Abstract
It has been known that the structural transition from PrPC to PrPSc leads to the prion formation. This putative conformational change challenges the central dogma of the protein folding theory—“one sequence, one structure.” Generally, scientists believe that there must be either a posttranslational modification or environmental factors involved in this event. However, all of the efforts to solve the mystery of the PrPC to PrPSc transition have ended in vain so far. Here we provide evidence linking O-linked glycosylation to the structural transition based on prion peptide studies. We find that the O-linked α-GalNAc at Ser-135 suppresses the formation of amyloid fibril formation of the prion peptide at physiological salt concentrations, whereas the peptide with the same sugar at Ser-132 shows the opposite effect. Moreover, this effect is sugar specific. Replacing α-GalNAc with β-GlcNAc does not yield the same effect.
The prion protein (PrP, 254 aa for hamster PrP) has been found to be associated with the prion infectivity. The normal product of the prion gene is expressed as a glycophosphatidyl inositol-anchored glycoprotein on the outer cell membrane (1–3). The solution NMR structure of recombinant syrian hamster prion protein rPrP(90–231) was determined in 1997 (4). The secondary structure consists of three α-helices (144–153, 172–194, 200–227) interdispersed between two short β-strands (129–131, 161–163); the N terminus (90–111) is largely unstructured (Fig. 1). A posttranslational process that converts the cellular prion protein (PrPC) to the abnormal, insoluble, pathogenic isoform (PrPSc) has been implicated in the prion formation during the development of prion diseases.
Figure 1.
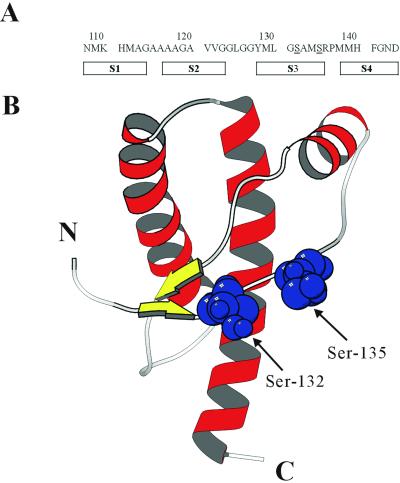
(A) The amino acid sequence of PrP (108–144). Two putative O-linked glycosylation sites, Ser-132 and Ser-135, are underlined. The predicted secondary-structure profile of PrPSc is also shown. (B) The solution structure of recombinant PrP (sequence 125–228) (PDB ID code 1B10) is produced by using the program molscript (31). β-Strands and α-helices are displayed in yellow and red, respectively. Ser-132 and Ser-135 are shown in blue by space-filled presentation.
CD spectroscopy and Fourier transform infrared spectroscopy studies have indicated that approximately half of the α-helical and coiled structure in PrPC is transformed into β-sheet in PrPSc (5). However, the thermodynamics of the α-to-β transition, including the factors that influence it, remain unknown. PrPC has a high turnover rate in vivo. This high turnover makes the purification of PrPC extremely difficult (6). The low yield of purified PrPC from animal brains or low level of expression in mammalian cells as well as the insolubility of PrPSc have remained a bottleneck in prion research. It has been difficult to discern whether there are chemical modifications in PrPC that are absent in PrPSc or vice versa. Although Edman sequencing data suggest that the amino acid sequence of PrPSc is identical with the translational product of the prion gene (7), we cannot rule out the possibility of low levels of unidentified chemical modifications in PrPC or PrPSc as the culprit of the medical malady. To date, the search for differential modifications between PrPC and PrPSc has focused on the two N-linked glycosylation sites at N181 and N197. However, based on the results of tunicamycin treatment, an N-linked glycosylation inhibitor (8), as well as experiments on the expression of unglycosylated mutated prion protein in neuroblastoma cells (9), N-linked glycans are no longer linked to the disease.
Animal experiments have also suggested that the so-called “species barriers” in the transmission of the prion diseases reside in differences in the primary structure of the PrP between the inoculum and the host, with prion propagation proceeding most efficiently when the sequences of the two are identical. Moreover, Kaneko and coworkers (10, 11) have noted that structural differences or changes in the peptide domain corresponding to human prion sequence 90–144 alone are directly relevant to the transmission of prion diseases. In light of these observations, most of the structural studies on prion peptides have focused on this region.
Spectroscopic studies indicate that the peptides derived from this region can form α-helical or β-sheet structures in response to variations in the solvent system. For example, the peptides 104–122, 129–141, and 109–141 can be induced to form α-helical structures in aqueous solution in the presence of helix-promoting organic solvents like hexafluoroisopropanol, trifluoroethanol, or detergents such as SDS and dodecyl phosphocholine. On the other hand, the peptide 90–145 forms β-structures when dissolved in aqueous solution at physiological salt concentrations, or in 50% acetonitrile (12). In light of the unique capability of this region of the prion protein toward α-to-β conversion, we believe that this region plays a key role in the posttranslational processes that induce PrPC to form PrPSc (13).
In contrast to the N-linked glycosylation, the possibility that an O-linked sugar might be involved in the PrPC–PrPSc conversion has not been discussed. This is because O-linked glycans have not been detected in isolated PrPC or in PrPSc. There could be a number of reasons for this. First, the purification of PrPC relies on the use of monoclonal antibodies, which do not necessarily catch all populations. Second, O-glycans are more vulnerable toward degradation during hydrazinolysis in the glycan assay. Third, the O-glycosylated form might exist as a minor population and could be easily missed. The sequence 108–144 of prion protein contains four putative β-strands (S1, S2, S3, and S4) in PrPSc (14). Within this sequence, there are two serines, Ser-132 and Ser-135, which could be glycosylated. Both residues are available as potential O-glycosylated sites because they reside in the exterior of PrPC (Fig. 1).
In this work, we have examined the effects of O-linked α-GalNAc, β-GalNAc, and β-GlcNAc at Ser-132 and Ser-135 on the conformation and solution properties of the prion peptide 108–144.
Materials and Methods
Peptide Synthesis.
The synthesized prion peptide 108–144, denoted PrP(108–144), was prepared by the batch fluorenylmethoxycarbonyl (fmoc)-polyamide method. The N-terminal end of the peptide was acetylated and the C-terminal end amidated to alleviate any electrostatic interactions involving the two ends. To synthesize the glycosylated peptides, Fmoc-Ser(Ac3-α- d-GalNHAc)-OH, Fmoc-Ser(Ac3-β-d-GlcNHAc)-OH and Fmoc-Ser(Ac3-β-d-GalNHAc)-OH were used instead of Fmoc-Ser(tBu)-OH. The peptide with an α-GalNAc at Ser-132 and Ser-135 were denoted S132-α-GalNAc and S135-α-GalNAc, respectively. The peptide with a β-GlcNAc at Ser-132 and Ser-135 were denoted S132-β-GlcNAc and S135-β-GlcNAc, respectively. The peptide with an unnatural β-GalNAc at Ser-135 was denoted S135-β-GalNAc. Fmoc-Ser(Ac3-α-d-GalNHAc)-OH, Fmoc-Ser(Ac3-β-d-GlcNHAc)-OH and Fmoc-Ser(Ac3-β-d-GalNHAc)-OH were prepared by modification of literature procedures (15–18).
CD Spectroscopy.
The peptides were dissolved in 20 mM NaOAc with 140 mM NaCl (pH 3.7) and incubated at room temperature. CD spectra were recorded in a π* CD spectrometer (Applied Photophysics, Surrey, U.K.). CD spectra were recorded in a 1-mm cell, between 190 and 250 nm, and at room temperature. A scan interval of 1 nm with an integration of 200,000 points was used. The spectrum of the same buffer was collected as a baseline and subtracted automatically.
NMR Spectroscopy.
All NMR spectra were recorded on a Bruker AM 500 NMR spectrometer. For one-dimensional spectra, peptides were dissolved in a buffer of 20 mM NaOAc-d3/140 mM NaCl to a final concentration of 0.88 mM. A 1/100 volume of sodium 3-(trimethylsilyl)-propionic-2, 2, 3, 3-d4 acid solution (0.75% in D2O) was added as an internal reference. The pH values were adjusted to 3.7 for higher solubility. Quoted pH values were not corrected for the D/H isotope effect. Samples were incubated at room temperature. Their solution 1H NMR spectra were recorded as a function of time at 298 K. For two-dimensional spectra, more concentrated samples were prepared (1 mM). Spectra were acquired with 2,000 data points in direct dimension and 512 increments in indirect dimension. 80-ms mixing time in total correlated spectroscopy (TOCSY) and 300-ms mixing time in NOESY were used. Data were processed by xwinnmr software (Bruker, Billerica, MA). The shifted square sine bell window functions in both dimensions were applied for all spectra. The ansig program (version 3.3) was used to assign the spectra (19).
Electron Microscopy (EM).
The peptides were dissolved in 20 mM NaOAc with 140 mM NaCl (pH 3.7 or 5). Except in the case of S132-α-GalNAc, the peptide solutions were incubated at room temperature in advance to promote the fibril formation. S132-α-GalNAc solution showed fibril structure under EM without advanced incubation. Peptide negative staining was performed on carbon-coated 300-mesh copper grids. Samples were absorbed for 1 min, stained with 2% uranyl acetate for another minute, and, after drying, were viewed in a ZEISS 902 electron microscope at 80 kV at standard magnification of ×50,000.
Results
Effects of α-GalNAc on Fibril Formation.
Not unexpectedly, EM of the PrP(108–144) revealed the formation of fibril structures (Fig. 2). The EM observation is corroborated by the negative ellipticity at 218 nm in the CD spectrum and a vibrational band at 1,627 cm−1 in the Fourier transform infrared spectrum, indicating that the PrP(108–144) had undergone β-structure formation after 14-day incubation (data not shown). As expected, fibril formation was enhanced at increasing peptide concentrations, indicating that the process is concentration dependent (Fig. 3). The solubility of the peptide decreases with the increase of the buffer pH from 3.0 to 6.0. To shorten the incubation time, the experiments were performed at pH 3.7.
Figure 2.
EM pictures of prion peptides. (A) PrP (108–144) at pH 5. (B) S135-α-GalNAc at pH 5. (C) S132-α-GalNAc at pH 3.7. (D) S135-β-GlcNAc at pH 3.7. The bars represent 150 nm in length.
Figure 3.
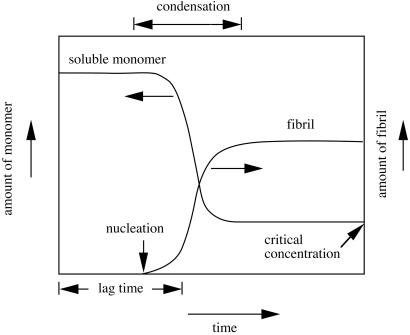
Concentration-dependent fibrillogenesis. Below the critical concentration, no fibril is formed. When the monomer concentration is higher than the critical concentration, the monomer nucleates and condenses to form the fibril. The fibrillogenesis continues until the equilibrium is achieved when the monomer concentration drops to or below the critical concentration. The length of time before fibril formation commences is called lag time.
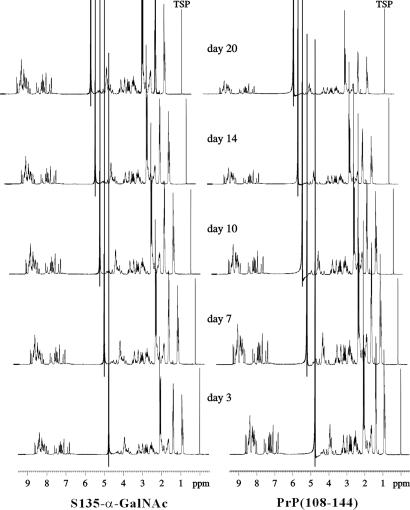
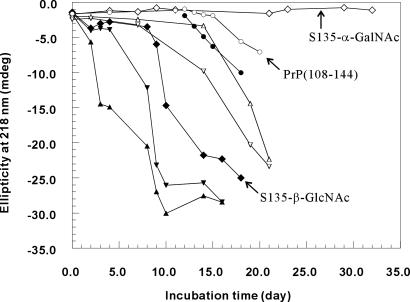
Surprisingly, the addition of an α-GalNAc group at Ser-135 suppressed the fibril formation. No fibril structure can be found under EM (Fig. 2). When the 1H NMR spectra of S135-α-GalNAc are compared closely with those of PrP(108–144) (Fig. 4), the most striking observation is the significant reduction in the signal intensity of the PrP(108–144) peptide, which becomes increasingly apparent for long incubation times. The intensity decreased with the propagation of amyloid formation and the concomitant reduction in the concentration of water-soluble or monomeric peptide, whereas that of S135-α-GalNAc remained the same. This result is consistent with irreversible aggregation of the PrP(108–144) peptide with time. The kinetics of interconversion between the water-soluble monomer and the aggregate is apparently slow so that only the monomeric peptide is observed in the high-resolution spectrum.
Figure 4.
The one-dimensional NMR spectra of PrP(108–144) and S135-α-GalNAc after different incubation times.
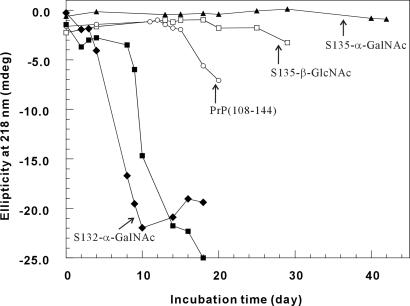
However, the inhibitory effect of α-GalNAc appears to be site-specific, because S132-α-GalNAc does not have the same effect. In fact, the fibrillization of S132-α-GalNAc is even faster than that of PrP(108–144) (Fig. 5). The addition of some fibril of S132-α-GalNAc to a solution of PrP(108–144) quickly initiates the fibrillogenesis. The similar heterogeneous seeding-effect is also noted for the other peptides (Fig. 6). Thus, the polymerization could be either a heterogeneously or homogeneously seeded process. In any case, that α-GalNAc at Ser-135 interrupts the fibril assembly, while the same glycosylation at Ser-132 accelerates the process, is startling indeed.
Figure 5.
The fibrillization time course as monitored by CD. ▴, 0.05 mM S135-α-GalNAc; □, 0.05 mM S135-β-GlcNAc; ○, 0.05 mM PrP(108–144); ■, 0.075 mM S135-β-GlcNAc; ⧫, 0.05 mM S132-α-GalNAc. On day 10, the sample for 0.05 mM S132-α-GalNAc had reached equilibrium.
Figure 6.
Heterogeneously seeded polymerization. ○, 0.05 mM PrP(108–144); ●, on day 12, S132-α-GalNAc fibril was added as a seed to the solution of 0.05 mM PrP(108–144); ◊, 0.075 mM S135-α-GalNAc; ▵, 0.075 mM S135-α-GalNAc + S135-β-GlcNAc fibril; ▿, 0.075 mM S135-α-GalNAc + PrP(108–144) fibril; ⧫, 0.075 mM S135-β-GlcNAc; ▴, 0.075 mM S135-β-GlcNAc + S132-α-GalNAc fibril; ▾, 0.075 mM S135-β-GlcNAc + PrP(108–144) fibril.
Effects of β-GlcNAc on Fibril Formation.
The apparent opposite effect of a single α-GalNAc when introduced to different positions suggests that the local conformation stability of this region is important in the nucleation step. To verify whether different sugar species can have the same effect, we synthesized another analog peptide with a β-GlcNAc at Ser-135 (S135-β-GlcNAc). Like α-GalNAc, β-GlcNAc is a common sugar unit for O-linked glycosylation. In contrast to S135-α-GalNAc, which remained a random-coil structure even after 6 months, the CD spectrum of S135-β-GlcNAc showed a strong negative ellipticity at 218 nm after 2 weeks of incubation (Fig. 7). Time course experiments indicate that the rate of fibril formation is as follows: S132-α-GalNAc > PrP(108–144) > S135-β-GlcNAc ≫ S135-α-GalNAc. These interesting contrasts suggest that the effects of glycosylation on the structural transition and fibrillization are site-specific and sugar-specific.
Figure 7.
Comparison of the fibrillization propensity between S135-α-GalNAc and S135-β-GlcNAc. The CD spectra of these peptides in 20 mM NaOAc buffer with 140 mM NaCl (pH 3.7) were recorded after different incubation times. Solid line, 0.1 mM S135-α-GalNAc incubated for 6 months; dashed line, 0.075 mM S135-β-GlcNAc incubated for 2 weeks only.
Sugar Specificity.
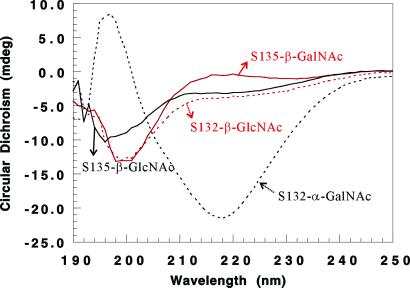
To gain more insights into the sugar specificity, the peptide with a β-GlcNAc at Ser-132, denoted S132-β-GlcNAc, was also prepared. Similar to S135-β-GlcNAc, S132-β-GlcNAc did not appear to exhibit any dramatically different behavior toward fibril formation compared with PrP(108–144) (Fig. 8). It seems that α-GalNAc has a greater effect on either promoting or attenuating fibrillization, than β-GlcNAc. The only differences between α-GalNAc and β-GlcNAc are the orientation of the glycosidic bond and the hydroxyl group at C-4 (Scheme S1).
Figure 8.
The effect of sugar configuration on fibrillization. The CD spectra of 0.05 mM peptides in 20 mM NaOAc buffer with 140 mM NaCl (pH 3.7) were recorded after different incubation times. Black solid line, S135-β-GlcNAc incubated for 4 weeks; red solid line, S135-β-GalNAc incubated for 4 weeks; black dashed line, S132-α-GalNAc incubated for 2 weeks only; red dashed line, 0.05 mM S132-β-GlcNAc incubated for 4 weeks.
Scheme 1.
β-GlcNAc (left) and α-GalNAc (right) as linked to the side-chain of a serine residue via the glycosidic bond.
To ascertain whether or not the sugar configuration affects the fibrillization, the peptide with a β-GalNAc at Ser-135, denoted S135-β-GalNAc, was synthesized. Surprisingly, S135-β-GalNAc did not form any fibrils even after the peptide solution was incubated for 4 weeks, the time course over which S135-β-GlcNAc has already started fibrillization (Fig. 8). Although we do not know for sure yet whether or not S135-β-GalNAc will ever undergo fibrillization and behave like S135-α-GalNAc, it is clear that the orientation of the hydroxyl group at C-4 is important in affecting the process.
The Solution Structures of the Peptides.
Although an α-GalNAc attached at Ser-135 dramatically affected the fibrillogenesis process of the peptide, there was no evidence from the NMR of these peptides that glycosylation affected their solution structures in a major way. For example, when the two-dimensional NMR spectrum of S135-GalNAc was compared with that of PrP (108–144), only the Hα of Ser-135 was downfield shifted by about 0.2 ppm because of the glycosylation. Except for the amide hydrogens of Ser-135 and Arg-136, which also exhibited downfield shifts of about 0.2 ppm, most of these amide protons were not shifted significantly (data not shown). Similarly, spectral changes were noted for the other glycosylated peptides only in the vicinity of the glycosylation site.
Discussion
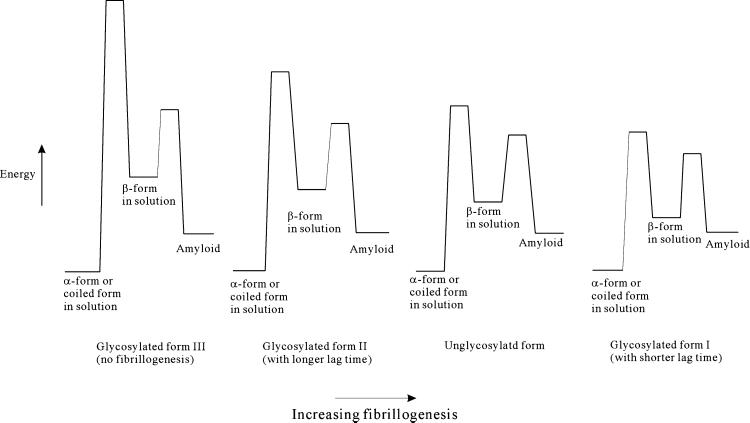
Although covalent attachment of the sugar moiety exerts only a minor influence on the solution structure of the water-soluble monomer prion peptide, glycosylation has a dramatic effect on peptide fibrillogenesis, as we have shown in this study. The structural perturbations induced by the glycosylation are primarily local. Thus, the effects of the glycosylation relate to the conformational properties of the peptides in solution (as opposed to their equilibrium structures in solution), specifically, the accessibility of other structural forms to the peptide under ambient conditions. Evidently, for fibrillization, the ensemble of conformational states associated with the β-structure must become thermally accessible, or become sufficiently accessible to populate these states to seed the nucleation of amyloid fibrils. Alternatively, the barrier for transition between the random-coil prion peptide and the β-structure must be lowered to facilitate the structural transition between the two forms (Fig. 9). Apparently, adding an α-GalNAc at Ser-132 stabilizes the β-structure relative to the random coil, rendering the structural conversion from the random coil to the β-structure kinetically more facile, or thermodynamically more favorable, whereas adding the same sugar at Ser-135 has the opposite effect. Prion peptides with different glycosylations show different lag-times of amyloid formation, indicating that glycosylation must affect the barriers of the nucleation processes that ultimately culminate in fibrillogenesis as well.
Figure 9.
The conformational energy diagrams of the prion peptides with and without glycosylation.
Thus, glycosylation alters the kinetic equilibrium between the water-soluble monomer (α-helix conformation in the case of prion protein, or random coil conformation in the case of prion peptide) and the prion-induction form (β-sheet conformation), as well as the kinetics of the nucleation processes that lead to fibrillogenesis. The stability and the solubility of the prion protein or peptide species, as well as their resistance to proteases, are critical to the kinetics of the nucleation process, as a critical concentration must be reached before nucleation could occur. The formation of these nucleation centers, which probably exist only in minor amounts and are difficult to detect, is critical to the initiation of the seeded polymerization. Fig. 10 depicts a noncatalytic nucleated polymerization model (20–22) that might be used to illustrate the nature of the seeded polymerization. Amyloid formation might be promoted by the seeding effect of a minor amount of nuclei. The nuclei might come from the local increase in the protein concentration over the critical concentration or the decrease of the critical concentration in response to the cellular environmental change (homogeneous seeding). The formation of these nuclei might be accelerated by the aberrant glycosylation machinery, or introduced from an external environment through transfection (heterogeneous seeding).
Figure 10.
A model for the noncatalytic seeded polymerization. This model supposes that the various glycosylated forms have different ability to go through the coil-to-β (or α-to-β) structural transition and fibrillization. The fibrillogenesis process can be homogeneously seeded, or heterogeneously seeded by the nuclei provided from the external environment or formed because of aberrant glycosylation.
It has been suggested that prion propagation is facilitated by homology within the central sequence of PrPC and PrPSc. The transmission of prion diseases between species is often much less efficient than transmission within the species. The requirements for sequence similarity might offer clues for the species-barrier during the disease transmission. In other words, in the case of prion infection, the species barrier depends on the efficiency difference between homogeneous seeding and heterogeneous seeding.
Studies of transgenic mice indicate that a factor designated “protein X” appears to interact with PrPC and is involved in the conversion of PrPC to PrPSc (23). Rudd and his coworkers (24) have compared the glycan composition between PrPC and PrPSc. They have found that PrPSc contains decreased levels of glycans with bisecting sugars and increased levels of tri- and tetra-antennary sugars, and have suggested that the glycosylation machinery might be perturbed during the PrPC-PrPSc conversion (24). Here we have shown that only one sugar unit can affect the formation of amyloid fibril. This finding raises the question as to whether glycosylation-related enzymes are the mysterious protein X that initiates the prion formation.
In this article, we report conformational effects on the prion peptide induced by the covalent attachment of two monosaccharides: GlcNAc and GalNAc. Although GalNAc shows a more dramatic influence on the coil (or α)-to-β structural conversion of the prion peptide, the effects associated with this sugar are probably not physiological. In contrast, the fact that O-GlcNAc glycosylation has been found in many proteins, including the tau protein and the β-amyloid precursor membrane protein related to the neuron degenerative Alzheimer's disease, emphasizes the physiological significance of GlcNAc (25–28). O-linked GalNAc generally occurs in mucin-type glycoproteins that contain repeating units of Ser, Thr, and Pro in short regions of the peptide chain (29). Thus, we do not normally expect GalNAc to be associated with the α-to-β conversion of the prion protein in vivo, because there are only two Ser and one Pro in this polymorphic region. However, when it does happen, the effects should be dramatic, if not catastrophic, as we have demonstrated here. At this juncture, it is not possible to rule out this possibility one way or another, as our knowledge about O-glycosylation remains pretty limited.
Recently, Peretz and coworkers (30) attempted to find a way to prevent PrPC-PrPSc conversion by using various antibodies recognizing different epitopes of PrPC. Their conclusions point to the 132–140 domain of PrPC as closely related to the prion propagation. Interestingly, this region overlaps with our O-linked glycosylation sites. In the present study, we show that glycosylation in this region can affect the thermodynamics of the coil-to-β (or α-to-β) structural conversion of the prion peptide. This result might implicate or link the O-linked glycosylation with prion formation.
Acknowledgments
The electron microscopy was obtained on a ZEISS 902 electron microscope in the Institute of Molecular Biology, Academia Sinica, and we thank Dr. Sue Lin-Chao and Ms. Sue-Ping Lee for access to this facility as well as their kind assistance in the use of microscope. This work was supported by a Program Project grant awarded by Academia Sinica as well as a grant from the National Scientific Council, Taiwan (NSC90-2119-M001-010).
Abbreviations
- PrP
prion protein
- PrPC
normal PrP
- PrPSc
disease-causing isoform of PrP
- EM
electron microscopy
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Prusiner S B. Science. 1991;252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner S B. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prusiner S B, Scott M R, DeArmond S J, Cohen F E. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 4.James T L, Liu H, Ulyanov N B, Farr-Jones S, Zhang H, Donne D G, Kaneko K, Groth D, Mehlhorn I, Prusiner S B, Cohen F E. Proc Natl Acad Sci USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan K-M, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R J, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan K-M, Stahl N, Prusiner S B. Protein Sci. 1992;1:1343–1352. doi: 10.1002/pro.5560011014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stahl N, Baldwin M A, Teplow D B, Hood L, Gibson B W, Burlingame A L, Prusiner S B. Biochemistry. 1993;32:1991–2002. doi: 10.1021/bi00059a016. [DOI] [PubMed] [Google Scholar]
- 8.Taraboulos A, Rogers M, Borchelt D R, McKinley M P, Scott M, Serban D, Prusiner S B. Proc Natl Acad Sci USA. 1990;87:8262–8266. doi: 10.1073/pnas.87.21.8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korth C, Kaneko K, Prusiner S B. J Gen Virol. 2000;81:1555–1563. doi: 10.1099/0022-1317-81-10-2555. [DOI] [PubMed] [Google Scholar]
- 10.Kaneko K, Ball H L, Wille H, Zhang H, Groth D, Torchia M, Tremblay P, Safar J, Prusiner S B, DeArmond S J, et al. J Mol Biol. 2000;295:997–1007. doi: 10.1006/jmbi.1999.3386. [DOI] [PubMed] [Google Scholar]
- 11.Kaneko K, Peretz D, Pan K-M, Blochberger T C, Wille H, Gabizon R, Griffith O H, Cohen F E, Baldwin M A, Prusiner S B. Proc Natl Acad Sci USA. 1995;92:11160–11164. doi: 10.1073/pnas.92.24.11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H, Kaneko K, Nguyen J T, Livshits T L, Baldwin M A, Cohen F E, James T L, Prusiner S B. J Mol Biol. 1995;250:514–526. doi: 10.1006/jmbi.1995.0395. [DOI] [PubMed] [Google Scholar]
- 13.Harrison P M, Bamborough P, Daggett V, Prusiner S B, Cohen F E. Curr Opin Struct Biol. 1997;7:53–59. doi: 10.1016/s0959-440x(97)80007-3. [DOI] [PubMed] [Google Scholar]
- 14.Huang Z, Prusiner S B, Cohen F E. Fold Des. 1995;1:13–19. [PubMed] [Google Scholar]
- 15.Kuduk S D, Schwarz J B, Chen X-T, Glunz P W, Sames D, Ragupathi G, Livingston P O, Danishefsky S J. J Am Chem Soc. 1998;120:12474–12485. [Google Scholar]
- 16.Lemieux R U, Ratcliffe R M. Can J Chem. 1979;57:1244–1251. [Google Scholar]
- 17.Nakahara Y, Nakahara Y, Ito Y, Ogawa T. Carbohydr Res. 1998;309:287–296. doi: 10.1016/s0008-6215(98)00142-6. [DOI] [PubMed] [Google Scholar]
- 18.Seitz O, Wong C-H. J Am Chem Soc. 1997;119:8766–8776. [Google Scholar]
- 19.Kraulis P J. J Magnet Res. 1989;84:627–633. [Google Scholar]
- 20.Horiuchi M, Caughey B. Structure (London) 1999;7:R231–R240. doi: 10.1016/s0969-2126(00)80049-0. [DOI] [PubMed] [Google Scholar]
- 21.Caughey B. Nat Med. 2000;6:751–754. doi: 10.1038/77476. [DOI] [PubMed] [Google Scholar]
- 22.Harper J D, Lansbury P T., Jr Annu Rev Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- 23.Telling G C, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen F E, DeArmond S J, Prusiner S B. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 24.Rudd P M, Endo T, Colominas C, Groth D, Wheeler S F, Harvey D J, Wormald M R, Serban H, Prusiner S B, Kobata A, Dwek R A. Proc Natl Acad Sci USA. 1999;96:13044–13049. doi: 10.1073/pnas.96.23.13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Comer F I, Hart G W. J Biol Chem. 2000;275:29179–29182. doi: 10.1074/jbc.R000010200. [DOI] [PubMed] [Google Scholar]
- 26.Haltiwanger R S, Busby S, Grove K, Li S, Mason D, Medina L, Moloney D, Philipsberg G, Scartozzi R. Biochem Biophys Res Commun. 1997;231:237–242. doi: 10.1006/bbrc.1997.6110. [DOI] [PubMed] [Google Scholar]
- 27.Arnold C H, Johnson G V W, Cole R N, Dong D L Y, Lee M, Hart G W. J Biol Chem. 1996;271:28741–28744. doi: 10.1074/jbc.271.46.28741. [DOI] [PubMed] [Google Scholar]
- 28.Griffith L S, Mathes M, Schmitz B. J Neurosci Res. 1995;41:270–278. doi: 10.1002/jnr.490410214. [DOI] [PubMed] [Google Scholar]
- 29.Van den Steen P, Rudd P M, Dwek R A, Opdenakker G. Crit Rev Biochem Mol Biol. 1998;33:151–208. doi: 10.1080/10409239891204198. [DOI] [PubMed] [Google Scholar]
- 30.Peretz D, Williamson R A, Kaneko K, Vergara J, Leclerc E, Schmitt-Ulms G, Mehlhorn I R, Legname G, Wormald M R, Rudd P M, et al. Nature (London) 2001;412:739–743. doi: 10.1038/35089090. [DOI] [PubMed] [Google Scholar]
- 31.Kraulis P J. J Appl Cryst. 1991;24:946–950. [Google Scholar]