Abstract
Water plays an important role in determining the folding, structure, dynamics, and, in turn, the function of proteins. We have utilized a combination of fluorescence approaches such as the wavelength-selective fluorescence approach to monitor the effect of varying degrees of hydration on the organization and dynamics of the functionally important tryptophan residues of gramicidin in reverse micelles formed by sodium bis(2-ethylhexyl) sulfosuccinate. Our results show that tryptophans in gramicidin, present in the single-stranded β6.3 conformation, experience slow solvent relaxation giving rise to red-edge excitation shift (REES). In addition, changes in fluorescence polarization with increasing excitation or emission wavelength reinforce that the gramicidin tryptophans are localized in motionally restricted regions of the reverse micelle. Interestingly, the extent of REES is found to be independent of the [water]/[surfactant] molar ratio (wo). We attribute this to heterogeneity in gramicidin tryptophan localization. Fluorescence intensity and mean fluorescence lifetime of the gramicidin tryptophans show significant reductions with increasing wo indicating sensitivity to increased polarity. Since the dynamics of hydration is related to folding, structure, and eventually function of proteins, we conclude that REES could prove to be a potentially sensitive tool to explore the dynamics of proteins under conditions of changing hydration.
INTRODUCTION
Maintenance of appropriate ion balance across a biological membrane is crucial for cellular integrity and function. Ion channels are crucial cellular components that are involved in the maintenance of such an ion balance. They represent an important class of transmembrane proteins and serve as key elements in signaling and sensing pathways and to connect the inside of the cell to its outside in a selective fashion. They are crucial for normal functioning of cells since a defective ion channel can lead to diseases (Cooper and Jan, 1999) such as cystic fibrosis (Stutts et al., 1995). In addition, ion channels are known to be specific targets for neuroactive toxins (Garcia, 2004). The recent success in crystallographic analyses of ion channels, starting with the Streptomyces lividans K+ channel (KcsA) (Doyle et al., 1998), constitutes an exciting development in contemporary membrane biology (Rees et al., 2000).
The linear peptide gramicidin forms prototypical ion channels specific for monovalent cations and has been extensively used to study the organization, dynamics, and function of membrane-spanning channels (Killian, 1992; Andersen and Koeppe, 1992; Koeppe and Andersen, 1996; Wallace, 2000; Miloshevsky and Jordan, 2004). Gramicidin serves as an excellent model for transmembrane channels due to its small size, ready availability, and the relative ease with which chemical modifications can be performed. This makes gramicidin unique among small membrane-active peptides and provides the basis for its use to explore the principles that govern the folding and function of membrane-spanning channels in particular, and membrane proteins in general.
The unique sequence of alternating l- and d-chirality renders gramicidin sensitive to the environment in which it is placed. Gramicidin therefore adopts a wide range of environment-dependent conformations (Urry, 1971; Ramachandran and Chandrasekaran, 1972; Veatch et al., 1974). In membranes, the initial conformation adopted by gramicidin has been reported to be influenced by the nature of the solvent in which it was dissolved before incorporation, i.e., gramicidin conformation in membranes depends on its “solvent history” (LoGrasso et al., 1988). The most preferred (thermodynamically stable) conformation in membranes is the single-stranded β6.3 conformation (Killian et al., 1988). The head-to-head (amino terminal-to-amino terminal) single-stranded β6.3 helical dimer form is the cation conducting channel conformation of gramicidin in membranes (O'Connell et al., 1990). In this conformation, the carboxy terminus is exposed to the membrane-water interface and the amino terminus is buried in the hydrophobic core of the membrane. This places the tryptophan residues clustered at the membrane-water interface at the entrance to the channel (O'Connell et al., 1990; Ketchem et al., 1993; Mukherjee and Chattopadhyay, 1994).
Hydration plays a key role in cellular structure and function and is crucial for lipid-protein interactions in membranes (Ho and Stubbs, 1992; Mentré, 2001). Water has a crucial role in determining the folding, structure, dynamics, and in turn, the function of proteins (Colombo et al., 1992; Xu and Cross, 1999; Fenimore et al., 2002; Mattos, 2002; Timasheff, 2002). It is estimated that a threshold level of hydration (<0.4 grams of water per gram of protein) is required to fully activate the dynamics and function of globular proteins (Bizzarri and Cannistraro, 2002). In addition, it has become increasingly clear that water molecules mediate lipid-protein interactions (Ho and Stubbs, 1992; Essen et al., 1998; McAuley et al., 1999) and hence the function of membrane proteins (Sankararamakrishnan and Sansom, 1995; Okada et al., 2002; Kouyama et al., 2004). Any alteration in the degree of hydration, particularly at the protein-lipid interface, could potentially lead to modifications of protein structure that could in turn modify its function. Interestingly, the conformational preference of gramicidin in a low dielectric solvent has previously been shown to be highly dependent on the concentration of water (Xu and Cross, 1999). In fact, water is found to accelerate the interconversion of double-stranded intertwined parallel dimers to antiparallel dimers probably via catalysis of hydrogen bond exchange. This becomes particularly relevant in situations where availability of water is limited as in the interior of a membrane bilayer (Chattopadhyay and Mukherjee, 1999).
Biological and model membranes (liposomes) and membrane-mimetic sytems such as micelles, however, are not appropriate for exploring the effect of hydration on the organization and dynamics of peptides and proteins incorporated in them since the controlled variation of water content is difficult to achieve in these systems. Reverse micelles represent a unique type of organized molecular assembly that offers the advantage of monitoring dynamics of molecules incorporated in them with varying states of hydration. Amphiphilic surfactants such as AOT (sodium bis(2-ethylhexyl) sulfosuccinate), self-assemble to form reverse (or inverted) micelles in nonpolar solvents in which the polar headgroups of the surfactant monomers cluster to form a micellar core and are directed toward the center of the assembly, and the hydrophobic tails extend outward into the bulk organic phase (Luisi and Magid, 1986; Luisi et al., 1988). Reverse micelles are relatively simple yet versatile systems. They provide an attractive model system for biomembranes since they mimic a number of important and essential features of biological membranes although lacking much of the complexity associated with them. It is known that the dynamics of liquids in confined spaces is different than that of their bulk counterparts (Granick, 1991; Brubach et al., 2001) and this constitutes one of the main reasons for the popularity that reverse micelles enjoy as a model system in studies of water dynamics (Levinger, 2002). The highly structured yet heterogeneous water molecules in reverse micelles represent interesting models for water molecules present in biological systems such as membranes, which are more difficult to analyze experimentally. The physical and chemical properties of the entrapped water are markedly different from the properties of bulk water but similar in several aspects to those of biological interfacial water as found in membranes or protein interfaces (Jain et al., 1989; Ikushima et al., 1997; Brubach et al., 2001; Venables et al., 2001). The interfacial water is crucial for the induction of secondary structure in peptides and proteins when bound to surfaces such as membranes or micelles, as well as for variation of their local internal motion.
Both experimental (Jain et al., 1989; Ikushima et al., 1997; Venables et al., 2001) and theoretical (Faeder and Ladanyi, 2000) approaches have shown that the key structural parameter of reverse micelles is the [water]/[surfactant] molar ratio (wo), which determines micellar size as well as the unique physicochemical properties of the entrapped water. In addition, reverse micelles enjoy certain advantages in spectroscopic and, in particular, fluorescence studies since they are small and optically transparent, have well-defined sizes, and are relatively scatter-free. They therefore represent model systems suitable for the study of peptides and proteins in membrane-mimetic, hydration-controlled environments (Souto and Ito, 2000; Valdez et al., 2001; Raghuraman and Chattopadhyay, 2003).
In this article, we have employed a combination of fluorescence approaches such as the wavelength-selective fluorescence approach to monitor the effect of varying degrees of hydration on the dynamics of gramicidin in AOT reverse micelles. The double chain anionic surfactant AOT has been extensively used to form reverse micelles in nonpolar solvents. One of the advantages of using AOT is that reverse micelles formed by AOT can solubilize a large quantity of water in a nonpolar solvent. In addition, reverse micelles formed by AOT retain a spherical shape over a wide range of wo. As a result of this, the radius of the entrapped water pool can be linearly related to wo (Eastoe et al., 1990).
Wavelength-selective fluorescence comprises a set of approaches based on the red-edge effect in fluorescence spectroscopy, which can be used to directly monitor the environment and dynamics around a fluorophore in an organized molecular assembly (Chattopadhyay, 2003; Raghuraman et al., 2003). A shift in the wavelength of maximum fluorescence emission toward higher wavelengths, caused by a shift in the excitation wavelength toward the red edge of the absorption band, is termed red-edge excitation shift (REES) (Demchenko, 2002; Chattopadhyay, 2003; Raghuraman et al., 2003). This effect is mostly observed with polar fluorophores in motionally restricted environments such as viscous solutions or condensed phases where the dipolar relaxation time for the solvent shell around a fluorophore is comparable to or longer than its fluorescence lifetime. REES arises due to slow rates of solvent relaxation (reorientation) around an excited-state fluorophore, which is dependent on the motional restriction imposed on the solvent molecules in the immediate vicinity of the fluorophore. Utilizing this approach, it becomes possible to probe the mobility parameters of the environment itself (which is represented by the relaxing solvent molecules) using the fluorophore merely as a reporter group. This makes the use of REES in particular and the wavelength-selective fluorescence approach in general very useful since, as mentioned earlier, hydration plays a crucial modulatory role in a large number of important cellular events including protein folding, lipid-protein interactions, and ion transport. The unique feature about REES is that whereas all other fluorescence techniques, such as fluorescence quenching, resonance energy transfer, and polarization measurements, yield information about the fluorophore itself, REES provides information about the relative rates of solvent (water in biological systems) relaxation dynamics, which is not possible to obtain by other techniques. Since the dynamics of hydration is directly associated with the functionality of proteins, REES could prove to be a novel and sensitive tool to explore the organization and dynamics of soluble and membrane proteins under varying degrees of hydration. An in-depth discussion of the photophysical framework for REES and wavelength-selective fluorescence approach is provided in recent reviews (Chattopadhyay, 2003; Raghuraman et al., 2003).
MATERIALS AND METHODS
Materials
Gramicidin A′ (from Bacillus brevis), AOT, and trifluoroethanol (TFE) were purchased from Sigma Chemical (St. Louis, MO). Gramicidin A′, as obtained, is a mixture of gramicidins A, B, and C. The purity of AOT was confirmed by good agreement of its UV absorption spectrum with a previously reported spectrum (Luisi and Magid, 1986). Water was purified through a Millipore (Bedford, MA) Milli-Q system and used throughout. The isooctane used was of spectroscopic grade.
Sample preparation
Reverse micelles of AOT containing gramicidin were prepared without the addition of any cosolvent as follows. Briefly, 48 nmol gramicidin (12 nmol in experiments involving measurements of fluorescence intensity or polarization) in TFE was dried under a stream of nitrogen while being warmed gently (∼35°C). After further drying under a high vacuum for at least 12 h, 1.5 ml of 50 mM AOT in isooctane was added, and samples were vortexed for 3 min. Appropriate amounts of water were subsequently added to make reverse micellar dispersions of different wo. The samples were kept in the dark for at least 10 h before any measurements were made. Incubation for this length of time avoids conformational heterogeneity (see Results). The optical density of the fluorescent samples used for quantitative fluorescence measurements (see Figs. 2, 4, 5, and 8) at the excitation wavelength was <0.2 in all cases. Background samples were prepared the same way except that gramicidin was not added to them. All experiments were done at 23°C.
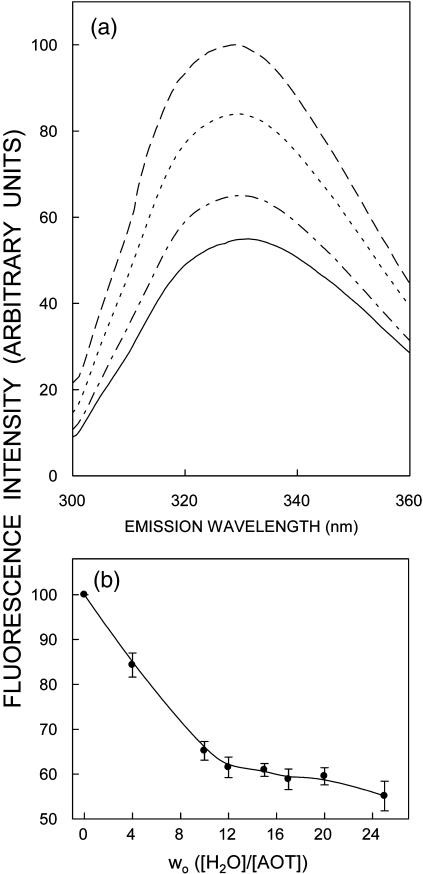
FIGURE 2.
Effect of increasing amounts of added water on (a) fluorescence emission spectra and (b) fluorescence intensity of gramicidin in AOT reverse micelles. Fluorescence emission spectra are shown in a as a function of [water]/[surfactant] molar ratio (wo) in order of decreasing intensity corresponding to wo = 0 (dashed line), 4 (dotted line), 10 (dash-dotted line), and 25 (solid line). Fluorescence intensity was monitored at 328 nm and is plotted as a function of wo in b. The data points shown are the mean ± standard error of five independent measurements. The excitation wavelength used was 280 nm. The ratio of gramicidin to surfactant (AOT) was 1:6250 (mol/mol) and the concentration of gramicidin was 8 μM in all cases. See Materials and Methods for other details.
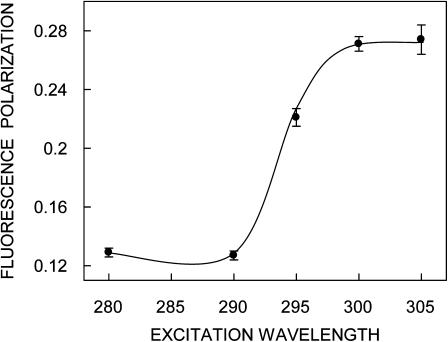
FIGURE 4.
Representative plot of fluorescence polarization of gramicidin in AOT reverse micelles as a function of excitation wavelength. The [water]/[surfactant] molar ratio (wo) was 15. The emission wavelength was 328 nm in all cases. The data points shown are the mean ± SE of at least three independent measurements. All other conditions are as in Fig. 2. See Materials and Methods for other details.
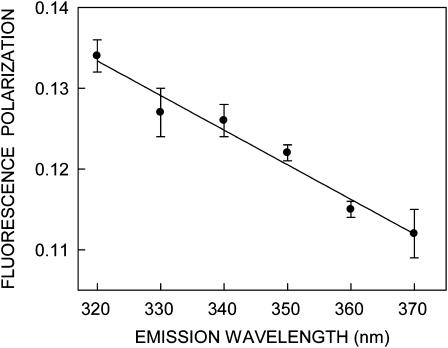
FIGURE 5.
Representative plot of fluorescence polarization of gramicidin in AOT reverse micelles as a function of emission wavelength. The [water]/[surfactant] molar ratio (wo) was 15. The excitation wavelength was 280 nm in all cases. The data points shown are the mean ± SE of at least three independent measurements. All other conditions are as in Fig. 2. See Materials and Methods for other details.
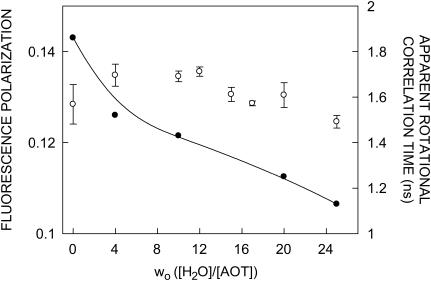
FIGURE 8.
Fluorescence polarization values (○) and apparent rotational correlation times (•) of gramicidin in AOT reverse micelles as a function of wo. The apparent rotational correlation times were calculated using Eq. 5 (see text for other details). The excitation wavelength was 280 nm and emission was monitored at 328 nm. Fluorescence polarization data shown are the mean ± SE of multiple measurements. All other conditions are as in Fig. 2. See Materials and Methods for other details.
The molar ratio of peptide to surfactant was carefully chosen to give an optimum signal/noise ratio with minimal perturbation to the micellar organization and negligible interprobe interactions. The maximum peptide concentration in the reverse micelles was 32 μM, whereas the concentration of AOT was 50 mM in all cases. This corresponds to a maximum molar ratio of peptide to surfactant of 1:1563 (mol/mol). At such a low gramicidin/surfactant molar ratio, not more than one peptide molecule would be present per reverse micelle on average. This rules out any aggregation effects, especially keeping in mind the aggregation number of AOT of ∼50–300 in the range of wo between 5 and 25 (Zhou et al., 2002).
Steady-state fluorescence measurements
Steady-state fluorescence measurements were performed with a Hitachi F-4010 spectrofluorometer using 1-cm path length quartz cuvettes. Excitation and emission slits with a nominal bandpass of 5 nm were used for all measurements. Background intensities of samples in which gramicidin was omitted were negligible in most cases and were subtracted from each sample spectrum to cancel out any contribution due to the solvent Raman peak and other scattering artifacts. The spectral shifts obtained with different sets of samples were identical in most cases, or were within ±1 nm of the ones reported. Fluorescence polarization measurements were performed using a Hitachi polarization accessory. Polarization values were calculated from the equation (Lakowicz, 1999),
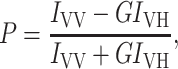 |
(1) |
where IVV and IVH are the measured fluorescence intensities (after appropriate background subtraction) with the excitation polarizer vertically oriented and emission polarizer vertically and horizontally oriented, respectively. G is the grating correction factor and is the ratio of the efficiencies of the detection system for vertically and horizontally polarized light, and is equal to IHV/IHH. All experiments were done with multiple sets of samples, and average values of polarization are shown in Figs. 4, 5, and 8.
Time-resolved fluorescence measurements
Fluorescence lifetimes were calculated from time-resolved fluorescence intensity decays using a Photon Technology International (London, Ontario, Canada) LS-100 luminescence spectrophotometer in the time-correlated single-photon counting mode. This machine uses a thyratron-gated nanosecond flash lamp filled with nitrogen as the plasma gas (17 ± 1 inches of mercury vacuum) and is run at 17–20 kHz. Lamp profiles were measured at the excitation wavelength using Ludox (colloidal silica) as the scatterer. To optimize the signal/noise ratio, 10,000 photon counts were collected in the peak channel. The excitation wavelength used was 297 nm and emission was set at 328 nm. All experiments were performed using excitation and emission slits with a bandpass of 10 nm or less. The sample and the scatterer were alternated after every 5% acquisition to ensure compensation for shape and timing drifts occurring during the period of data collection. This arrangement also prevents any prolonged exposure of the sample to the excitation beam, thereby avoiding any possible photodamage to the fluorophore. The data stored in a multichannel analyzer was routinely transferred to an IBM PC for analysis. Fluorescence intensity decay curves so obtained were deconvoluted with the instrument response function and analyzed as a sum of exponential terms,
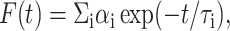 |
(2) |
where F(t) is the fluorescence intensity at time t and αi is a preexponential factor representing the fractional contribution to the time-resolved decay of the component with a lifetime τi such that Σiαi = 1. The decay parameters were recovered using a nonlinear least squares iterative fitting procedure based on the Marquardt algorithm (Bevington, 1969). The program also includes statistical and plotting subroutine packages (O'Connor and Phillips, 1984). The goodness of the fit of a given set of observed data and the chosen function was evaluated by the reduced χ2 ratio, the weighted residuals (Lampert et al., 1983), and the autocorrelation function of the weighted residuals (Grinvald and Steinberg, 1974). A fit was considered acceptable when plots of the weighted residuals and the autocorrelation function showed random deviation about zero with a minimum χ2 value generally not more than 1.5. Mean (average) lifetimes 〈τ〉 for biexponential decays of fluorescence were calculated from the decay times and preexponential factors using the following equation (Lakowicz, 1999):
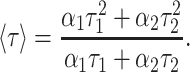 |
(3) |
Circular dichroism measurements
Circular dichroism (CD) measurements were carried out at room temperature (23°C) on a JASCO (Tokyo, Japan) J-715 spectropolarimeter which was calibrated with (+)-10-camphorsulfonic acid (Chen and Yang, 1977). The spectra were scanned in a quartz optical cell with a path length of 0.1 cm. All spectra were recorded in 0.5 nm wavelength increments with a 4-s response and a band width of 1 nm. For monitoring changes in secondary structure, spectra were scanned from 210 to 280 nm at a scan rate of 100 nm/min. Each spectrum is the average of 12 scans with a full-scale sensitivity of 10 mdeg. All spectra were corrected for background by subtraction of appropriate blanks and were smoothed, making sure that the overall shape of the spectrum remained unaltered. Data are represented as mean residue ellipticities and were calculated using the formula
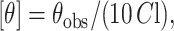 |
(4) |
where θobs is the observed ellipticity in mdeg, l is the path length in cm, and C is the concentration of peptide bonds in mol/L.
RESULTS
Gramicidin conformation in reverse micelles monitored by circular dichroism spectroscopy
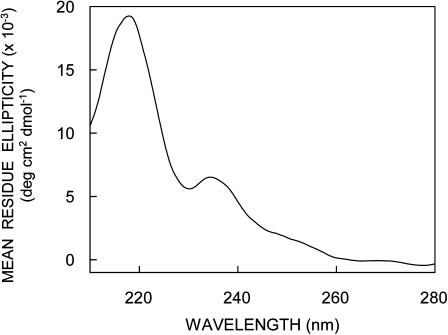
Circular dichroism spectroscopy has been extensively used to characterize gramicidin conformations (Killian et al., 1988; LoGrasso et al., 1988; Salom et al., 1992; Rawat et al., 2004). The single-stranded β6.3 conformation has been earlier shown to be the preferred conformation in AOT reverse micelles, and the extent of interconversion is found to be dependent on the length of incubation (Salom et al., 1992). To avoid any conformational heterogeneity in our samples, we chose to use TFE, which would allow direct incorporation of gramicidin in the single-stranded β6.3 conformation. Nonetheless, to further ensure that there is no conformational heterogeneity, we incubated the samples in the dark for 10 h before measurements were made (see Materials and Methods). The CD spectra of gramicidin incorporated into AOT reverse micelles is shown in Fig. 1. The spectral characteristics are typical of the single-stranded β6.3 conformation with two characteristic peaks of positive ellipticity at ∼218 and 235 nm and a valley at ∼230 nm.
FIGURE 1.
Far-UV CD spectra of gramicidin in AOT reverse micelles. The [water]/[surfactant] molar ratio (wo) was 25. The ratio of gramicidin to surfactant (AOT) was 1:1563 (mol/mol) and the concentration of gramicidin was 32 μM. See Materials and Methods for other details.
Fluorescence characteristics of gramicidin in reverse micelles
The fluorescence emission spectra and intensity of gramicidin in AOT reverse micelles as a function of increasing [water]/[surfactant] ratio (wo) are shown in Fig. 2. Fig. 2 a shows that the emission maximum of gramicidin in the absence of any added water is at 328 nm. In membranes of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine the emission maximum of gramicidin has been reported by us to be at 333 nm (Rawat et al., 2004). Therefore, in reverse micelles the emission maximum of gramicidin is blue-shifted by only 5 nm, indicating that the overall polarity in the vicinity of the gramicidin tryptophans in reverse micelles is not significantly different from that experienced in membranes. Importantly, the emission maximum undergoes a small red shift of 3 nm (328 to 331 nm) when wo is increased up to 25 (see Table 1). This is in sharp contrast to the change in emission maximum observed for interfacially localized tryptophans such as the sole tryptophan of tryptophan octyl ester (Sengupta and Sengupta, 2000) and melittin (Raghuraman and Chattopadhyay, 2003) in AOT reverse micelles. We have recently shown that the extent of the dependence of the emission maximum of a reverse micelle-bound fluorophore on wo is not uniform and is related to the position (location) of the fluorophore in the reverse micellar assembly (Kelkar and Chattopadhyay, 2004). Thus the weak dependence of the emission maximum of the gramicidin tryptophans on wo may be due to the relatively deep location of the gramicidin tryptophans in the acyl chain region of the reverse micelle. However, the peak fluorescence intensity shows a progressive reduction with increasing wo, indicating increased polarity around the gramicidin tryptophans with increasing water content of the reverse micelle (see Fig. 2 b). There is an ∼55% decrease in peak fluorescence intensity when wo is increased from 0 to 25. Interestingly, the extent of decrease in peak fluorescence intensity is more pronounced for low values of wo (<10).
TABLE 1.
Fluorescence emission maxima and REES of gramicidin in AOT reverse micelles as a function of wo*
| [Water]/[surfactant] molar ratio (wo) | Fluorescence emission maximum† (nm) | REES (nm) |
|---|---|---|
| 0 | 328 | 6 |
| 4 | 330 | 7 |
| 10 | 330 | 6 |
| 15 | 330 | 7 |
| 20 | 330 | 8 |
| 25 | 331 | 7 |
The ratio of gramicidin to AOT was 1:1563 (mol/mol).
The excitation wavelength was 280 nm.
Red-edge excitation shift (REES) of gramicidin in reverse micelles
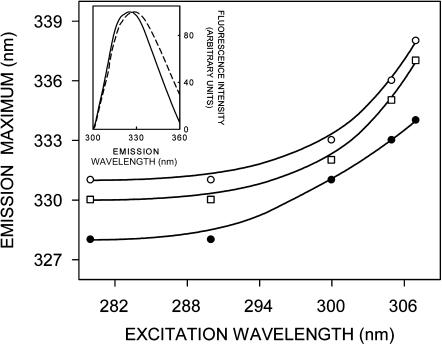
The shifts in the maxima of fluorescence emission of the tryptophan residues of gramicidin bound to AOT reverse micelles of varying wo as a function of excitation wavelength are shown in Fig. 3. (We have used the term maximum of fluorescence emission in a somewhat wider sense here. In every case, we have monitored the wavelength corresponding to maximum fluorescence intensity, as well as the center of mass of the fluorescence emission. In most cases, both these methods yielded the same wavelength. In cases where minor discrepancies were found, the center of mass of emission has been reported as the fluorescence maximum.) As the excitation wavelength is changed from 280 to 307 nm, the emission maxima of gramicidin shift toward longer wavelengths in all cases. The emission maxima are shifted from 328 to 334 nm (for wo = 0), 330 to 337 nm (wo = 15), and 331 to 338 nm (wo = 25), which correspond to REES of 6–7 nm in these cases. The magnitude of REES obtained for gramicidin bound to AOT reverse micelles with varying wo is shown in Table 1. It is possible that there could be further red shift if excitation is carried out beyond 307 nm. We found it difficult to work in this wavelength range due to low signal/noise ratio and artifacts due to the solvent Raman peak that sometimes remained even after background subtraction. Such dependence of the emission maximum on excitation wavelength is characteristic of the red-edge excitation shift. This implies that the tryptophans in the gramicidin single-stranded β6.3 conformation, on the average, are localized in a motionally restricted region of the reverse micelle. Earlier work from our group (Mukherjee and Chattopadhyay, 1994; Rawat et al., 2004) and others (Koeppe et al., 2003) have shown that gramicidin tryptophans in the single-stranded β6.3 conformation are localized in a motionally restricted region of the membrane, consistent with the interfacial localization of these tryptophans in the membrane (O'Connell et al., 1990; Ketchem et al., 1993). Our results show that the gramicidin tryptophans, on the average, are localized in a region of slow solvent relaxation (reorientation) in the reverse micellar assembly, possibly at the reverse micellar interface. The interfacial region of reverse micelles is associated with bound water, which displays characteristic dynamics (Jain et al., 1989; Ikushima et al., 1997; Faeder and Ladanyi, 2000; Brubach et al., 2001; Venables et al., 2001). Since REES arises due to the dynamics of reorientational motion of solvent molecules, these results assume significance in the context of recent reports of slow (∼nanoseconds) water relaxation in reverse micelles (Bhattacharyya and Bagchi, 2000; Hazra and Sarkar, 2001; Bhattacharyya, 2003).
FIGURE 3.
Effect of changing excitation wavelength on the wavelength of maximum emission of gramicidin in AOT reverse micelles corresponding to wo = 0 (•), 15 (□), and 25 (○). All other conditions are as in Fig. 1. The inset shows intensity-normalized fluorescence emission spectra of melittin (solid line) and gramicidin (dashed line) in AOT reverse micelles at wo = 0. The experimental conditions for melittin are as in Fig. 1 in Raghuraman and Chattopadhyay (2003). See Materials and Methods for other details.
The extent of REES appears to be more or less independent of wo and corresponds to 6–8 nm in all cases (see Table 1). Gramicidin is a multitryptophan peptide, and therefore the red-edge excitation shift may be indicative of the average environment experienced by the tryptophans. The locations of these tryptophans would therefore be heterogenous in the reverse micelle. The presence of tryptophans at various locations would contribute to spectral heterogeneity and hence broadening. The inset in Fig. 3 shows this to be true since the emission spectrum of gramicidin bound to AOT reverse micelles is relatively broad compared to that of a single tryptophan-containing peptide, melittin, under similar conditions. Fig. 3 also shows that gramicidin displays REES even without any added water, i.e., at wo = 0. It is interesting to speculate about the origin of REES in this case. This could possibly be due to the presence of tightly bound water molecules in AOT reverse micelles even at wo = 0 (Luisi and Magid, 1986). Another contributing factor could be the polar headgroup of AOT, which may change dipole moment upon excitation giving rise to REES.
Fluorescence polarization of gramicidin in reverse micelles is dependent on excitation and emission wavelength
In addition to the shift in emission maximum on red-edge excitation, fluorescence polarization is also known to be dependent on excitation wavelength in motionally restricted media (Mukherjee and Chattopadhyay, 1995, and references therein). Due to strong dipolar interactions with the surrounding solvent molecules, there is a decreased rotational rate of the fluorophore in the solvent-relaxed state. Red-edge excitation results in selective excitation of this subclass of fluorophore. Because of strong interactions with the polar solvent molecules in the excited state, one may expect these “solvent-relaxed” fluorophores to rotate more slowly, thereby increasing the polarization. A representative excitation polarization spectrum (i.e., a plot of steady-state polarization versus excitation wavelength) of gramicidin tryptophans in AOT reverse micelles is shown in Fig. 4. The polarization of gramicidin tryptophans undergoes considerable change upon altering the excitation wavelength, with a sharp increase toward the red edge of the absorption band and a characteristic dip at 290 nm. Such an increase in polarization upon red-edge excitation for peptides and proteins containing tryptophans, especially in media of reduced mobility, has been previously reported (Valeur and Weber, 1978; Mukherjee and Chattopadhyay, 1994).
It is known that tryptophan has two overlapping So → S1 electronic transitions (1La and 1Lb) that are almost perpendicular to each other (Callis, 1997). Both So → 1La and So → 1Lb transitions occur in the 260–300 nm range. In nonpolar solvents, 1La has higher energy than 1Lb. However, in polar solvents, the energy level of 1La is lowered, making it the lowest energy state. This inversion is believed to occur because 1La transition has higher dipole moment (as it is directed through the ring –NH group), and can have dipole-dipole interactions with polar solvent molecules. Irrespective of whether 1La or 1Lb is the lowest S1 state, equilibration between these two states is believed to be very fast (of the order of 10−12 s), so that only emission from the lower S1 state is observed (Ruggiero et al., 1990). In a motionally restricted polar environment, absorption at the red edge photoselects the lowest energy S1 (1La in this case), and thus the polarization is high since depolarization only due to small angular differences between the absorption and emission transition moments and solvent reorientation, if any, occurs. Excitation at the shorter wavelengths, however, populates both 1La and 1Lb states. Equilibration between these two states produces a depolarization due to the ∼90° angular difference between 1La and 1Lb moments. Thus, near 290 nm, there is a dip in polarization due to maximal absorption by the 1Lb state. Fig. 4 shows such a characteristic dip around 290 nm in the excitation polarization spectrum of gramicidin tryptophans. Thus, the sharp increase in polarization toward the red edge of the absorption band is probably because the extent of depolarization in gramicidin tryptophans is reduced at the red edge not only due to decreased rotational rate of the fluorophore in the solvent-relaxed state, but also due to photoselection of the predominantly 1La transition, which in turn reduces the contribution to depolarization because of 1Lb → 1La equilibration.
For fluorophores incorporated in motionally restricted media, fluorescence polarization is also known to be dependent on emission wavelength. Under such conditions, a steady and significant decrease in polarization is observed with increasing emission wavelength (Mukherjee and Chattopadhyay, 1995, and references therein). Fig. 5 shows representative variation in steady-state polarization of gramicidin tryptophans in AOT reverse micelles as a function of wavelength across its emission spectrum. As seen from the figure, there is a considerable reduction in polarization with increasing emission wavelength. The lowest polarization is observed toward the red edge where the solvent-relaxed emission predominates. Taken together, the changes in fluorescence polarization of gramicidin in AOT reverse micelles as a function of excitation and emission wavelengths reinforce the presence of motionally restricted environment in the vicinity of the gramicidin tryptophans.
Fluorescence lifetime and polarization of gramicidin tryptophans in reverse micelles of varying hydration
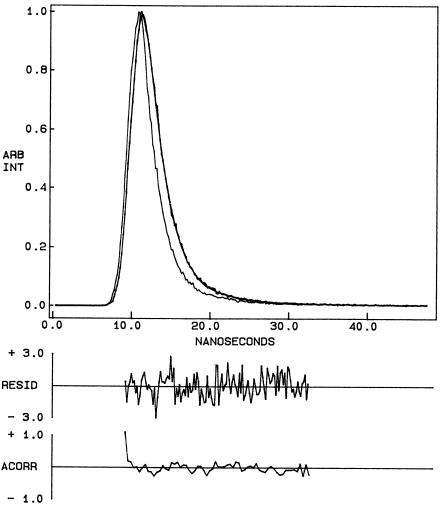
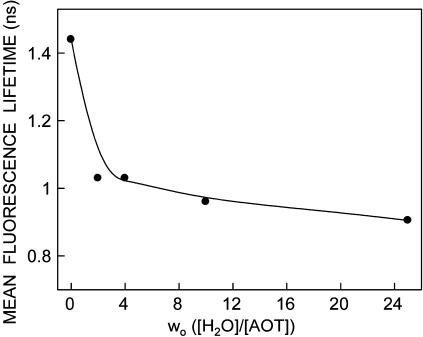
Fluorescence lifetime serves as a sensitive indicator for the local environment and polarity in which a given fluorophore is placed (Prendergast, 1991). A typical decay profile of gramicidin tryptophans in AOT reverse micelles with its biexponential fitting and the various statistical parameters used to check the goodness of the fit is shown in Fig. 6. The fluorescence lifetimes of gramicidin tryptophans in AOT reverse micelles obtained as a function of wo are shown in Table 2. As seen from the table, all fluorescence decays could be fitted well with a biexponential function. We chose to use the mean fluorescence lifetime as an important parameter for describing the behavior of gramicidin tryptophans in AOT reverse micelles since it is independent of the number of exponentials used to fit the time-resolved fluorescence decay. The mean fluorescence lifetimes of gramicidin tryptophans in AOT reverse micelles calculated using Eq. 3 are plotted as a function of wo in Fig. 7. In general, an increase in the polarity of the tryptophan environment is known to reduce the lifetime of tryptophans due to fast deactivating processes in polar environments (Kirby and Steiner, 1970). The increased polarity around the gramicidin tryptophans in AOT reverse micelles with increasing wo is reflected in the decrease in the mean lifetime (∼36%) of gramicidin tryptophans with increase in the reverse micellar water content. Interestingly, the reduction in mean fluorescence lifetime is more pronounced at very low values of wo.
FIGURE 6.
Time-resolved fluorescence intensity decay of gramicidin in AOT reverse micelles at wo = 10. Excitation wavelength was at 297 nm which corresponds to a peak in the spectral output of the nitrogen lamp. Emission was monitored at 328 nm. The sharp peak on the left is the lamp profile. The relatively broad peak on the right is the decay profile, fitted to a biexponential function. The two lower plots show the weighted residuals and the autocorrelation function of the weighted residuals. All other conditions are as in Fig. 1. See Materials and Methods for other details.
TABLE 2.
Fluorescence lifetimes of gramicidin in AOT reverse micelles as a function of wo*
| [Water]/[surfactant] molar ratio (wo) | α1 | τ1 (ns) | α2 | τ2 (ns) |
|---|---|---|---|---|
| 0 | 0.39 | 1.87 | 0.61 | 0.73 |
| 2 | 0.45 | 1.28 | 0.55 | 0.57 |
| 4 | 0.11 | 1.96 | 0.89 | 0.71 |
| 10 | 0.63 | 1.09 | 0.37 | 0.58 |
| 20 | 0.63 | 1.05 | 0.37 | 0.58 |
| 25 | 0.06 | 2.21 | 0.94 | 0.64 |
The excitation wavelength was 297 nm; emission was monitored at 328 nm. All other conditions are as in Fig. 1. See Materials and Methods for other details.
FIGURE 7.
Effect of increasing amounts of water on mean fluorescence lifetime of gramicidin in AOT reverse micelles. The excitation wavelength used was 297 nm and emission wavelength was set at 328 nm. Mean fluorescence lifetimes were calculated from Table 2 using Eq. 3. All other conditions are as in Fig. 1. See Materials and Methods for other details.
The steady-state polarization of gramicidin in AOT reverse micelles as a function of increasing wo is shown in Fig. 8. As can be seen from the figure, there appears to be no systematic variation in polarization values with wo. Since it is known that fluorescence polarization values may be influenced by the change in fluorescence lifetime with increasing wo (see Fig. 7), we calculated the apparent (average) rotational correlation times for gramicidin tryptophans in AOT reverse micelles with increasing wo using Perrin's equation (Lakowicz, 1999):
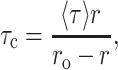 |
(5) |
where ro is the limiting anisotropy of tryptophan, r is the steady-state anisotropy (derived from the polarization values using r = 2P/(3 − P)), and 〈τ〉 is the mean fluorescence lifetime taken from Fig. 7. The values of the apparent rotational correlation times, calculated this way using a value of ro of 0.16 (Eftink et al., 1990), are shown in Fig. 8. As can be seen from the figure, the apparent rotational correlation times of gramicidin tryptophans in AOT reverse micelles show a progressive reduction with increasing water content of the reverse micelles. This is in contrast to the apparent invariance of the fluorescence polarization values with increasing wo. This implies that the apparent rotational correlation times, and not fluorescence polarization, are true indicators of rotational mobility in this case, since the latter may be influenced by the alteration in fluorescence lifetime. Taken together, there is an increase in the overall rotational mobility of gramicidin tryptophan with increasing water content in the reverse micellar system.
DISCUSSION
Knowledge of the dynamics of hydration at the molecular level is of considerable importance in understanding cellular structure and function since water plays a crucial role in the formation and maintenance of organized molecular assemblies such as proteins and membranes (Mentré, 2001). In particular, hydration has been shown to be a crucial parameter in the folding of proteins and it has been suggested that water-mediated interactions could guide the folding process even before the formation of native contacts (Papoian et al., 2004). Water has been shown to act as a catalyst for hydrogen bond exchange in protein folding thereby acting as a “foldase” (Xu and Cross, 1999). However, the membrane environment does not promote hydrogen bond exchange, making it possible to trap thermodynamically unstable conformations in the membrane environment (Arumugam et al., 1996). In such a scenario, it becomes important to study the effects of hydration on the conformation and dynamics of membrane proteins. Gramicidin is particularly suitable for such hydration studies due to its high surface/volume ratio and environment-sensitive conformational preference.
It is interesting to note that gramicidin represents a useful model for the realistic determination of conformational preference in a lipid bilayer environment despite the alternating sequence of l-d chirality generally not encountered in naturally occurring peptides and proteins. This is due to the fact that the dihedral angle combinations generated in the conformation space by various gramicidin conformations are “allowed” according to the Ramachandran plot (Andersen et al., 1996). Importantly, gramicidin channels share important structural features with other naturally occurring channel proteins like the bacterial KcsA K+ channel. These features include membrane interfacial localization of tryptophan residues, the channel interior being made of the peptide backbone, and ion selectivity arising out of backbone interactions (Wallace, 2000).
In this study, we have utilized a combination of fluorescence approaches such as the wavelength-selective fluorescence approach to monitor the effect of varying degrees of hydration on the organization and dynamics of the functionally important tryptophan residues of gramicidin in AOT reverse micelles. Our results show that tryptophans in gramicidin, present in the single-stranded β6.3 conformation, experience slow solvent relaxation giving rise to REES. Further, corresponding changes in fluorescence polarization with increasing excitation or emission wavelength reinforce the localization of gramicidin tryptophan in motionally restricted regions of the reverse micelle. Interestingly, the extent of REES does not appear to be influenced by the water content of the reverse micelle (wo), implying heterogeneity in tryptophan localization. However, the gramicidin tryptophans exhibit sensitivity to increased polarity generated due to increasing wo.
The contributions of individual tryptophan residues of gramicidin to the observed fluorescence deserve comment. Earlier work using fluorescence (Mukherjee and Chattopadhyay, 1994) and molecular dynamics simulations (Allen et al., 2003) have clearly indicated the heterogeneity of the contributing tryptophan residues. Whereas fluorescence measurements provide evidence for stacking interactions among Trp-9 and 15 at least in the fluorescence timescale (∼nanosecond) (Mukherjee and Chattopadhyay, 1994), molecular dynamics simulations point out motional flexibility giving rise to conformational heterogeneity of Trp-9 (Allen et al., 2003). Such conformational heterogeneity of Trp-9 could be present when gramicidin is bound to AOT reverse micelles. In the absence of additional data on conformational heterogeneity in the reverse micellar system, it is difficult to estimate how such heterogeneity would affect the observed fluorescence.
The overall invariance of REES with water content in the reverse micelle is somewhat surprising since the extent of REES has previously been shown to decrease with increasing wo for probes and peptides incorporated at the reverse micellar interface (Hof et al., 1997; Chattopadhyay et al., 2002; Raghuraman and Chattopadhyay, 2003) and in the water pool (Sarkar et al., 1996). This indicates that addition of water to the reverse micellar system in these cases leads to a reduction in the overall motional restriction experienced by the reorienting solvent molecules in the region of localization of the fluorophore. However, this has been shown to be not true for probes localized in the deeper acyl chain regions of the reverse micellar assembly. Thus, in case of the cholesterol analog 25-[N-[(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-methyl]amino]-27-norcholesterol (NBD-cholesterol) in which the fluorescent NBD moiety is positioned at a deeper acyl chain location in the reverse micellar assembly, the extent of REES increases with increasing wo (Kelkar and Chattopadhyay, 2004). This implies that the rate of solvent relaxation (reorientation) varies with probe location in the reverse micellar assembly. In the background of these results, our result of relative invariance of the magnitude of REES with increasing wo in the case of gramicidin in AOT reverse micelles, presents an interesting case. Gramicidin is a multitryptophan protein (Trp-9, 11, 13, and 15) and the location of these tryptophan residues would be heterogeneous in the reverse micelle. Whereas the carboxy terminal tryptophan (Trp-15) would occupy an interfacial position, the tryptophan residue at position 9 (Trp-9) would be placed in a relatively deep acyl chain region of the reverse micelle in the single-stranded β6.3 conformation. The overall variation in the extent of REES with increasing wo would then be dependent on the average of the variations with individual tryptophans. This could possibly explain the apparent insensitivity of the magnitude of REES to increasing wo for gramicidin in AOT reverse micelles.
It should be mentioned here that the apparent rotational correlation times reported by us (Fig. 8) are not exact since Perrin's equation is strictly applicable only in case of isotropic rotors (Lakowicz, 1999). Nonetheless, it is assumed here that this equation will apply to a first approximation. The presence of multiple tryptophans could be an additional complication. However, this would be minimized since we have used mean fluorescence lifetimes for calculating apparent rotational correlation times. Interestingly, a comparison with the reported global correlation times for the gramicidin channel (Lee et al., 1993) show that the rotational correlational times reported here are shorter by several orders of magnitude. This could be due to the fact that the global correlation times correspond to gramicidin in a membrane bilayer which offers greater motional restriction than reverse micelles. Another contributing factor could be the relatively high peptide/lipid ratio in the case of gramicidin incorporated in the membrane bilayer.
In conclusion, gramicidin bound to reverse micelles represents a convenient system to monitor the effect of graded hydration on the conformation and dynamics of this important ion channel peptide. These results are significant in understanding the interaction of gramicidin with membrane-mimetic media under conditions of varying hydration. More importantly, our results are relevant in the general context of ion channel dynamics with altered hydration since gramicidin remains the best characterized ion channel (Andersen and Koeppe, 1992).
Acknowledgments
D.A.K. thanks the Council of Scientific and Industrial Research for the award of a Senior Research Fellowship. We thank H. Raghuraman for helpful discussion, Y. S. S. V. Prasad and G. G. Kingi for technical help, and members of our laboratory for critically reading the manuscript.
This work was supported by the Council of Scientific and Industrial Research, Government of India.
References
- Andersen, O. S., and R. E. Koeppe. 1992. Molecular determinants of channel function. Physiol. Rev. 72:89–158. [DOI] [PubMed] [Google Scholar]
- Andersen, O. S., G. Saberwal, D. V. Greathouse, and R. E. Koeppe. 1996. Gramicidin channels—a solvable membrane “protein” folding problem. Indian J. Biochem. Biophys. 33:331–342. [PubMed] [Google Scholar]
- Arumugam, S., S. Pascal, C. L. North, W. Hu, K.-C. Lee, M. Cotten, R. R. Ketchem, F. Xu, M. Brenneman, F. Kovacs, F. Tian, A. Wang, S. Huo, and T. A. Cross. 1996. Conformational trapping in a membrane environment: a regulatory mechanism for protein activity? Proc. Natl. Acad. Sci. USA. 93:5872–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, T. W., O. S. Andersen, and B. Roux. 2003. Structure of gramicidin A in a lipid bilayer environment determined using molecular dynamics simulations and solid-state NMR data. J. Am. Chem. Soc. 125:9868–9877. [DOI] [PubMed] [Google Scholar]
- Bevington, P. R. 1969. Data Reduction and Error Analysis for the Physical Sciences. McGraw-Hill, New York.
- Bhattacharyya, K. 2003. Solvation dynamics and proton transfer in supramolecular assemblies. Acc. Chem. Res. 36:95–101. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya, K., and B. Bagchi. 2000. Slow dynamics of constrained water in complex geometries. J. Phys. Chem. A. 104:10603–10613. [Google Scholar]
- Bizzarri, A. R., and S. Cannistraro. 2002. Molecular dynamics of water at the protein-solvent interface. J. Phys. Chem. B. 106:6617–6633. [Google Scholar]
- Brubach, J.-B., A. Mermet, A. Filabozzi, A. Gerschel, D. Lairez, M.P. Krafft, and P. Roy. 2001. Dependence of water dynamics upon confinement size. J. Phys. Chem. B. 105:430–435. [Google Scholar]
- Callis, P. R. 1997. 1La and 1Lb transitions of tryptophan: applications of theory and experimental observations to fluorescence of proteins. Methods Enzymol. 278:113–150. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay, A. 2003. Exploring membrane organization and dynamics by the wavelength-selective fluorescence approach. Chem. Phys. Lipids. 122:3–17. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay, A., and S. Mukherjee. 1999. Red edge excitation shift of a deeply embedded membrane probe: implications in water penetration in the bilayer. J. Phys. Chem. B. 103:8180–8185. [Google Scholar]
- Chattopadhyay, A., S. Mukherjee, and H. Raghuraman. 2002. Reverse micellar organization and dynamics: a wavelength-selective fluorescence approach. J. Phys. Chem. B. 106:13002–13009. [Google Scholar]
- Chen, G. C., and J. T. Yang. 1977. Two-point calibration of circular dichrometer with d-10-camphorsulphonic acid. Anal. Lett. 10:1195–1207. [Google Scholar]
- Colombo, M. F., D. C. Rau, and V. A. Parsegian. 1992. Protein solvation in allosteric regulation: a water effect on hemoglobin. Science. 256:655–659. [DOI] [PubMed] [Google Scholar]
- Cooper, E. C., and L. Y. Jan. 1999. Ion channel genes and human neurological disease: recent progress, prospects and challenges. Proc. Natl. Acad. Sci. USA. 96:4759–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demchenko, A. P. 2002. The red-edge effects: 30 years of exploration. Luminescence. 17:19–42. [DOI] [PubMed] [Google Scholar]
- Doyle, D. A., J. M. Cabral, R. A. Pfuetzner, A. Kuo, J. M. Gulbis, S. L. Cohen, B. T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. [DOI] [PubMed] [Google Scholar]
- Eastoe, J., W. K. Young, B. H. Robinson, and D. C. Steytler. 1990. Scattering studies of microemulsions in low-density alkanes. J. Chem. Soc. Faraday Trans. 86:2883–2889. [Google Scholar]
- Eftink, M. R., L. A. Selvidge, P. R. Callis, and A. A. Rehms. 1990. Photophysics of indole derivatives: experimental resolution of La and Lb transitions and comparison with theory. J. Phys. Chem. 94:3469–3479. [Google Scholar]
- Essen, L.-O., R. Siegert, W. D. Lehmann, and D. Oesterhelt. 1998. Lipid patches in membrane protein oligomers: crystal structure of the bacteriorhodopsin-lipid complex. Proc. Natl. Acad. Sci. USA. 95:11673–11678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faeder, J., and B. M. Ladanyi. 2000. Molecular dynamics simulations of the interior of aqueous reverse micelles. J. Phys. Chem. B. 104:1033–1046. [Google Scholar]
- Fenimore, P. W., H. Frauenfelder, B. H. McMohan, and F. G. Parak. 2002. Slaving: solvent fluctuations dominate protein dynamics and function. Proc. Natl. Acad. Sci. USA. 99:16047–16051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, M. L. 2004. Gate expectations. Nature. 430:153–155. [DOI] [PubMed] [Google Scholar]
- Granick, S. 1991. Motions and relaxations of confined liquids. Science. 253:1374–1379. [DOI] [PubMed] [Google Scholar]
- Grinvald, A., and I. Z. Steinberg. 1974. On the analysis of fluorescence decay kinetics by the method of least-squares. Anal. Biochem. 59:583–598. [DOI] [PubMed] [Google Scholar]
- Hazra, P., and N. Sarkar. 2001. Intramolecular charge transfer processes and solvation dynamics of coumarin 490 in reverse micelles. Chem. Phys. Lett. 342:303–311. [Google Scholar]
- Ho, C., and C. D. Stubbs. 1992. Hydration at the membrane protein-lipid interface. Biophys. J. 63:897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof, M., P. Lianos, and A. Laschewsky. 1997. An amphiphilic hemicyanine dye employed as a sensitive probe of water in reverse AOT micelles. Langmuir. 13:2181–2183. [Google Scholar]
- Ikushima, Y., N. Saito, and M. Arai. 1997. The nature and structure of water/AOT/ethane (W/O) microemulsion under supercritical conditions studied by high-pressure FT-IR spectroscopy. J. Colloid Interface Sci. 186:254–263. [DOI] [PubMed] [Google Scholar]
- Jain, T. K., M. Varshney, and A. Maitra. 1989. Structural studies of aerosol OT reverse micellar aggregates by FT-IR spectroscopy. J. Phys. Chem. 93:7409–7416. [Google Scholar]
- Kelkar, D. A., and A. Chattopadhyay. 2004. Depth-dependent solvent relaxation in reverse micelles: a fluorescence approach. J. Phys. Chem. B. 108:12151–12158. [Google Scholar]
- Ketchem, R. R., W. Hu, and T. A. Cross. 1993. High-resolution conformation of gramicidin A in a lipid bilayer by solid-state NMR. Science. 261:1457–1460. [DOI] [PubMed] [Google Scholar]
- Killian, J. A. 1992. Gramicidin and gramicidin-lipid interactions. Biochim. Biophys. Acta. 1113:391–425. [DOI] [PubMed] [Google Scholar]
- Killian, J. A., K. U. Prasad, D. Hains, and D. W. Urry. 1988. The membrane as an environment of minimal interconversion. A circular dichroism study on the solvent dependence of the conformational behavior of gramicidin in diacylphosphatidylcholine model membranes. Biochemistry. 27:4848–4855. [DOI] [PubMed] [Google Scholar]
- Kirby, E. P., and R. F. Steiner. 1970. The influence of solvent and temperature upon the fluorescence of indole derivatives. J. Phys. Chem. 74:4480–4490. [Google Scholar]
- Koeppe, R. E., and O. S. Andersen. 1996. Engineering the gramicidin channel. Annu. Rev. Biophys. Biomol. Struct. 25:231–258. [DOI] [PubMed] [Google Scholar]
- Koeppe, R. E., H. Sun, P. C. A. van der Wel, E. M. Scherer, P. Pulay, and D. V. Greathouse. 2003. Combined experimental/theoretical refinement of indole ring geometry using deuterium magnetic resonance and ab initio calculations. J. Am. Chem. Soc. 125:12268–12276. [DOI] [PubMed] [Google Scholar]
- Kouyama, T., T. Nishikawa, T. Tokuhisa, and H. Okumura. 2004. Crystal structure of the L intermediate of bacteriorhodopsin: evidence for vertical translocation of a water molecule during the proton pumping cycle. J. Mol. Biol. 335:531–546. [DOI] [PubMed] [Google Scholar]
- Lakowicz, J. R. 1999. Principles of Fluorescence Spectroscopy. Kluwer-Plenum Press, New York.
- Lampert, R. A., L. A. Chewter, D. Phillips, D. V. O'Connor, A. J. Roberts, and S. R. Meech. 1983. Standards for nanosecond fluorescence decay time measurements. Anal. Chem. 55:68–73. [Google Scholar]
- Lee, K.-C., W. Hu, and T. A. Cross. 1993. 2H NMR determination of the global correlation time of the gramicidin channel in a lipid bilayer. Biophys. J. 65:1162–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinger, N. E. 2002. Water in confinement. Science. 298:1722–1723. [DOI] [PubMed] [Google Scholar]
- LoGrasso, P. V., F. Moll, and T. A. Cross. 1988. Solvent history dependence of gramicidin A conformations in hydrated lipid bilayers. Biophys. J. 54:259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisi, P. L., M. Giomini, M. P. Pileni, and B. H. Robinson. 1988. Reverse micelles as hosts for proteins and small molecules. Biochim. Biophys. Acta. 947:209–246. [DOI] [PubMed] [Google Scholar]
- Luisi, P. L., and L. J. Magid. 1986. Solubilization of enzymes and nucleic acids in hydrocarbon micellar solutions. CRC Crit. Rev. Biochem. 20:409–475. [DOI] [PubMed] [Google Scholar]
- Mattos, C. 2002. Protein-water interactions in a dynamic world. Trends Biochem. Sci. 27:203–208. [DOI] [PubMed] [Google Scholar]
- McAuley, K. E., P. K. Fyfe, J. P. Ridge, N. W. Isaacs, R. J. Cogdell, and M. R. Jones. 1999. Structural details of an interaction between cardiolipin and an integral membrane protein. Proc. Natl. Acad. Sci. USA. 96:14706–14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentré, P. 2001. Water in the cell. Cell. Mol. Biol. 47:709–970. [PubMed] [Google Scholar]
- Miloshevsky, G. V., and P. C. Jordan. 2004. Permeation in ion channels: the interplay of structure and theory. Trends Neurosci. 27:308–314. [DOI] [PubMed] [Google Scholar]
- Mukherjee, S., and A. Chattopadhyay. 1994. Motionally restricted tryptophan environments at the peptide-lipid interface of gramicidin channels. Biochemistry. 33:5089–5097. [DOI] [PubMed] [Google Scholar]
- Mukherjee, S., and A. Chattopadhyay. 1995. Wavelength-selective fluorescence as a novel tool to study organization and dynamics in complex biological systems. J. Fluoresc. 5:237–246. [DOI] [PubMed] [Google Scholar]
- O'Connell, A. M., R. E. Koeppe, and O. S. Andersen. 1990. Kinetics of gramicidin channel formation in lipid bilayers: transmembrane monomer association. Science. 250:1256–1259. [DOI] [PubMed] [Google Scholar]
- O'Connor, D. V., and D. Phillips. 1984. Time-Correlated Single Photon Counting. Academic Press, London. 180–189.
- Okada, T., Y. Fujiyoshi, M. Silow, J. Navarro, E. M. Landau, and Y. Shichida. 2002. Functional role of internal water molecules in rhodopsin revealed by x-ray crystallography. Proc. Natl. Acad. Sci. USA. 99:5982–5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoian, G. A., J. Ulander, M. P. Eastwood, Z. Luthey-Schulten, and P. G. Wolynes. 2004. Water in protein structure prediction. Proc. Natl. Acad. Sci. USA. 101:3352–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast, F. G. 1991. Time-resolved fluorescence techniques: methods and applications in biology. Curr. Opin. Struct. Biol. 1:1054–1059. [Google Scholar]
- Raghuraman, H., and A. Chattopadhyay. 2003. Organization and dynamics of melittin in environments of graded hydration: a fluorescence approach. Langmuir. 19:10332–10341. [Google Scholar]
- Raghuraman, H., D. A. Kelkar, and A. Chattopadhyay. 2003. Novel insights into membrane protein structure and dynamics utilizing the wavelength-selective fluorescence approach. Proc. Ind. Natl. Sci. Acad. A. 69:25–35. [Google Scholar]
- Ramachandran, G. N., and R. Chandrasekaran. 1972. Conformation of peptide chains containing both L- and D-residues: part I – helical structures with alternating L- and D- residues with special reference to the LD-ribbon and the LD-helices. Ind. J. Biochem. Biophys. 9:1–11. [PubMed] [Google Scholar]
- Rawat, S. S., D. A. Kelkar, and A. Chattopadhyay. 2004. Monitoring gramicidin conformations in membranes: a fluorescence approach. Biophys. J. 87:831–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees, D. C., G. Chang, and R. H. Spencer. 2000. Crystallographic analyses of ion channels: lessons and challenges. J. Biol. Chem. 275:713–716. [DOI] [PubMed] [Google Scholar]
- Ruggiero, A. J., D. C. Todd, and G. R. Fleming. 1990. Subpicosecond fluorescence anisotropy studies of tryptophan in water. J. Am. Chem. Soc. 112:1003–1014. [Google Scholar]
- Salom, D., C. Abad, and L. Braco. 1992. Characterization of gramicidin A in an inverted micellar environment. A combined high-performance liquid chromatographic and spectroscopic study. Biochemistry. 31:8072–8079. [DOI] [PubMed] [Google Scholar]
- Sankararamakrishnan, R., and M. S. P. Sansom. 1995. Water-mediated conformational transitions in nicotinic receptor M2 helix bundles: a molecular dynamics study. FEBS Lett. 377:377–382. [DOI] [PubMed] [Google Scholar]
- Sarkar, M., J. G. Ray, and P. K. Sengupta. 1996. Luminescence behaviour of 7-hydroxyflavone in aerosol OT reverse micelles: excited-state proton transfer and red-edge excitation effects. J. Photochem. Photobiol. A. 95:157–160. [Google Scholar]
- Sengupta, B., and P. K. Sengupta. 2000. Influence of reverse micellar environments on the fluorescence emission properties of tryptophan octyl ester. Biochem. Biophys. Res. Commun. 277:13–19. [DOI] [PubMed] [Google Scholar]
- Souto, A. L. C. F., and A. S. Ito. 2000. Tryptophan fluorescence studies of melanotropins in the amphiphile-water interface of reversed micelles. Eur. Biophys. J. 29:38–47. [DOI] [PubMed] [Google Scholar]
- Stutts, M. J., C. M. Canessa, J. C. Olsen, M. Hamrick, J. A. Cohn, B. C. Rossier, and R. C. Boucher. 1995. CFTR as a cAMP-dependent regulator of sodium channels. Science. 269:847–850. [DOI] [PubMed] [Google Scholar]
- Timasheff, S. N. 2002. Protein hydration, thermodynamic binding and preferential hydration. Biochemistry. 41:13473–13482. [DOI] [PubMed] [Google Scholar]
- Urry, D. W. 1971. The gramicidin A transmembrane channel: a proposed π(L,D) helix. Proc. Natl. Acad. Sci. USA. 68:672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez, D., J.-Y. Le Huérou, M. Gindre, W. Urbach, and M. Waks. 2001. Hydration and protein folding in water and in reverse micelles: compressibility and volume changes. Biophys. J. 80:2751–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valeur, B., and G. Weber. 1978. A new red-edge effect in aromatic molecules: anomaly of apparent rotation revealed by fluorescence polarization. J. Chem. Phys. 69:2393–2400. [Google Scholar]
- Veatch, W. R., E. T. Fossel, and E. R. Blout. 1974. The conformation of gramicidin A. Biochemistry. 13:5249–5256. [DOI] [PubMed] [Google Scholar]
- Venables, D. S., K. Huang, and C. A. Schmuttenmaer. 2001. Effect of reverse micellar size on the librational band of confined water and methanol. J. Phys. Chem. B. 105:9132–9138. [Google Scholar]
- Wallace, B. A. 2000. Common structural features in gramicidin and other ion channels. Bioessays. 22:227–234. [DOI] [PubMed] [Google Scholar]
- Xu, F., and T. A. Cross. 1999. Water: foldase activity in catalyzing polypeptide conformational rearrangements. Proc. Natl. Acad. Sci. USA. 96:9057–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, G.-W., G.-Z. Li, and W. J. Chen. 2002. Fourier transform infrared investigation on water states and the conformations of aerosol-OT in reverse microemulsions. Langmuir. 18:4566–4571. [Google Scholar]