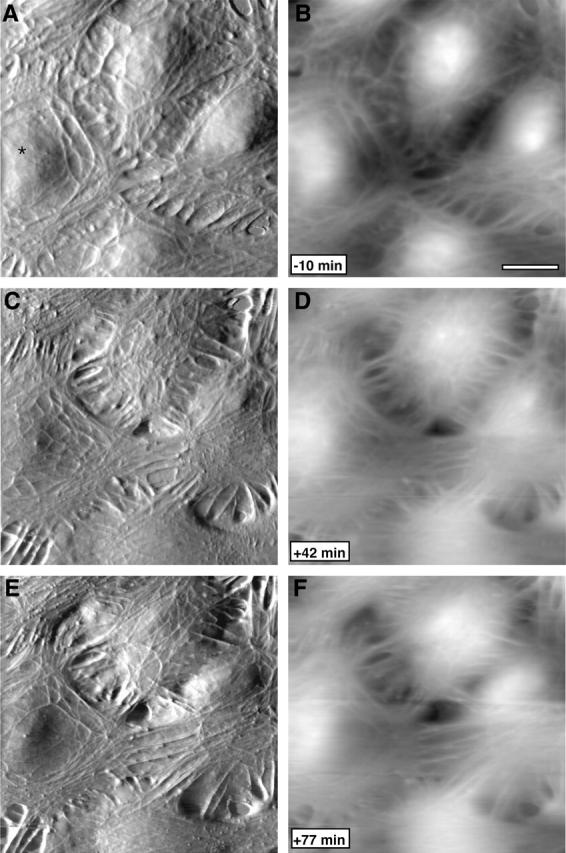
Nocodazole treatment of BPAECs produces an increase in filamentous structures. Time series of AFM (A–C) deflection images and (D–F) corresponding height images collected before and after exposure to nocodazole. Cells were treated with ∼50 μM nocodazole at time 0 min. There is an increase in the filamentous structures by 42 min, followed by a lateral reorganization. For example, the cell marked with an asterisk redistributes the filamentous features to the cell periphery by 77 min. Scale bar is 5 μm.
FIGURE 9.
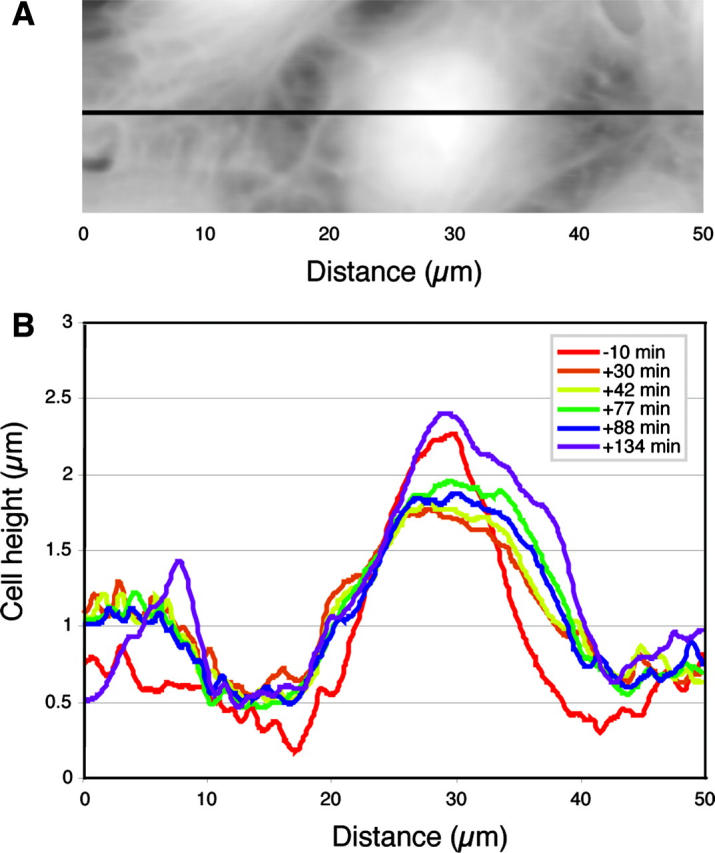
Changes in height of BPAEC due to nocodazole treatment. Cells were treated with ∼50 μM nocodazole at time 0 min. (A) AFM height image of BPAEC. The black line shows position of the cross section. (B) A cross-section profile is collected at the same position at different time points. Cell height decreases by 30 min and stays low for at least 1 h. Cells regain their height by 134 min.
DISCUSSION
A significant barrier to understanding the micromechanical and cortical architecture of living cells are the methods currently available. Mechanical measurements of living cells have largely been confined to measurements of whole cells or large regions of cells, using methods such as cell poking, micropipette aspiration, and stretching (Zahalak et al., 1990; Hochmuth, 2000; Basso and Heersche, 2002). Small area measurements of mechanics by magnetic twisting or optical tweezers are too sparse to provide a detailed mechanical picture (Wang and Ingber, 1994; MacKintosh and Schmidt, 1999). Spatially resolved AFM force measurements offer a more detailed mechanical map of living cells (Radmacher et al., 1996; Hofmann et al., 1997; A-Hassan et al., 1998; Vinckier and Semenza, 1998). However, thinking about micromechanical organization of cells is often based substantially on images of the cytoskeleton produced by immunofluorescence and EM, and the independent cell mechanics measurements described above. But it is clear that the organization of cytoskeleton per se does not provide a complete mechanical picture; one also needs to know where and how strongly different cytoskeletal components are connected, among other things. Thus, in the absence of a complete biochemical description of the system, there is a need for a high-resolution mechanical picture.
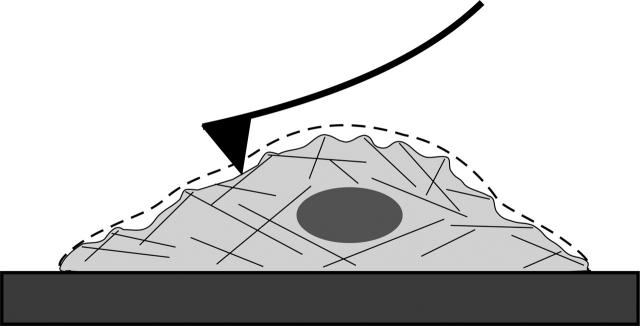
Here we have found that the AFM imaging offers an approach to directly examine micromechanical organization of the cell cortex at very high resolution. This work follows a number of other AFM studies of living cells (e.g., Henderson et al., 1992; Barbee et al., 1994; Hoh and Schoenenberger, 1994; Putman et al., 1994; Braet et al., 1997; Quist et al., 2000) but differs in that we conclude that for a certain class of features in BPAECs the contrast is almost entirely mechanical in origin (Fig. 10). Although there is no formal way of uncoupling topographic features from mechanical features in AFM images of living cells, evidence here is that virtually all fine structure seen in AFM images derives from differential mechanical properties. This conclusion is based on the mechanical nature of live cell AFM imaging and a comparison of scanning EM or AFM of fixed cells with live cell AFM images. The images of fixed cells show a smooth surface with no filamentous features, whereas the live cell AFM images show a highly complex mesh of filaments. This is consistent with an extensive body of scanning EM images of cells prepared using a variety of methods; we are not familiar with a single instance of scanning EM where such features are seen at the plasma membrane (from the extracellular side).
FIGURE 10.
Schematic illustrating contrast mechanism for micromechanical AFM imaging of cells. Contour of the cell before imaging is shown as a dashed line. During AFM imaging the tip moves along the apical surface of the cell and interacts with structures in the cortex by differential deformation. Soft structures such as the lipid membrane are easily deformed, whereas stiff structures such as cytoskeleton resist the applied force. As a result, the contrast derives in large part from local mechanical properties of the sample.
The view of the micromechanical architecture of BPAECs that results from the AFM imaging is far more detailed than apparent from confocal microscopy, although consistent with deep etch electron microscopy of similar cells that shows a complex network of cortical cytoskeleton. We conclude that the filamentous features in the AFM images represent the cytoskeleton based on three lines of reasoning. To begin with, the observed structures are highly reminiscent of cytoskeletal morphology. Further, the correlated AFM and immunofluorescence data demonstrate that some of the filaments are actin and vimentin. Finally, treatment with anticytoskeletal drugs produces corresponding changes in the AFM images. This interpretation is also consistent with previous AFM imaging of living cells, where similar features have been attributed to the cytoskeleton using immunological or pharmacological approaches (Henderson et al., 1992; Rotsch and Radmacher, 2000). This conclusion is also consistent with the generally accepted finding that cytoskeleton is a major determinant of cellular mechanics and has physical properties that can give rise to mechanical features seen in the images. Because the AFM is a surface imaging tool, only structures on the apical side of cells interact with the probe, and the imaging is limited to the cell cortex. Here we define the cortex as what is accessible to the AFM in a typical imaging experiment. The maximum indentation depth of ∼600 nm (at an imaging force of ∼1 nN) sets an upper limit for the amount of deformation. A lower limit for the layer thickness is estimated assuming that only the membrane, not the underlying cytoskeleton, deforms during imaging. In typical images this value is in the range 10–50 nm. For a variety of reasons the deformation is unlikely to be near either of these limits, and we estimate the thickness of this region to be a few hundred nanometers. Thus, our working hypothesis is that the contrast in our AFM images results from the variations in the micromechanical organization of the cell surface, which in turn reflects the organization of the CS of these vascular endothelial cells.
The correlated AFM-CFM and pharmacology experiments demonstrate that actin contributes to the micromechanical architecture in the BPAEC cortex. This is consistent with previous studies that have shown actin to be a significant contributor to the CS (Lazarides, 1975; Heuser and Kirschner, 1980; Condeelis, 1981; Bretscher, 1991). We were also able to identify features in the AFM images that correlate with vimentin staining but not tubulin. However, many features seen in the AFM images could not be assigned to any of the proteins for which we stained. Nonetheless, polygonal shape and branch angles suggest that many of the unidentified features are actin. Further, it is exceptionally difficult to visualize CS by CFM; on the other hand, the AFM provides very high contrast and suboptical resolution images of the cell cortex. Thus, it is likely that very thin cortical filaments of actin, vimentin, or microtubules are seen in the AFM but cannot be visualized by CFM. There are also other candidate molecules for which we have not yet stained, and there may also be unexpected molecules that contribute to cortical mechanics. Finally, the data collection may complicate the image correlation; in particular the time for fixation after AFM imaging allows for minor changes to cytoskeleton. Identification of all the features seen in the AFM images will require further work and possibly novel labeling approaches. The results from pharmacological treatments of BPAEC CS further support the finding that actin is a central mechanical component of the cortex in these cells. Cytochalasin B led to disruption of the filamentous network, whereas nocodazole resulted in an increase of filamentous structures. The latter result is attributed to the fact that nocodazole has been shown to increase the number of actin filaments (Ballestrem et al., 2000).
The organization of the cortical mesh reported here has interesting implications for the mechanical properties of cells. Both the intertwined mesh and layered mesh models present a unique mechanical picture of the cell. A cortex composed solely of a fine mesh would produce a cell that is very soft, whereas a coarse mesh would result in a more rigid cell, but leave large soft spots on the surface. Combining the two by intertwining a fine mesh with a coarse mesh, produces a mechanically stable structure on both long and short length scales. An intertwined mesh would allow for mechanically coupled responses to external forces acting on the cell, whereas layered meshes could in principle respond more independently. In terms of remodeling and the level of coordination in the CS, an intertwined mesh would suggest that cortical remodeling of the fine mesh and the coarse mesh is highly coordinated. Although the data do not exclude the layered mesh model, in our view it is unlikely that the two meshes are fully separated and we favor the intertwined mesh model.
A general concern with AFM imaging of living cells is that the imaging process in some way perturbs the cells. In our experiments, it is clear that extended imaging does cause the cells to respond; imaging for more than 1.5–2 h results in a significant enlargement of fenestrae and eventually causes the cells to detach. On shorter time scales we do not see any obvious effects that are tip induced. When we vary the time interval between images the rate of movement remains constant, suggesting that the cell is not responding to the repeated imaging. However, we can not exclude the possibility that the initial contact between tip and cell initiates some of the events described here, in particular since endothelial cells sense and respond to mechanical forces (Helmke and Davies, 2002; Ingber, 2002).
Mechanical properties of isolated cytoskeletal components have been studied in significant detail. Mechanics of whole living cells have also been studied, although generally with modest spatial resolution relative to dimensions of the cell. However, the connection between molecular mechanics and cellular mechanics, which depends on the micromechanical organization of the cell remains poorly understood. The results presented here reveal the micromechanical architecture for length scales on the order of 100 nm to 100 μm, which bridges the length scales of macromolecular assemblies with whole cells and small cell assemblies. Thus, the capability to visualize micromechanical architecture of the endothelial cortex at high resolution presents the opportunity to further connect molecular mechanics with cellular mechanics.
Acknowledgments
We thank Dr. Michael Edidin, Dr. Joseph Garcia, Dr. Susan Craig, and Dr. Douglas Robinson for helpful discussions. We also acknowledge Michael Delannoy and the Johns Hopkins School of Medicine Microscopy Facility for assistance with the scanning electron microscopy. We are also very grateful to Marlene Carlyle and Digital Instruments/Veeco for loan of the BioScope AFM used in the work described here.
This work was supported in part by a grant from the National Institutes of Health (HL076241).
References
- A-Hassan, E., W. F. Heinz, M. D. Antonik, N. P. D'Costa, S. Nageswaran, C. A. Schoenenberger, and J. H. Hoh. 1998. Relative microelastic mapping of living cells by atomic force microscopy. Biophys. J. 74:1564–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts, B., D. Bray, J. Lewis, M. Raff, K. Roberts, and J. D. Watson. 1994. Molecular Biology of the Cell. Garland, New York.
- Ballestrem, C., B. Wehrle-Harler, B. Hinz, and B. A. Imhof. 2000. Actin-dependent lamellipodia formation and microtubule-dependent tail retraction control-directed cell migration. Mol. Biol. Cell. 11:2999–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee, K. A., P. F. Davies, and R. Lal. 1994. Shear stress-induced reorganization of the surface topography of living endothelial cells imaged by atomic force microscopy. Circ. Res. 74:163–171. [DOI] [PubMed] [Google Scholar]
- Basso, N., and J. N. Heersche. 2002. Characteristics of in vitro osteoblastic cell loading models. Bone. 30:347–351. [DOI] [PubMed] [Google Scholar]
- Boal, D. 2002. Mechanics of the Cell. Cambridge University Press, Cambridge, UK.
- Braet, F., R. De Zanger, and E. Wisse. 1997. Drying cells for SEM, AFM and TEM by hexamethyldisilazane: a study on hepatic endothelial cells. J. Microsc. 186:84–87. [DOI] [PubMed] [Google Scholar]
- Bretscher, A. 1991. Microfilament structure and function in the cortical cytoskeleton. Annu. Rev. Cell Biol. 7:337–374. [DOI] [PubMed] [Google Scholar]
- Charras, G. T., P. P. Lehenkari, and M. A. Horton. 2001. Atomic force microscopy can be used to mechanically stimulate osteoblasts and evaluate cellular strain distributions. Ultramicroscopy. 86:85–95. [DOI] [PubMed] [Google Scholar]
- Chazov, E. I., A. V. Alexeev, A. S. Antonov, V. E. Koteliansky, V. L. Leytin, E. V. Lyubimova, V. S. Repin, D. D. Sviridov, V. P. Torchilin, and V. N. Smirnov. 1981. Endothelial cell culture on fibrillar collagen: model to study platelet adhesion and liposome targeting to intercellular collagen matrix. Proc. Natl. Acad. Sci. USA. 78:5603–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis, J. S. 1981. Reciprocal interactions between the actin lattice and cell membrane. Neurosci. Res. Program Bull. 19:83–99. [PubMed] [Google Scholar]
- Costa, K. D., and F. C. P. Yin. 1999. Analysis of indentation: implications for measuring mechanical properties with atomic force microscopy. J. Biomech. Eng. 121:462–471. [DOI] [PubMed] [Google Scholar]
- Dimitriadis, E. K., F. Horkay, J. Maresca, B. Kachar, and R. S. Chadwick. 2002. Determination of elastic moduli of thin layers of soft material using the atomic force microscope. Biophys. J. 82:2798–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, C., R. Skalak, and K. L. Sung. 1991. Cytoplasmic rheology of passive neutrophils. Biorheology. 28:557–567. [DOI] [PubMed] [Google Scholar]
- Dudek, S. M., and J. G. Garcia. 2001. Cytoskeletal regulation of pulmonary vascular permeability. J. Appl. Physiol. 91:1487–1500. [DOI] [PubMed] [Google Scholar]
- Elson, E. L. 1988. Cellular mechanics as an indicator of cytoskeletal structure and function. Annu. Rev. Biophys. Biophys. Chem. 17:397–430. [DOI] [PubMed] [Google Scholar]
- Evans, E. A. 1989. Structure and deformation properties of red blood cells: concepts and quantitative methods. Methods Enzymol. 173:3–35. [DOI] [PubMed] [Google Scholar]
- Flanagan, L. A., J. Chou, H. Falet, R. Neujahr, J. H. Hartwig, and T. P. Stossel. 2001. Filamin A, the Arp2/3 complex, and the morphology and function of cortical actin filaments in human melanoma cells. J. Cell Biol. 155:511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith, C. G., R. Skalak, and S. Chien. 1998. Shear stress induces spatial reorganization of the endothelial cell cytoskeleton. Cell Motil. Cytoskeleton. 40:317–330. [DOI] [PubMed] [Google Scholar]
- Gittes, F., B. Mickey, J. Nettleton, and J. Howard. 1993. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J. Cell Biol. 120:923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, J., R. Skalak, S. Chien, and A. Hoger. 1997. Spectrin properties and the elasticity of the red blood cell membrane skeleton. Biorheology. 34:327–348. [DOI] [PubMed] [Google Scholar]
- Hartwig, J. H., and P. Shevlin. 1986. The architecture of actin filaments and the ultrastructural location of actin-binding protein in the periphery of lung macrophages. J. Cell Biol. 103:1007–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidemann, S. R., P. Lamoureaux, and R. E. Buxbaum. 2000. Opposing views on tensegrity as a structural framework for understanding cell mechanics. J. Appl. Physiol. 89:1670–1678. [PubMed] [Google Scholar]
- Helmke, B. P., and P. F. Davies. 2002. The cytoskeleton under external fluid mechanical forces: hemodynamic forces acting on the endothelium. Ann. Biomed. Eng. 30:284–296. [DOI] [PubMed] [Google Scholar]
- Henderson, E., P. G. Haydon, and D. S. Sakaguchi. 1992. Actin filament dynamics in living glial cells imaged by atomic force microscopy. Science. 257:1944–1946. [DOI] [PubMed] [Google Scholar]
- Heuser, J. 2000. The production of ‘cell cortices’ for light and electron microscopy. Traffic. 1:545–552. [DOI] [PubMed] [Google Scholar]
- Heuser, J. E., and M. W. Kirschner. 1980. Filament organization revealed in platinum replicas of freeze-dried cytoskeletons. J. Cell Biol. 86:212–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochmuth, R. M. 2000. Micropipette aspiration of living cells. J. Biomech. 33:15–22. [DOI] [PubMed] [Google Scholar]
- Hofmann, U. G., C. Rotsch, W. J. Parak, and M. Radmacher. 1997. Investigating the cytoskeleton of chicken cardiocytes with the atomic force microscope. J. Struct. Biol. 119:84–91. [DOI] [PubMed] [Google Scholar]
- Hoh, J. H., and C. A. Schoenenberger. 1994. Surface morphology and mechanical properties of MDCK monolayers by atomic force microscopy. J. Cell Sci. 107:1105–1114. [DOI] [PubMed] [Google Scholar]
- Howard, J. 2001. Mechanics of Motor Proteins and the Cytoskeleton. Sinauer Press, Sunderland, MA.
- Ingber, D. E. 2002. Mechanical signaling and the cellular response to extracellular matrix in angiogenesis and cardiovascular physiology. Circ. Res. 91:877–887. [DOI] [PubMed] [Google Scholar]
- Ingber, D. E., S. R. Heidemann, P. Lamoureux, and R. E. Buxbaum. 2000. Opposing views on tensegrity as a structural framework for understanding cell mechanics. J. Appl. Physiol. 89:1663–1670. [DOI] [PubMed] [Google Scholar]
- Janmey, P. A., U. Euteneuer, P. Traub, and M. Schliwa. 1991. Viscoelastic properties of vimentin compared with other filamentous biopolymer networks. J. Cell Biol. 113:155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey, P. A., S. Hvidt, J. Kas, D. Lerche, A. Maggs, E. Sackmann, M. Schliwa, and T. P. Stossel. 1994. The mechanical-properties of actin gels - elastic-modulus and filament motions. J. Biol. Chem. 269:32503–32513. [PubMed] [Google Scholar]
- Janssen, K. P., L. Eichinger, P. A. Janmey, A. A. Noegel, M. Schliwa, W. Witke, and M. Schleicher. 1996. Viscoelastic properties of F-actin solutions in the presence of normal and mutated actin-binding proteins. Arch. Biochem. Biophys. 325:183–189. [DOI] [PubMed] [Google Scholar]
- Joy, D. C. 2002. SMART—a program to measure SEM resolution and imaging performance. J. Microsc. 208:24–34. [DOI] [PubMed] [Google Scholar]
- Karcher, H., J. Lammerding, H. Huang, R. T. Lee, R. D. Kamm, and M. R. Kaazempur-Mofrad. 2003. A three-dimensional viscoelastic model for cell deformation with experimental verification. Biophys. J. 85:3336–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, H., A. Ishijima, and T. Yanagida. 1994. Direct measurement of stiffness of single actin filaments with and without tropomyosin by in vitro nanomanipulation. Proc. Natl. Acad. Sci. USA. 91:12962–12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurachi, M., M. Hoshi, and H. Tashiro. 1995. Buckling of a single microtubule by optical trapping forces: direct measurement of microtubule rigidity. Cell Motil. Cytoskeleton. 30:221–228. [DOI] [PubMed] [Google Scholar]
- Lazarides, E. 1975. Immunofluorescence studies on the structure of actin filaments in tissue culture cells. J. Histochem. Cytochem. 23:507–528. [DOI] [PubMed] [Google Scholar]
- Lee, J. S., and A. I. Gotlieb. 2002. Microtubule-actin interactions may regulate endothelial integrity and repair. Cardiovasc. Pathol. 11:135–140. [DOI] [PubMed] [Google Scholar]
- MacKintosh, F. C., and C. F. Schmidt. 1999. Microrheology. Curr. Opin. Colloid Interface Sci. 4:300–307. [Google Scholar]
- Mathur, A. B., A. M. Collinsworth, W. M. Reichert, W. E. Kraus, and G. A. Truskey. 2001. Endothelial, cardiac muscle and skeletal muscle exhibit different viscous and elastic properties as determined by atomic force microscopy. J. Biomech. 34:1545–1553. [DOI] [PubMed] [Google Scholar]
- Mathur, A. B., G. A. Truskey, and W. M. Reichert. 2000. Atomic force and total internal reflection fluorescence microscopy for the study of force transmission in endothelial cells. Biophys. J. 78:1725–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunrinade, O., G. T. Kameya, and G. A. Truskey. 2002. Effect of fluid shear stress on the permeability of the arterial endothelium. Ann. Biomed. Eng. 30:430–446. [DOI] [PubMed] [Google Scholar]
- Pollard, T. D., L. Blanchoin, and R. D. Mullins. 2000. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29:545–576. [DOI] [PubMed] [Google Scholar]
- Pollard, T. D., and G. G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. [DOI] [PubMed] [Google Scholar]
- Putman, C. A. J., K. O. Vanderwerf, B. G. Degrooth, N. F. Vanhulst, and J. Greve. 1994. Viscoelasticity of living cells allows high-resolution imaging by tapping mode atomic-force microscopy. Biophys. J. 67:1749–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quist, A. P., S. K. Rhee, H. Lin, and R. Lal. 2000. Physiological role of gap-junctional hemichannels. Extracellular calcium-dependent isosmotic volume regulation. J. Cell Biol. 148:1063–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radmacher, M., M. Fritz, and P. K. Hansma. 1995. Imaging soft samples with the atomic force microscope: gelatin in water and propanol. Biophys. J. 69:264–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radmacher, M., M. Fritz, C. M. Kacher, J. P. Cleveland, and P. K. Hansma. 1996. Measuring the viscoelastic properties of human platelets with the atomic force microscope. Biophys. J. 70:556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotsch, C., and M. Radmacher. 2000. Drug-induced changes of cytoskeletal structure and mechanics in fibroblasts: an atomic force microscopy study. Biophys. J. 78:520–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satcher, R., C. F. Dewey, Jr., and J. H. Hartwig. 1997. Mechanical remodeling of the endothelial surface and actin cytoskeleton induced by fluid flow. Microcirculation. 4:439–453. [DOI] [PubMed] [Google Scholar]
- Satcher, R. L., Jr., and C. F. Dewey, Jr. 1996. Theoretical estimates of mechanical properties of the endothelial cell cytoskeleton. Biophys. J. 71:109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer, R. C., Jr., F. Gong, M. S. Bitrick, Jr., and T. L. Smith. 1993. Thrombin and bradykinin initiate discrete endothelial solute permeability mechanisms. Am. J. Physiol. 264:H1798–H1809. [DOI] [PubMed] [Google Scholar]
- Tsukita, S., and S. Yonemura. 1999. Cortical actin organization: lessons from ERM (ezrin/radixin/moesin) proteins. J. Biol. Chem. 274:34507–34510. [DOI] [PubMed] [Google Scholar]
- Vinckier, A., and G. Semenza. 1998. Measuring elasticity of biological materials by atomic force microscopy. FEBS Lett. 430:12–16. [DOI] [PubMed] [Google Scholar]
- Wakatsuki, T., B. Schwab, N. C. Thompson, and E. L. Elson. 2001. Effects of cytochalasin D and latrunculin B on mechanical properties of cells. J. Cell Sci. 114:1025–1036. [DOI] [PubMed] [Google Scholar]
- Wang, N., and D. E. Ingber. 1994. Control of cytoskeletal mechanics by extracellular matrix, cell shape, and mechanical tension. Biophys. J. 66:2181–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, I., J. Niewohner, and J. Faix. 1999. Cytoskeletal protein mutations and cell motility in Dictyostelium. Biochem. Soc. Symp. 65:245–265. [PubMed] [Google Scholar]
- Yamada, S., D. Wirtz, and S. C. Kuo. 2000. Mechanics of living cells measured by laser tracking microrheology. Biophys. J. 78:1736–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung, A., and E. Evans. 1989. Cortical shell-liquid core model for passive flow of liquid-like spherical cells into micropipets. Biophys. J. 56:139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahalak, G. I., W. B. McConnaughey, and E. L. Elson. 1990. Determination of cellular mechanical properties by cell poking, with an application to leukocytes. J. Biomech. Eng. 112:283–294. [DOI] [PubMed] [Google Scholar]