Abstract
G protein-coupled receptors (GPCRs) mediate our sense of vision, smell, taste, and pain. They are also involved in cell recognition and communication processes, and hence have emerged as a prominent superfamily for drug targets. Unfortunately, the atomic-level structure is available for only one GPCR (bovine rhodopsin), making it difficult to use structure-based methods to design drugs and mutation experiments. We have recently developed first principles methods (MembStruk and HierDock) for predicting structure of GPCRs, and for predicting the ligand binding sites and relative binding affinities. Comparing to the one case with structural data, bovine rhodopsin, we find good accuracy in both the structure of the protein and of the bound ligand. We report here the application of MembStruk and HierDock to β1-adrenergic receptor, endothelial differential gene 6, mouse and rat I7 olfactory receptors, and human sweet receptor. We find that the predicted structure of β1-adrenergic receptor leads to a binding site for epinephrine that agrees well with the mutation experiments. Similarly the predicted binding sites and affinities for endothelial differential gene 6, mouse and rat I7 olfactory receptors, and human sweet receptor are consistent with the available experimental data. These predicted structures and binding sites allow the design of mutation experiments to validate and improve the structure and function prediction methods. As these structures are validated they can be used as targets for the design of new receptor-selective antagonists or agonists for GPCRs.
Keywords: GPCR‖olfactory receptor‖β-adrenergic receptor‖endothelial differentiation gene‖taste receptor
G protein-coupled receptors (GPCRs) mediate senses such as odor, taste, vision, and pain (1) in mammals. In addition, important cell recognition and communication processes often involve GPCRs. Indeed, many diseases involve malfunction of these receptors (2), making them important targets for drug development. Unfortunately, despite their importance there is insufficient structural information on GPCRs for structure-based drug design. This is because these membrane-bound proteins are difficult to crystallize, and the atomic-level structure has been solved only for bovine rhodopsin (3, 4). Consequently, it is important to develop theoretical methods to predict the structure and function of GPCRs (5, 6).
Experimental data relevant to the function of GPCRs is available for ligand activation of GPCRs (7–15) and site-directed mutagenesis (16–18). This data has led to information about structural features in the ligand-binding regions of GPCRs (refs. 5 and 19, and references therein). Protein sequence analyses on GPCRs reveals a common protein topology consisting of a membrane-spanning seven-helix bundle, which likely accommodates the binding site for low-molecular-weight ligands. Structurally, GPCRs can be classified as (i) GPCRs with short N terminus (5–80 residues) and (ii) GPCRs with a long N-terminal ectodomain (≈80–600 residues). The long N terminus of class II GPCRs may be involved in the ligand recognition (8), but ultimately the bound ligand probably moves into the transmembrane (TM) region to activate the G protein.
To provide structural and ligand-binding information on GPCRs, we have been developing computational strategies and techniques for predicting:
the tertiary (three-dimensional) structure of GPCRs by using only the amino acid sequence (MembStruk), and
binding site and binding energy of various ligands to GPCRs (HierDock).
The first report on these developments (20, 21) focused on olfactory receptors (ORs) because ligand-binding data were available for 24 simple organic molecules to 14 different ORs (14). In this article we report the structure and function prediction of GPCRs for four classes for which there is no three-dimensional structure:
β1-adrenergic receptor (β1AR),
endothelial differentiation gene (EDG6),
rat and mouse I7 ORs, and
human sweet receptor.
In addition we validate our techniques by predicting the three-dimensional structure of bacteriorhodopsin and bovine rhodopsin for which crystal structure data are available. The function prediction technique is validated for retinal bound to bovine rhodopsin.
The predicted ligand binding sites and the ordering of binding energies for the other GPCRs calculated here are consistent with the experimental data on ligand activation measurements and also site-directed mutation studies. Thus, the predicted binding site of cis-retinal when using just the protein crystal structure of bovine rhodopsin leads to a rms deviation in coordinates (CRMS) error of 0.6 Å from the crystal structure of the retinal/rhodopsin complex. The system with the most complete site-directed mutagenesis experiments to characterize the active site is epinephrine/β-adrenergic receptor (βAR). We find that our predicted binding site is in excellent agreement with the conclusions from experiment.
Thus, the predicted structures should be useful in
designing mutagenesis experiments to validate the structure of the binding site,
predicting the natural ligands binding to specific GPCRs,
designing specific drugs for GPCRs, and
predicting mutations to make the GPCRs specific for new ligands.
The next section summarizes various details of the MembStruk and HierDock methods. In addition, we validate these methods by comparing to experimental data for rhodopsin and bacteriorhodopsin. This is followed by results and discussion for the five GPCRs and by conclusions in the last section.
Computational Methods
Force Fields.
For the protein, we have used the DREIDING FF (22) with charges from CHARMM22 (23) or charge equilibration (QEq) method (24). For the ligands we used the DREIDING FF with Gasteiger charges (25). For the lipids we used the DREIDING FF with QEq charges. The calculations treated the solvent (water) by using the continuum solvation methods such as Surface Generalized Born (SGB) (26) or the Analytical Volume Generalized Born (AVGB) (27) methods.
The MembStruk Protocol for Predicting Structures of GPCRs.
The MembStruk protocol for predicting structures of TM proteins consists of the following steps:
- 1.
Prediction of TM regions.
- 2.
Construction and optimization of individual helices.
- 3.
Assembly of the seven-helical TM bundle.
- 4.
Coarse grain optimization of the TM bundle.
- 5.
Addition of interhelical loops and optimization of the full structure.
Step 1.
We developed the TM2NDS program that determines TM regions in GPCRs using hydropathicity analysis (28, 29) calculated using the Eisenberg hydrophobicity scale (29), combined with input from multisequence profiles. The multisequence alignment profile is obtained from the sequence analysis for a particular family of GPCRs. For example, the sequence analysis for ORs used 23 rat and mouse ORs reported by Singer et al. (30). TM2NDS analyzes the hydrophobicity profile of all of the sequences used in the alignment and assigns the TM regions by using capping rules for helices.
Step 2.
The canonical right-handed α-helices were then built with extended side chains and optimized using the NEIMO torsional molecular dynamics (MD) method (31–33), with fixed bonds and angles. This allows for sequence-specific distortions in the helix, such as for proline, and also the optimization of the side chain conformations.
Step 3.
Each helical axis was oriented according to the 7.5-Å electron density map of bovine rhodopsin (34). The hydrophobic moments of the NEIMO-optimized helical bundle were aligned so that the net hydrophobic moment of each helix would be pointing outward toward the membrane from the center of mass.
Step 4.
We have developed a dynamics program, coarserot, that performs coarse-grain rotation of the helical orientations, starting with the directions of the net hydrophobic moment of each helix from Step 3. Each helix is rotated through a grid of rotation angles about its helical axis. The total energy of this helix in the field of all of the other helices (fixed) is minimized using conjugate gradients. After finding the optimum configuration for each specific helix, we then go through a second cycle (seven such optimizations) and continue until the energy converges. We then add two layers of explicit lipid molecules (52 molecules of dilauroylphosphatidyl choline lipid) that were optimized with the current configuration of the seven helices. Then, to achieve proper packing of the TM helices, the seven-helix-bilayer complex is further optimized with rigid-body MD of the seven helices and lipid for 100 ps.
Step 5.
Following the rigid-body dynamics, loops were added to the helices by using whatif software (35). We then identified the possible disulfide linkages among the conserved cysteines across all GPCRs (36, 37) and added disulfide bonds where there are close in the predicted structure. It is plausible to consider the disulfide linkages among conserved cysteines in the TM region earlier in the protocol, but the disulfide linkages among the loops are added at this step to constrain the loop conformation. After the addition of loops and disulfide linkages, we used SCWRL (38) to add the side chains for all of the residues and then performed a full-atom MD optimization of the structure by using MPSim (39), with the explicit lipids.
Structure Validation.
Structure prediction for bacteriorhodopsin.
MembStruk was first tested on bacteriorhodopsin, which is not a GPCR but a seven-helical transmembrane protein for which crystal structures have been solved at various activated states and mutants with resolutions varying from 3.5 Å (40) to 1.55 Å (41).
We started from the sequence of bacteriorhodopsin and predicted the structure by using MembStruk without any information from the crystal structure studies. We used diphosphatidyl glycerophosphate (the lipid present in the purple membrane from Halobacterium halobium) bilayers to describe the membrane. The CRMS of Cα atoms in the predicted structure for the residues in the TM region is 3.3 Å and 2.9 Å compared with the crystal structures (2BRD and 1C3W) with resolutions of 3.5 Å and 1.55 Å, respectively. Including the loops, the overall CRMS for all 221 amino acids is 8.6 Å and 6.2 Å. Of course, the loop regions are much less well defined in the crystal structure studies and may be affected by packing forces in the crystal.
Structure prediction for bovine rhodopsin.
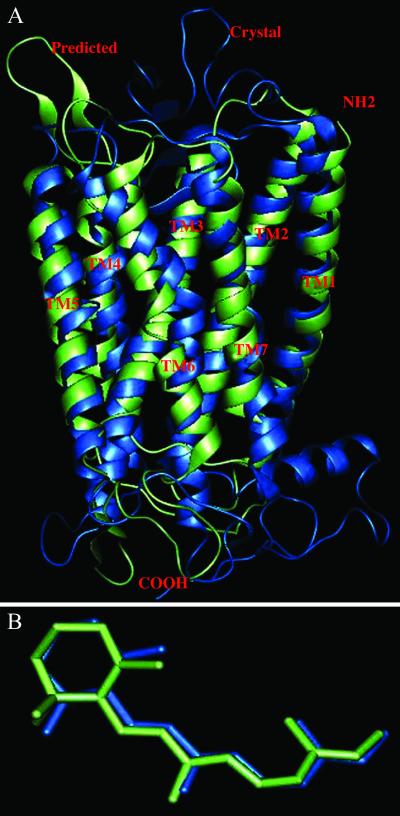
The only GPCR for which the crystal structure is available is bovine rhodopsin (3, 4), with a resolution of 2.8 Å. To predict this structure with MembStruk, we carried out a multiple sequence alignment using clustalw (42), considering the sequence of rhodopsin plus eighty-five other sequences, having sequence identity between 99% and 40%, picked from a blast search (ref. 43; this computation was performed at the Swiss Institute of Bioinformatics by using the blast network service). The assigned TM regions differ by an average of two residues from the crystal structure. This alignment implied a disulfide linkage between residues Cys-110 and Cys-187. We then used MembStruk to predict structure for rhodopsin. This differs by CRMS = 3.1 Å from the Cα atoms of the crystal (Fig. 1A). The individual helices differ from the crystal structures by CRMS of 1.0 Å for helix 1, 2.1 Å for helix 2, 1.2 Å for helix 3, 1.1 Å for helix 4, 1.8 Å for helix 5, 2.2 Å for helix 6, and 1.6 Å for helix 7. The crystal structure is missing 10 residues in loop regions, and 13 residues are missing their side-chain atoms. Of course, the calculated structure has all atoms of all residues. Thus, to compare with experiment we ignore the missing residues and atoms. The remaining residues (including loops) of the predicted structure differ from the crystal structure by 8.3 Å CRMS. The major contribution to this CRMS is the low-resolution loop region. We will next consider binding of the ligand to this predicted structure to determine whether 3 Å accuracy in the TM region is adequate for predicting the ligand binding site.
Figure 1.
(A) Comparison of predicted structure for bovine rhodopsin (green) with the x-ray crystal structure (blue). The TM regions have a CRMS of 3.1 Å. (B) Comparison of the HierDock predicted structure of cis-retinal/Rhodopsin to the crystal structure.
Function Prediction for GPCRs.
With the exception of rhodopsin, our only test on the validity of the predicted GPCR structures will be to compare with experimental ligand activation data. Thus, it is essential that we have a reliable and efficient procedure for predicting binding-site and relative-binding affinities of ligands in GPCRs. Because the ligand binding site is not known in many GPCRs, the entire protein should be scanned to identify likely binding sites and then the relative binding energies of the ligands calculated in these sites. For this purpose we use the HierDock protocol (20, 44), which has been applied successfully to both globular and membrane proteins (20, 45, 46).
HierDock Protocol.
The HierDock ligand screening protocol (44) follows a hierarchical strategy for examining ligand binding conformations and calculating their binding energies. This involves the use of coarse-grain docking methods to quickly scan the entire GPCR to locate the most plausible protein/ligand complexes, followed by molecular mechanics/dynamics (MM/MD) simulations (including continuum solvation) of these good structures by using more accurate scoring functions. The steps in HierDock are as follows.
Coarse-grain docking—level 1.
First we carried out a coarse-grain docking procedure [currently using DOCK 4.0 (47)] in which a number of ligand conformations are sampled in the void regions within the receptor. The void regions described using spheres generated over the whole receptor (using the sphgen program in DOCK4.0). No assumptions were made on the nature or the location of the binding site in these receptors. We then generated and scored (using the DOCK scoring function) 1,000 configurations for each box, keeping the 10% (100) of the best scoring structures.
Fine-grain optimization—level 2.
We then minimized the energy of the ligand (with protein fixed) for the 100 structures from level 1 by using the Dreiding FF and QEq charges. These 100 minimized structures are ranked by using both energy and solvation.
All-atom optimization—level 3.
Ten percent of the best structures from level 1 are further minimized with all atoms both protein and ligand movable. Each structure was scored using the binding energies: BE = Energy (free ligand) + Energy (free protein) − Energy (ligand–protein complex), where the system is solvated for each case by using AVGB solvation method.
Scanning the Entire Receptor for Binding Sites.
We used the molecular surface of the protein to define potential binding regions within the receptor. We then partitioned the void region in the entire receptor into 10–15 overlapping docking boxes, each with a volume of (10 Å)3. We excluded regions in contact with the membrane or near the intracellular region likely to be involved in binding to the G protein. Steps 1 and 2 of the HierDock protocol were applied first to a ligand known to activate the GPCR, scanning the entire GPCR structure to locate the most favorable ligand binding site(s). We then defined the “binding region” as the protein site where most of the best scoring ligands clustered. If there is more than one site with clusters of good ligand binding energies, we treated them equally as potential binding regions. Then we defined the binding region as a cube having 10 Å on each side centered at this site(s).
Docking of the Library of Ligands in the Binding Region.
We then docked the entire library of potential ligands for the receptor to this binding region and calculated their relative binding energies (steps 2 and 3 of HierDock). The ligands were then ranked by binding energy. This procedure assumes that the same binding site is used for all ligands that bind to the receptor. An ambiguity here is that we cannot be sure that binding to this site will lead to G-protein release, which is usually the basis of the experimental measurements (rather than binding affinity).
Validation for Function Prediction Protocol for cis-Retinal/Bovine Rhodopsin.
To validate the HierDock protocol for GPCRs, we predicted the binding of cis-retinal to the crystal minimized structure of bovine rhodopsin. First we extracted the bovine rhodopsin structure plus bound rhodopsin from the 2.8-Å-resolution crystal structure and added all of the missing residues in the loops and also the missing side-chain atoms by using PolyGraf. Hydrogens and counterions Na+ and Cl− were added energy of the protein–ligand complex was minimized. We refer to this structure as “crystal minimized.” Then cis-retinal was removed from this “crystal minimized” rhodopsin protein structure for docking. The cis-retinal used for docking was built and minimized with the DREIDING FF and Gasteiger charges with the AVGB continuum solvation method for the free ligand. Although this ligand is covalently bound to Lys-296 in the crystal structure of rhodopsin, for docking we used the full cis-retinal ligand (replacing the C—N bond in the crystal with CHO and adding an H to Lys-296). The rhodopsin from the “crystal minimized” structure was partitioned into 13 overlapping regions for step 1 of HierDock. The final optimized best binding structure for the retinal/rhodopsin complex from step 3 of HierDock is compared with the crystal structure in Fig. 1B. The docked cis-retinal/rhodopsin leads to a CRMS difference of 1.2 Å with the crystal structure. The docked structure has a distance of 2.8 Å between the C atom of —CHO group of retinal C and the side-chain N of Lys-296 (“CN bond,” to which it should bond covalently after eliminating H2O to form the Schiff base). We then made this covalent CN bond to Lys-296 and reminimized the ligand–protein structure. This leads to CRMS difference of 0.62 Å between the cis-retinal of the docked and crystals structures. We consider that these results validate the HierDock protocol. This test is a blind prediction of the binding site of cis-retinal without using any experimental information on the binding site of cis-retinal in bovine rhodopsin.
For the GPCRs considered in this paper, we do not have an experimental protein structure, and it is important to know how well HierDock can predict ligand binding by using the predicted structure from MembStruk. Thus, we applied HierDock (steps 1–3) to determine the binding site of cis-retinal in the predicted structure of rhodopsin. Without covalent attachment of the ligand to Lys-296, the final optimized structure of cis-retinal is 2.8 Å CRMS from the crystal structure with the CN bond being 8.1 Å. A second criteria for validity in the predicted binding site is identifying the residues close to the ligand. We find that residues G114, A117, T118, G121, E122, L125, H211, Y268, A292, and A295 are within 5 Å of both the ligand binding sites. Thus, we conclude that the MembStruk predicted structure can be used for predicting binding sites sufficiently well to direct mutation studies.
Results and Discussion
In this section we present the structure and function prediction using MembStruk and HierDock for β1AR, EDG6, mammalian I7 ORs, and human sweet receptor.
β1 Adrenergic Receptor.
The βAR family plays an extremely important role in mediating the sympathetic nervous response. All members of the βAR subfamily respond to similar native catecholamine ligands (epinephrine and norepinephrine). Genes exist in most animals for nine different subclasses of three α1 receptors (α1A, α1B, α1D), three α2 receptors (α2A, α2B, α2C), and three β receptors (β1, β2, β3) expressed in different tissues and leading to different responses. β1AR is expressed primarily in cardiac tissue, where it regulates blood pressure and heart rate in responses to stress. A major problem in designing drugs for the βAR family is the cross interaction of drugs among these subtypes. For example, β-blockers meant to act on only β1AR, also activate other subtypes of βAR. Hence, to design subtype receptor-specific drugs it is essential to have the structure of each of the subtypes of βAR.
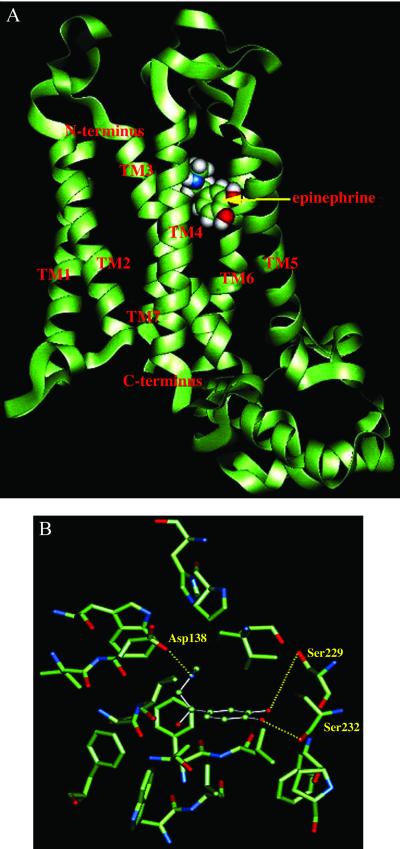
Deletion mutagenesis and proteolytic cleavage studies of the βAR show that most of the connecting hydrophilic regions can be deleted without affecting ligand binding properties, suggesting that the ligand binding site is in the barrel of the TM region (48). Mutation of Asp-138 reduces the binding to both agonists and antagonists (16, 17), suggesting that this residue is involved in binding and that agonists and antagonists might have similar binding sites. Mutation experiments show clearly that the two ortho catechol —OH groups make hydrogen bond contacts with Ser-229 and Ser-232 (5). The amine group of epinephrine makes a hydrogen bond with Asp-138 (16). Our structure also predicts that Asp-138 forms a hydrogen bond with the OH group of the alkyl chain in epinephrine.
We used MembStruk to predict the structure of β1AR. For the TM region predictions, the sequences from β1 to β4 subtypes were aligned. The third intracellular loop of β1AR is 74 residues long. Scanning the entire β1AR receptor led to three possible binding regions. We then minimized the structure of the receptor with epinephrine for each of these three sites and carried out a second HierDock calculation by using the optimized sites. One of the three regions (shown in Fig. 2A) emerged clearly as the best binding site for epinephrine. We find that TMs 3, 5, and 6 are involved in the binding of epinephrine (as suggested experimentally). Fig. 2B shows the residues involved in the binding of epinephrine. Indeed, we find hydrogen bond contacts of epinephrine with residues Asp-138, Ser-229, and Ser-232. In addition, we find a hydrophobic interaction with Phe-341 (seen above the catechol ring). These four residues were all identified experimentally as necessary for ligand binding to the βAR (12, 13, 15). We find that Ser-229 and Ser-232 form hydrogen bonds with meta and para OH groups of the catechol ring, respectively, just as found experimentally.
Figure 2.
(A) Predicted binding site of epinephrine in the predicted structure of β1-adrenergic receptor. (B) Residues within 5 Å of epinephrine bound to β1-adrenergic receptor. Shown in bold are the three residues Asp-138, Ser-229, and Ser-232 (deduced from mutation experiments to be involved in binding).
In summary, the binding conformation predicted with HierDock matches precisely the results from all experimental studies. This validates both the MembStruk and HierDock protocols, suggesting that accuracy of ≈3 Å for structure prediction in the TM regions is adequate to identify binding site and structure.
EDG6 Receptor.
The EDG receptor subfamily of GPCRs is implicated in diverse biological processes such as cell proliferation, differentiation, and migration, making it important for clinical applications. Based on sequence homology the EDG receptor family is partitioned into two major subgroups. The s1p subgroup (comprised of EDG1, EDG3, EDG5, EDG6, and EDG8) responds to sphingosine-1-phosphate (s1p), whereas the lysophosphatidic acid (lpa) subgroup (comprised of EDG2, EDG4, and EDG7) responds to lpa. For EDG6 we consider the ligands s1p as a positive and lpa as a negative (12) ligand.
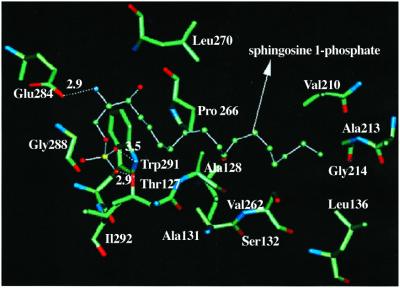
Using MembStruk, we predicted the structure of EDG6 receptor and then applied HierDock scanning on EDG6 with s1p to determine its binding site. This leads to the structure in Fig. 3, where the s1p binding site lies between TM3 and TM7. Fig. 3 shows that residues W291, E284, and T127 are important in recognizing the s1p ligand and shows other residues within 3 Å of the ligand. We propose that mutation of residues W291, E284, and T127 will affect the binding of s1p to EDG6. The calculated binding energy for s1p is 7.6 kcal/mol more favorable than for lpa, which is consistent with the experimental data on the radiolabeled binding assay measurements for s1p and lpa to the EDG6 receptor (12).
Figure 3.
Residues within 5 Å of the sphingosine-1-phosphate in the predicted structure of EDG6.
Mammalian ORs.
ORs form one of the largest gene families in the genome, with ≈1,000 different proteins believed to interact with a range of odorant molecules. Unlike βAR and EDG receptors, each OR is broadly tuned to recognize many odorants. Each odorant elicits a response from a combination of ORs (14) so that olfactory perception depends on a complex set of ligand-recognition interactions.
Predicted Binding Site of Aldehydes for Mouse I7 and Rat I7 OR.
The mouse and rat I7 ORs differ by only 15 residues, four of which are in the TM domains. But their odorant binding profile is thought to be different. Thus, Krautwurst et al. reported (11) that rat I7 recognizes n-octanal and n-heptanal, whereas mouse I7 recognizes only n-heptanal. On the other hand, Bozza et al. recently reported (7) that both mouse and rat I7 are activated by both n-octanal and n-heptanal. Using MembStruk, we predicted independent structures for both rat and mouse I7. The CRMS difference between all atoms in the TM region is 1.7 Å. This is consistent with the 95.4% homology between the two sequences. The calculated binding energies show a difference in binding energy of 0.2 kcal/mol between octanal and heptanal in rat I7 and a difference of 0.3 kcal/mol in mouse I7. Hence, we find that there is little or no preference for n-octanal over n-heptanal for these receptors, as predicted by the model of Singer (30) and in agreement with the experiments of Bozza et al. Of course, these minor differences might lead in a difference in the activation of G protein second-messenger pathway (the process observed experimentally).
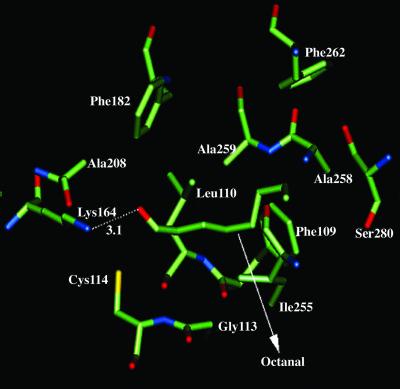
Fig. 4 shows the predicted binding site for n-octanal in rat I7 (very similar to that for heptanal). TM helices 3, 4, and 6 form the binding cavity. As indicated by the dotted line in Fig. 4, Lys-164 forms a hydrogen bond and is critical in recognizing the aldehyde functional group for both octanal and heptanal, in agreement with previous modeling of rat I7 by Singer et al. (30). Mutation of this residue should switch receptor specificity toward other functional groups. Other residues important in binding are Phe-109, Cys-114, Cys-117, and Ile-255, which we suggest for mutational studies. We find that none of the sequence differences between rat I7 and mouse I7 are located near the binding pocket, supporting the similarity of binding profiles found by Bozza et al. (7).
Figure 4.
Binding site of octanal in rat I7 OR.
Human Sweet Receptor.
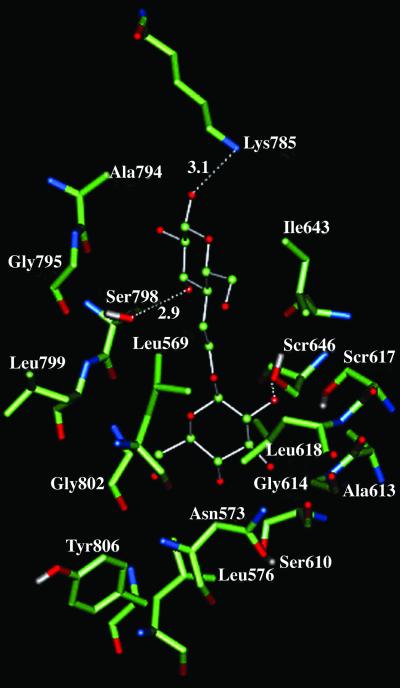
The sweet and bitter receptors have been identified (9, 13, 49, 50) to be GPCRs, with fewer sweet receptors than bitter receptors. J. Liao and P. G. Schultz (private communication) recently identified the sequence for human sweet receptor. This sequence has an extracellular N terminus of ≈600 residues. Using MembStruk, we predicted the structure of this receptor without the N terminus. Using HierDock, we calculated the binding energies for a library of 65 tastants (J. Liao and P. G. Schultz, private communication), including a variety of sugars, amino acids, artificial sweeteners, bitter tastants, and other tastants. Fig. 5 shows the predicted binding site of trehalose. It involves residues in TM domains 1, 2, 3, and 7 plus Lys-785 [located in extracellular loop (EC) 3]. We found all 12 sugars to be anchored to Ser-646 and Ser-798 through hydrogen bonds, suggesting them for mutation studies. the top 15 tastants selected using the dual criteria of strong binding and contacts to Ser-646 and Ser-798 are sweet molecules, supporting our predictions. A table for these results can be viewed at www.wag.caltech.edu/GPCR. It is possible that the N terminus (not considered here) also plays a role in recognition sugars to be exposed to the TM regions.
Figure 5.
Binding site of trehalose in human sweet receptor. This shows residues within 3 Å of trehalose and includes the hydrogen bonds to Ser-798, Ser-646, and Lys-785.
Comparison of Binding-Site Location in Various GPCRs.
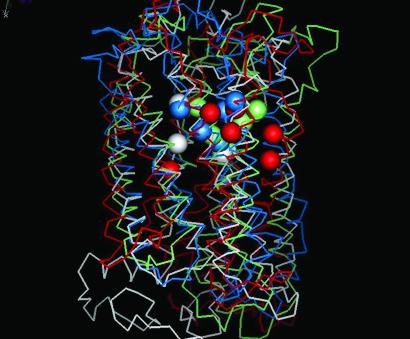
Fig. 6 shows the location of the important residues involved in ligand binding for four of the five GPCRs studied here. We find that TM 2, 3, 4, 5, and 7 are involved in the recognition of ligand. The spheres in white show important residues for binding of cis-retinal in bovine rhodopsin, whereas those in red shows the residues found in the binding region for β-adrenergic receptors. The other spheres show residues involved in binding to the other GPCRs. We find that the residues involved in binding are in similar spatial location for most of the GPCRs and agree well with known experimental results. The strictly conserved D(E)RY sequence in all these GPCRs is present in the intracellular loop 2 (IC2) connecting TM3 and TM4.
Figure 6.
Comparison of the predicted binding sites for GPCRs: white, bovine rhodopsin; green, rat I7 OR; blue, mouse I7 OR; red, β1AR.
The function of GPCRs is to couple ligand binding in the extracellular region to G-protein activation in the intracellular region. Based on these results we propose the following model for the initiation of signaling. After the ligand is bound to the GPCR, the extracellular loop 2 may close down over the barrel, perhaps by recognizing the exposed side of the ligand or of a part of the TM region that responds to the binding of ligand. The crystal structure of bovine rhodopsin shows just such a closed loop. The dramatic movement of EC2 in response to ligand binding may cause helix 3 to translate in the cytoplasmic direction, exposing the D(E)RY sequence to the cytoplasmic region near the G protein. This might initiate the signal transduction pathway. We hope to use our predicted structures in dynamical studies to test such ideas.
Summary
We predict the structure and function for four classes of GPCRs, β1AR, EDG6, human sweet receptor, and mouse/rat I7 olfactory receptors, for which there are no known experimental structures.
We validated our predicted protein structures by comparing to the crystal structure for bovine rhodopsin, with an accuracy of ≈3.0 Å for the TM regions. HierDock prediction of the binding site of cis-retinal in bovine rhodopsin gives an accuracy of 0.6 Å for the crystal rhodopsin structure and 2.8 Å for the predicted rhodopsin structure.
The binding site for epinephrine in β1AR is in excellent agreement with mutation experiments. The structure and function prediction for all GPCRs are in good agreement with the experimental ligand activation data currently available. These structures suggest additional site-directed mutagenesis studies to test the predicted structure and function of GPCR.
Our GPCR structures can also be used to predict new ligands that would bind to specific GPCRs, providing additional tests of our predicted structures. Such experiments will be useful to further refine the predicted structure and function. Our predicted GPCR structures should also be useful for predicting the function of orphan GPCRs.
Acknowledgments
This research was initiated with support from Army Research Office–Multidisciplinary University Research Initiative (MURI) (Grant DAAG55-98-1-0266) and continued with National Institutes of Health support (Grants R01-GM62253-01, R01-AI40567, and R01-CA85779). We particularly want to thank IBM for a Shared University Research grant that provided the computational facilities that made this work possible. The facilities of Materials and Process Simulation Center are also supported by grants from the Department of Energy–Accelerated Strategic Compliance Initiative, Army Research Office/Defense University Research Instrumentation Program, Army Research Office/MURI, the National Institutes of Health, the National Science Foundation, Chevron–Texaco, General Motors, 3M, Avery–Dennison, Seiko–Epson, Kellogg's, Beckman Institute, and Asahi Kasei.
Abbreviations
- βAR
β-adrenergic receptor
- β1AR
β1-adrenergic receptor
- CRMS
rms deviation in coordinates
- EDG
endothelial differentiation gene
- GPCR
G protein-coupled receptor
- OR
olfactory receptor
- TM
transmembrane
References
- 1.Dong X, Han S K, Zylka M J, Simon M I, Anderson D J. Cell. 2001;106:619–632. doi: 10.1016/s0092-8674(01)00483-4. [DOI] [PubMed] [Google Scholar]
- 2.Wilson S, Bergsma D. Pharmaceutical News. 2000;7:3–16. [Google Scholar]
- 3.Palczewski K, Kumasaka T, Hori T, Behnke C A, Motoshima H, Fox B A, Le Trong I, Teller D C, Okada T, Stenkamp R E, et al. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 4.Teller D C, Okada T, Behnke C A, Palczewski K, Stenkamp R E. Biochemistry. 2001;40:7761–7772. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strader C D, Fong T M, Tota M R, Underwood D, Dixon R A F. Annu Rev Biochem. 1994;63:101–132. doi: 10.1146/annurev.bi.63.070194.000533. [DOI] [PubMed] [Google Scholar]
- 6.Shacham S, Topf M, Avisar N, Glaser F, Marantz Y, Bar-Haim S, Noiman S, Naor Z, Becker O M. Med Res Rev. 2001;21:472–483. doi: 10.1002/med.1019. [DOI] [PubMed] [Google Scholar]
- 7.Bozza T, Feinstein P, Zheng C, Mombaerts P. J Neurosci. 2002;22:3033–3043. doi: 10.1523/JNEUROSCI.22-08-03033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Max M, Gopishanker Y, Huang L, Rong M, Liu Z, Campagne F, Weinstein H, Damak S, Margolskee R F. Nat Genet. 2001;28:58–63. doi: 10.1038/ng0501-58. [DOI] [PubMed] [Google Scholar]
- 9.Nelson G, Hoon M A, Chandrasekhar J, Zhang Y, Ryba N J P, Zuker C S. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- 10.Buck L B. Cell. 2000;100:611–618. doi: 10.1016/s0092-8674(00)80698-4. [DOI] [PubMed] [Google Scholar]
- 11.Krautwurst D, Yau K W, Reed R R. Cell. 1998;95:917–926. doi: 10.1016/s0092-8674(00)81716-x. [DOI] [PubMed] [Google Scholar]
- 12.Van Brocklyn J R, Graler M H, Bernhardt G, Hobson J P, Lipp M, Spiegel S. Blood. 2000;95:2624–2629. [PubMed] [Google Scholar]
- 13.Adler E, Hoon M A, Mueller K L, Chandrasekhar J, Ryba N J P, Zuker C S. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- 14.Malnic B, Hirono J, Sato T, Buck L B. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 15.Mombaerts P. Science. 1999;286:707–711. doi: 10.1126/science.286.5440.707. [DOI] [PubMed] [Google Scholar]
- 16.Dixon R A F, Sigal I S, Cadelore M R, Register R B, Scattergood W, Rands E, Strader C D. EMBO J. 1987;6:3269–3275. doi: 10.1002/j.1460-2075.1987.tb02645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strader C D, Sigal I S, Candelore M R, Rands E, Hill W S, Dixon R A F. J Biol Chem. 1988;231:10267–10269. [PubMed] [Google Scholar]
- 18.Parrill A L, Wang D, Bautista D L, Van Brocklyn J R, Lorincz Z, Fischer D J, Baker D L, Liliom K, Spiegel S, Tigyi G. J Biol Chem. 2000;275:39379–39384. doi: 10.1074/jbc.M007680200. [DOI] [PubMed] [Google Scholar]
- 19.Mierke D F, Giragossian C. Med Res Rev. 2001;21:450–471. doi: 10.1002/med.1018. [DOI] [PubMed] [Google Scholar]
- 20.Floriano W B, Vaidehi N, Singer M, Shepherd G, Goddard W A., III Proc Natl Acad Sci USA. 2000;97:10712–10716. doi: 10.1073/pnas.97.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vaidehi, N., Floriano, W. B., Singer, M. S., Shepherd, G. & Goddard, W. A., III (2001) Patent CIT 3191.
- 22.Mayo S L, Olafson B D, Goddard W A., III J Phys Chem. 1990;94:8897–8909. [Google Scholar]
- 23.MacKerell A D, Bashford D, Bellott M, Dunbrack R L, Evanseck J D, Field M J, Fischer S, Gao J, Guo H, Ha S, et al. J Phys Chem. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 24.Rappé A K, Goddard W A., III J Phys Chem. 1991;95:3358–3363. [Google Scholar]
- 25.Gasteiger J, Marsili M. Tetrahedron. 1980;36:3219–3228. [Google Scholar]
- 26.Ghosh A, Rapp C S, Friesner R A. J Phys Chem. 1998;102:10983–10990. [Google Scholar]
- 27.Zamanakos G. Ph.D. thesis. Pasadena, CA: California Institute of Technology; 2001. [Google Scholar]
- 28.Donnelly D. Biochem Soc Trans. 1993;21:36–39. doi: 10.1042/bst0210036. [DOI] [PubMed] [Google Scholar]
- 29.Eisenberg D, Weiss R M, Terwilliger T C. Proc Natl Acad Sci USA. 1984;8:140–144. doi: 10.1073/pnas.81.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singer M S, Weisinger-Lewin Y, Lancet D, Shepherd G M. Recept Channels. 1995;4:141–147. [PubMed] [Google Scholar]
- 31.Jain A, Vaidehi N, Rodriguez G. J Comp Phys. 1993;106:258–268. [Google Scholar]
- 32.Mathiowetz A M, Jain A, Karasawa N, Goddard W A., III Proteins. 1994;20:227–247. doi: 10.1002/prot.340200304. [DOI] [PubMed] [Google Scholar]
- 33.Vaidehi N, Jain A, Goddard W A., III J Phys Chem. 1996;100:10508–10517. [Google Scholar]
- 34.Schertler G F X. Eye. 1998;12:504–510. doi: 10.1038/eye.1998.138. [DOI] [PubMed] [Google Scholar]
- 35.Vriend G. J Mol Graphics. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 36.Okada T, Ernst O P, Palczewski K, Hofmann K P. Trends Biochem Sci. 2001;26:318–324. doi: 10.1016/s0968-0004(01)01799-6. [DOI] [PubMed] [Google Scholar]
- 37.Schoneberg T, Schulz A, Gudermann T. Rev Phys Biochem Pharm. 2002;144:145–227. [PubMed] [Google Scholar]
- 38.Bower M, Cohen F E, Dunbrack R L., Jr J Mol Biol. 1997;267:1268–1282. doi: 10.1006/jmbi.1997.0926. [DOI] [PubMed] [Google Scholar]
- 39.Lim K-T, Brunett S, Iotov M, McClurg R B, Vaidehi N, Dasgupta S, Taylor S, Goddard W A., III J Comput Chem. 1997;18:501–521. [Google Scholar]
- 40.Grigorieff N, Ceska T A, Downing K H, Baldwin J M, Henderson R. J Mol Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 41.Luecke H, Schobert B, Richter H T, Cartailler J P, Lanyi J K. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 42.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 43.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Floriano, W. B., Vaidehi, N. & Goddard, W. A., III (2001) Patent CIT 3192.
- 45.Datta D, Vaidehi N, Xu X, Goddard W A., III Proc Natl Acad Sci USA. 2002;99:2636–2641. doi: 10.1073/pnas.052709399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Datta, D., Vaidehi, N., Floriano, W. B., Kim, K. S., Prasadarao, N. V. & Goddard, W. A., III (2002) Proteins Struct. Funct. Genet., in press. [DOI] [PubMed]
- 47.Ewing T A, Kuntz I D. J Comput Chem. 1997;18:1175–1189. [Google Scholar]
- 48.Dixon D A F, Sigal I S, Randa E, Register R B, Candelore M R, Blake A D, Strader C D. Nature (London) 1987;326:73–77. doi: 10.1038/326073a0. [DOI] [PubMed] [Google Scholar]
- 49.Lindemann B. Curr Biol. 1996;6:1234–1237. doi: 10.1016/s0960-9822(96)00704-x. [DOI] [PubMed] [Google Scholar]
- 50.Hoon M A, Adler E, Lindemeier J, Battey J F, Ryba N J P, Zuker C S. Cell. 1999;96:541–551. doi: 10.1016/s0092-8674(00)80658-3. [DOI] [PubMed] [Google Scholar]