Abstract
DNA restriction fragments that are stably curved are usually identified by polyacrylamide gel electrophoresis because curved fragments migrate more slowly than normal fragments containing the same number of basepairs. In free solution, curved DNA molecules can be identified by transient electric birefringence (TEB) because they exhibit rotational relaxation times that are faster than those of normal fragments of the same size. In this article, the results observed in free solution and in polyacrylamide gels are compared for a highly curved 199-basepair (bp) restriction fragment taken from the VP1 gene in Simian Virus 40 (SV40) and various sequence mutants and insertion derivatives. The TEB method of overlapping fragments was used to show that the 199-bp fragment has an apparent bend angle of 46 ± 2° centered at sequence position 1922 ± 2 bp. Four unphased A- and T-tracts and a mixed A3T4-tract occur within a span of ∼60 bp surrounding the apparent bend center; for brevity, this 60-bp sequence element is called a curvature module. Modifying any of the A- or T-tracts in the curvature module by site-directed mutagenesis decreases the curvature of the fragment; replacing all five A- and T-tracts by random-sequence DNA causes the 199-bp mutant to adopt a normal conformation, with normal electrophoretic mobilities and birefringence relaxation times. Hence, stable curvature in this region of the VP1 gene is due to the five unphased A- and T- tracts surrounding the apparent bend center. Discordant solution and gel results are observed when long inverted repeats are inserted within the curvature module. These insertion derivatives migrate anomalously slowly in polyacrylamide gels but have normal, highly flexible conformations in free solution. Discordant solution and gel results are not observed if the insert does not contain a long inverted repeat or if the long inverted repeat is added to the 199-bp fragment outside the curvature module. The results suggest that long inverted repeats can form hairpins or cruciforms when they are located within a region of the helix backbone that is intrinsically curved, leading to large mobility anomalies in polyacrylamide gels. Hairpin/cruciform formation is not observed in free solution, presumably because of rapid conformational exchange. Hence, DNA restriction fragments that migrate anomalously slowly in polyacrylamide gels are not necessarily stably curved in free solution.
INTRODUCTION
The sequence-dependent curvature of DNA is thought to play an important role in many processes in the cell, including transcription (Marilley and Pasero, 1996; Perez-Martin and de Lorenzo, 1997; McGill and Fisher, 1998; Davis et al., 1999), replication (Eckdahl and Anderson, 1990; Zahn and Blattner, 1987), and recombination (Hizver et al., 2001). DNA curvature also appears to be involved in nucleosome positioning (Drew and Travers, 1985; Fitzgerald et al., 1994; Sivolob and Khrapunov, 1995) and the maintenance of nucleosome structure (Shrader and Crothers, 1990; Ohki et al., 1998). Studies of synthetic oligomers with various target sequences repeated in phase with the helix screw have shown that phased runs of 4–6 contiguous adenine residues (A-tracts) lead to stable curvature of the helix axis (reviewed in Crothers et al., 1990; Hagerman, 1990; Olson and Zhurkin, 1996; Schatzky-Schwartz et al., 1997; Allemann and Egli, 1997). The apparent bend angle ranges from 17° to 22° per A5-6-tract, depending on the length of the A-tract and the method of measurement (Levene et al., 1986; Koo et al., 1990; Ross et al., 1999; Podtelezhnikov et al., 2000; MacDonald et al., 2001; Barbič et al., 2003; Tchernaenko et al., 2003). Phased GGGCCC-tracts cause bends of similar magnitude in the opposite direction if the solution contains Mg2+ ions (Satchwell et al., 1986; Brukner et al., 1993, 1994; Dlakić and Harrington, 1995, 1996; Merling et al., 2003). Other sequence motifs can also lead to curvature of the helix backbone when repeated in phase with the helix screw, although the magnitude of curvature is relatively small (Bolshoy et al., 1991; McNamara and Harrington, 1991).
The rules that govern curvature in naturally occurring, mixed-sequence DNA are less well understood, because phased A- and/or G-tracts are relatively rare in genomic DNA. Curvature is often identified by the anomalously slow mobilities observed for certain restriction fragments in polyacrylamide gels (e.g., Marini et al., 1982; Stellwagen, 1983). The locus of curvature within a restriction fragment can be determined by the circular permutation assay (Wu and Crothers, 1984). In this assay, the sequence of a target DNA is circularly permuted with respect to its ends. If the target DNA is intrinsically curved, or contains a region of anisotropic (hinge-like) flexibility, the relative position of the curved region will vary in different permuted sequence isomers. The sequence isomer with the fastest mobility is the one with its locus of curvature located at or near one end of the fragment. For brevity, this sequence position will be described as the apparent bend center, even though the curvature of the helix axis might be delocalized due to the juxtaposition of several small, closely spaced bends.
Although the circular permutation assay is usually applied to DNA restriction fragments containing a few hundred basepairs, it can also be used to analyze kilobase-sized DNA molecules if the mobilities are measured in large-pore polyacrylamide gels (Stellwagen, 1995; Strutz and Stellwagen, 1996). The observed mobility patterns are independent of arbitrary gel running conditions, independent of pH over the range 6.5–8.6, and independent of the presence or absence of monovalent and some divalent ions in the buffer, indicating that the mobility patterns reflect the intrinsic curvature of the DNA molecules (Strutz and Stellwagen, 1996; Stellwagen, 2003). However, if the solution contains Mg2+ or Ca2+ ions the mobility maxima can be shifted by ∼100 basepairs (bp) and certain sequence isomers can exhibit multiple subbands, indicating the presence of conformational isomers in slow exchange (Stellwagen, 2000, 2003). Hence, the flexibility of large DNA molecules and the exact location(s) of the apparent bend center(s) can depend on the ionic composition of the solution.
In this work, the circular permutation assay in large-pore polyacrylamide gels was used to identify regions of curvature in the SV40 minichromosome. Restriction fragments containing these curved regions were then characterized by polyacrylamide gel electrophoresis and transient electric birefringence (TEB), two techniques that are sensitive to DNA conformation. TEB is particularly suitable to quantitate DNA curvature because it measures rotational relaxation times, which are approximately proportional to the third power of macromolecular length (Fredericq and Houssier, 1973). Curved DNA molecules have shorter end-to-end lengths than normal fragments containing the same number of basepairs and exhibit shorter rotational relaxation times (Hagerman, 1984; Koo et al., 1990; Stellwagen, 1991; Vacano and Hagerman, 1997; Lu et al., 2003b). Comparison of the relaxation times (τ) of normal and curved restriction fragments of the same size allows the apparent bend angle to be determined by the τ-ratio method (see Materials and Methods), if the locus of curvature is known (García Bernal and García de la Torre, 1980; Mellado and García de la Torre, 1982; Stellwagen, 1991; Vacano and Hagerman, 1997; Lu et al., 2003b). Global fitting of the τ-ratios of a series of overlapping restriction fragments allows the apparent bend angle and its sequence position to be separately determined (Nickol and Rau, 1992; Lu et al., 2003b).
As shown below, the SV40 genome contains four regions of curvature that can be detected by the circular permutation assay. TEB studies showed that the relaxation times observed for a 199-bp restriction fragment taken from the middle of the VP1 gene were relatively independent of the ionic composition of the solution, suggesting that the DNA helix is intrinsically curved at this site. Further analysis by the method of overlapping fragments (Lu et al., 2003b) showed that an apparent bend of 46 ± 2° is located at sequence position 1922 ± 2 bp. The sequence surrounding the apparent bend center contains four unphased A- and T-tracts and a mixed A3T4 sequence within a span of 59 bp; for brevity, this sequence element is called the curvature module. Modifying any of the A- or T-tracts in the curvature module by site-directed mutagenesis decreases the apparent curvature of the modified 199-bp fragment; replacing all of the A- and T-tracts by mixed-sequence DNA causes the modified fragment to exhibit normal polyacrylamide gel mobilities and TEB relaxation times. Hence, the curvature in this region of the VP1 gene can be attributed to the five unphased A- and T-tracts surrounding the apparent bend center.
Surprisingly, however, discordant gel and solution results were observed when long inverted repeats (palindromes) were inserted into the curvature module. All constructs with such inserts migrated anomalously slowly in polyacrylamide gels; however, they exhibited normal TEB relaxation times that varied somewhat from one buffer to another. Hence, these constructs appear to have normal, although relatively flexible, conformations in free solution. Discordant solution and gel results were not observed if long sequences without inverted repeats were inserted into the curvature module, or if long inverted repeats were inserted into the parent 199-bp fragment outside the curvature module. The results suggest that long inverted repeats form hairpins or cruciforms when they occur within a region of the helix backbone that is stably curved. Such branched structures would exhibit anomalously slow mobilities in polyacrylamide gels, but could have normal TEB relaxation times if the various conformational isomers were in fast exchange. Hence, DNA restriction fragments that exhibit anomalously slow electrophoretic mobilities are not necessarily curved in free solution.
MATERIALS AND METHODS
DNA samples
For the circular permutation studies, SV40 DNA (Gibco BRL, Invitrogen Life Technologies, Carlsbad, CA) was linearized by digestion with a series of single-cut restriction enzymes for 90–180 min, using the manufacturers' recommended temperatures and buffer conditions. The enzymes were denatured by heating the solutions at 65°C for 15 min and the DNAs were ethanol precipitated, redissolved in T0.1E buffer (10 mM Tris-HCl, pH 8.1, plus 0.1 mM EDTA) and stored at −20°C. Control experiments showed that identical results were obtained with and without phenol extracting the denatured restriction enzymes before ethanol precipitation; therefore, the phenol extraction step was usually omitted.
To obtain the relatively large quantities of DNA needed for the TEB experiments, SV40 DNA was digested by suitable restriction enzymes and the desired fragments isolated by agarose gel electrophoresis, subcloned into the polylinker of pUC19, transformed into Escherichia coli strain DH5 (Life Technologies) via the heat shock method, plated on Luria broth media containing ampicillin, isopropylthio-d-galactoside and 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal), and incubated at 37°C overnight, using standard methods (Sambrook and Russell, 2001). White colonies were cultured and large-scale plasmid preparations were carried out using methods described previously (Lu et al., 2002). After suitable restriction enzyme digestion, the desired fragment(s) were isolated by electrophoresis in 1% agarose gels and the desired band(s) excised from the gel. The agarose was dissolved with a chaotropic salt (QIAquick gel extraction kit, Qiagen, Valencia, CA), and the DNA was concentrated and desalted by adsorption on small diethylaminoethyl cellulose columns, as described previously (Stellwagen, 1981). The restriction fragments were eluted by a solution of 1.5 M Na acetate plus 0.5 M Tris-Cl, pH 7.4, ethanol precipitated, redissolved in the desired buffer and stored at −20°C until needed. All fragments were sequenced after cloning to verify their identity.
The A- and T-tracts in the 199-bp restriction fragment chosen for detailed analysis were modified by site-directed mutagenesis, using standard methods (Sambrook and Russell, 2001). The forward primers for the PCR reactions contained 35–45 nucleotides with 1–3 mutated residues near their centers; the reverse primers were the reverse complements of the forward primers. After 17 rounds of amplification with Pfu polymerase, the PCR products were purified by digesting the template DNA with DpnI (which digests only methylated cytosine residues), transformed into one-shot MAX efficiency DH5α-T1 cells, and spread on Luria broth/ampicillin plates. Individual colonies were screened by amplification in Terrific Broth containing 50 μg/mL ampicillin. Plasmid DNA was extracted from the cells using the Qiaprep spin miniprep kit (Qiagen), digested with appropriate restriction enzymes and analyzed by agarose gel electrophoresis to identify desirable mutants. After sequencing to verify the identification, the PCR-modified fragments were subcloned into pUC19 and large-scale plasmid preparations carried out as described above. Subcloning the mutants was necessary to eliminate molecular weight heterogeneities in the PCR-amplified fragments (data not shown).
The 199-bp restriction fragment was also modified by inserting one or more ClaI linkers (CCATCGATGG) into the BsmFI site at position 1913, the ApoI site at position 1948, and/or the PstI site at position 1988. As an example of the procedure used, the pUC19 construct containing the 199-bp SV40 fragment was cut at the unique BsmFI site, the sticky ends were filled in, ClaI linkers (New England Biolabs, Beverly, MA) were added, and the plasmid was recircularized by ligase, using standard procedures (Sambrook and Russell, 2001). After transformation as described above, excess linker was removed by digesting with ClaI and the resulting plasmid recircularized and sequenced. The desired fragments were isolated by restriction enzyme digestion and purified as described above.
The normal DNA fragments used to generate the standard curves of birefringence relaxation times as a function of molecular weight were isolated from plasmids pUC19, pBR322, or Litmus 28, contain 171–543 bp, have ∼50% (A + T) residues with no or only a few isolated An- or Tn-tracts (n ≥ 4), exhibit normal electrophoretic mobilities in polyacrylamide gels and normal birefringence relaxation times, and can be described by the same persistence length (Lu et al., 2002). The preparation of these fragments and the determination of their TEB relaxation times are described in detail elsewhere (Lu et al., 2002, 2003b).
The DNA fragments used in this study were dissolved in three different buffers, called B, C, and E for brevity. All buffers contained 1 mM Tris-Cl, pH 8.0, plus 0.1 mM EDTA (buffer B), 0.01 mM EDTA (buffer C), or 1 mM NaCl and 0.01 mM EDTA (buffer E). All buffers were prepared with deionized water (Nanopure II, Barnstead, Dubuque, IA).
Electrophoresis in analytical polyacrylamide gels
DNA restriction fragments were characterized by electrophoresis in analytical polyacrylamide gels containing 6.9% T (total acrylamide, w/v acrylamide + N,N′-methylenebisacrylamide (Bis)) and 3% C (cross-linker, w/w Bis/total acrylamide), using methods described previously (Holmes and Stellwagen, 1991). The gels were cast and run in TAE buffer (40 mM Tris/1 mM EDTA brought to pH 8.0 with glacial acetic acid) at room temperature, using E = 5.0 V/cm. All gels were aged overnight to allow the polymerization reaction to go to completion and preelectrophoresed for 2 h before the samples were loaded, to eliminate polar impurities remaining from the polymerization reaction. The 50-bp ladder (Invitrogen) was run in each gel as a normal mobility marker. Mobilities of the various DNA fragments were calculated from Eq. 1:
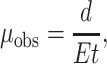 |
(1) |
where μobs is the observed mobility, d is the distance migrated in the gel in centimeters, E is the applied electric field strength in V/cm, and t is the migration time in seconds. All gels were run in triplicate; the average standard deviation of the mobilities was ±3%.
Mobility ratios, μR = μcurved/μnormal, and mobility decrements, (100 (μR − 1)), were calculated for each of the putatively curved fragments in each of the gels. Here, μcurved is the mobility of the potentially curved fragment and μnormal is the mobility of a normal fragment of exactly the same size, calculated from the equation of the line describing the mobility of the fragments in the 50-bp ladder run in the same gel. Note that the mobility ratios defined here are not the same as the molecular weight ratios used by others (e.g., Hagerman, 1985; Koo et al., 1986). The mobility decrements may be thought of as shape factors characterizing the apparent curvature of a target fragment, as determined by gel electrophoresis. Because the mobility of a normal fragment is larger than that of a curved fragment, μR < 1, and the mobility decrements are negative.
Circular permutation assay in large-pore polyacrylamide gels
The linearized, permuted sequence isomers of SV40 were electrophoresed in adjacent lanes in large-pore polyacrylamide gels containing 3.5–5.7% T and 1% C, cast and run in TBE buffer (0.032 M boric acid, 0.05 M Tris base, 1 mM EDTA, pH 8.6) as described previously (Strutz and Stellwagen, 1996; Stellwagen, 2003). The gels were aged overnight in a cold box at 4°C and preelectrophoresed for 2 h before the samples were loaded. The applied electric field was 8.3 V/cm, which caused the temperature in the gel to rise to ∼7°C. The gels were usually run for 16–20 h; the DNA migrated 4–10 cm into the gel during this time. Control experiments showed that the mobilities and mobility patterns were independent of the distance the DNA migrated into the gel. The mobilities of the various permuted sequence isomers were calculated from Eq. 1.
Transient electric birefringence
The apparatus and procedures used for the TEB measurements have been described previously (Stellwagen, 1981, 1991; Lu et al., 2002). Briefly, the birefringence instrument included a Spectra Physics (Oroville, CA) model 117A helium neon laser, quartz Glan-Thompson polarizer and analyzer (Karl Lambrecht Corp., Chicago, IL), a mica quarter-wave plate typically rotated −3° from the crossed position, and a photodiode-based optical amplifier. The time constant of the detecting circuit was ∼0.23 ± 0.06 μs. Square wave pulses ranging from 1 μs to several milliseconds in duration and from 100 V to 2 kV in amplitude, with 10–90% rise and fall times of ∼60 ns, were generated by a Cober (Stamford, CT) high-power pulse amplifier, Model 605P. The Kerr cell was a shortened 1-cm pathlength quartz spectrophotometer cell chosen for its negligible strain birefringence, thermostated at 20°C. A 400-Ω low impedance resistor was connected in parallel with the cell to keep the current constant. The electrodes were parallel platinum plates with a 1.5-mm separation, mounted on a Teflon support of standard design (Pytkowicz and O'Konski, 1959). The stray light constant, with the DNA solution and electrodes in the cell, was typically (0.5–1.0) × 10−5.
Each DNA sample in each buffer solution was pulsed 15 times in the single shot mode, and the field-free decay of each TEB signal was analyzed by fitting the decay curves individually using a nonlinear least-squares fitting program (CURVEFIT) adapted from the program designed by Bevington (1969). This rather laborious procedure was adopted because small but detectable decreases in the relaxation times were observed when the decay curves were averaged before analysis. The decay curves of fragments containing ∼250 bp or less could be fitted with a single relaxation time; longer fragments required two relaxation times, as described previously (Stellwagen, 1981; Lu et al., 2002). In such cases, the slower relaxation time was assigned to the end-over-end rotation of the DNA molecules and was used to calculate the τ-ratios and τ-decrements. The faster relaxation time, which comprised a small part of the total signal (typically 20% or less), can be attributed to bending motions (Pörschke, 1991) and was not analyzed in this study. The single relaxation time observed for the smaller DNA fragments and the slower relaxation time observed for fragments larger than ∼250 bp are described collectively as the terminal relaxation times. Because the terminal relaxation times were independent of the duration of the applied pulse between 3 and 30 μs, independent of electric field strength from 1.0 to 9.0 kV/cm, and independent of DNA concentration from 5 to 25 μg/mL (data not shown), pulse durations of 8 μs, electric field strengths of 5 kV/cm, and DNA concentrations of 14 μg/mL were used for all measurements. The reproducibility of the relaxation times (τ-values) was excellent; the average standard deviation of the τ-values was typically <±2%, even when using different independently prepared DNA stock solutions.
TEB relaxation times are a sensitive measure of DNA curvature, because they are approximately proportional to the third power of macromolecular length, as shown for rigid rods by Eq. 2:
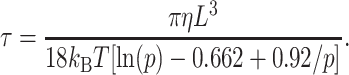 |
(2) |
Here, L is the apparent end-to-end length of the macromolecule in solution, η is the viscosity of the solvent, kB is Boltzmann's constant, T is the absolute temperature, and p is the axial ratio (Tirado and García de la Torre, 1980; Tirado et al., 1984). The relaxation times were used to calculate τ-ratios, τR, defined as τcurved/τnormal, where τcurved is the relaxation time of a target, possibly curved, fragment and τnormal is the relaxation time of a normal fragment containing the same number of basepairs. The values of τnormal were calculated from log-log plots of the relaxation times of a series of normal fragments as a function of the number of basepairs in each fragment, measured in each of the buffers used in this study (Lu et al., 2002, 2003b). This procedure eliminates the necessity of creating control fragments of exactly the same size as each of the target fragments and, more importantly, ensures that the relaxation times of the target fragments are compared with the relaxation times of “average” normal fragments of the same size. From the τ-ratios, τ-decrements, defined as (100 (τR − 1)), were calculated for each of the target fragments. The τ-decrements can be considered as shape factors characterizing the curvature of a target fragment in free solution, as determined by TEB. The τ-decrements are negative, since τcurved < τnormal.
If the location of the bend in a target fragment is known or can be assumed, the τ-ratios or τ-decrements can be compared with theoretical curves to estimate the apparent bend angle (García Bernal and García de la Torre, 1980; Mellado and García de la Torre, 1982; Stellwagen, 1991; Vacano and Hagerman, 1997; Lu et al., 2003b). If the location of the bend is not known, the apparent bend angle and sequence position can be separately determined by the method of overlapping fragments (Lu et al., 2003b). Briefly, several restriction fragments are generated with sequences somewhat offset from the sequence of the target fragment. Global fitting of the τ-ratios of the overlapping fragments to theoretical curves calculated as a function of apparent bend angle and fractional distance, S, of the bend from one end of the molecule allows the apparent bend angle and its sequence position to be separately determined, assuming only that the apparent bend angle does not depend on sequence position (Mellado and García de la Torre, 1982; Vacano and Hagerman, 1997). The method of overlapping fragments has been used to measure the apparent bend angle in the M13 origin of replication, which has been cloned into the plasmid Litmus 28 (Lu et al., 2003b). The magnitude of the bend and its sequence location have been confirmed by atomic force microscopy (Lu et al., 2003a), verifying the accuracy of the TEB method of overlapping fragments.
RESULTS AND DISCUSSION
Detection of curvature in the SV40 genome using the circular permutation assay and large-pore polyacrylamide gels
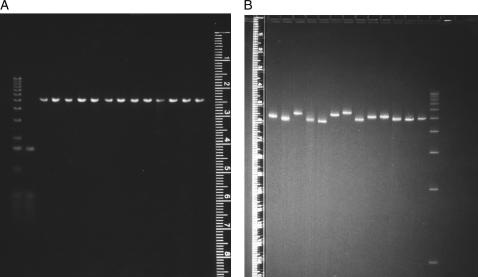
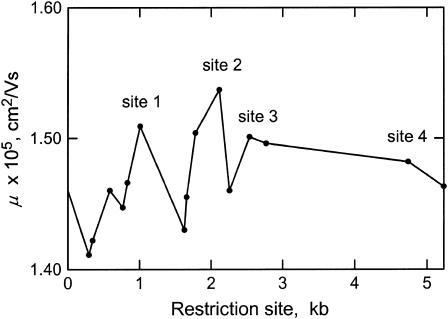
The electrophoretic mobilities observed for linearized, permuted sequence isomers of SV40 in agarose and large-pore polyacrylamide gels are illustrated in Fig. 1. In agarose gels (Fig. 1 A), the permuted sequence isomers migrate with identical mobilities, as expected for molecules of identical molecular weights in this gel matrix (Stellwagen, 2003). However, despite their identical molecular weights, the permuted sequence isomers migrate with variable mobilities in large-pore polyacrylamide gels (Fig. 1 B), indicating that the helix backbone is intrinsically curved. The mobilities of the various permuted sequence isomers, which are plotted as a function of the location of the restriction enzyme cut site in Fig. 2, suggest that SV40 contains four regions of curvature. Site 1 is located near the start of the gene for the minor capsid protein VP3; site 2 occurs within the gene for the major capsid protein VP1; site 3 is located near the bidirectional terminus of transcription; and site 4 is located near the origin of replication. The mobility maxima at sites 3 and 4 were confirmed by subcloning these sites into the polylinker of plasmid pUC19 and analyzing the mobility of linear, permuted sequence isomers of the resultant chimeras (data not shown). However, a precise determination of the location of the center of curvature at any site is not possible using this assay, because of the limited availability of suitable restriction enzyme cut sites.
FIGURE 1.
Electrophoretic mobility of the permuted sequence isomers of SV40 in: (A) 1% agarose and (B) 5.7% T, 1% C polyacrylamide gels. The leftmost lanes in panel A contain the 1-kb ladder and the HinfI digest of pBR322 as mobility markers; the rightmost lane in panel B contains the 1-kb ladder. The other lanes contain SV40 linearized with BanI, HpaII, PpuMI, EcoRV, HaeII, DrdI, AccI, AflII, EcoRI, XcmI, Bsp120I, BamHI, and BclI, from left to right.
FIGURE 2.
Mobilities of the permuted sequence isomers of SV40 plotted as a function of the restriction enzyme cut site, in kilobases. From left to right, the solid circles correspond to the mobilities of sequence isomers linearized by BanI, HpaII, PpuMI, EcoRV, HaeII, DrdI, AccI, AflII, EcoRI, XcmI, Bsp120I, BamHI, BclI, TaqI, and BglI, electrophoresed in a 3.5% T, 1% C polyacrylamide gel cast and run in TBE buffer at 4°C. The mobility maxima corresponding to the four sites of curvature discussed in the text are indicated.
Three of the four sites of curvature detected by the circular permutation assay have been identified previously by other investigators. Curvature within the VP1 gene (site 2) has been inferred from the anomalously slow electrophoretic mobilities of restriction fragments taken from this region (Mertz and Berg, 1974; Anderson, 1986; Milton and Gesteland, 1988; Milton et al., 1990). Curvature near the transcription terminus (site 3) has been identified by polyacrylamide gel electrophoresis (Poljak and Gralla, 1987) and electron microscopy (Griffith et al., 1986; Hsieh and Griffith, 1988). In addition, curvature near the origin of replication (site 4) has been detected by polyacrylamide gel electrophoresis (Ryder et al., 1986; Pauly et al., 1992). Hence, the circular permutation assay in large-pore polyacrylamide gels can accurately detect stable, sequence-dependent curvature in large, kilobase-sized DNA molecules.
Characterization of restriction fragments containing the four curvature sites
Restriction fragments containing the four sites of curvature in the SV40 genome were characterized by polyacrylamide gel electrophoresis and transient electric birefringence. The sizes of the various restriction fragments, their A + T contents, sequence locations, μ-decrements, terminal TEB relaxation times, and τ-decrements in buffers B and C are summarized in Table 1. As shown in column 5 of this table, the μ-decrements of restriction fragments derived from curvature sites 1, 2 and 3 ranged from −11 to −39, suggesting that these fragments are stably curved and/or anisotropically flexible (i.e., bent preferentially in a single direction). Surprisingly, however, the μ-decrements observed for the 205- and 263-bp fragments containing site 4, near the origin of replication, ranged from −2 to −6; i.e., the mobilities were virtually indistinguishable from those observed for normal uncurved DNA fragments of the same size. It seems likely that the apparent bend centers in the 205- and 263-bp fragments were located relatively close to the ends of these fragments (Ryder et al., 1986; Pauly et al., 1992), causing the fragments to migrate with normal mobilities in polyacrylamide gels. Similar results have been observed for other DNA fragments (Stellwagen, 1983). Hence, the circular permutation assay appears to be a more accurate indicator of curvature in this region of the SV40 genome than the electrophoretic mobilities of the particular restriction fragments chosen for study.
TABLE 1.
Mobility decrements, birefringence relaxation times, and τ-decrements observed for restriction fragments containing the four sites of curvature in the SV40 genome
| Buffer B
|
Buffer C
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Site | N, bp | A+T, % | Sequence location | μ-decrement* | τ, μs | τ-decrement† | τ, μs | τ-decrement† |
| 1 | 357 | 59.7 | 832–1171 | −11 | 17.0 | −2 | 16.2 | −3 |
| 2 | 199 | 57.8 | 1826–2017 | −24 | 3.7 | −16 | 3.7 | −13 |
| 2 | 340 | 59.7 | 1782–2117 | −39 | 14.4 | −7 | 12.8 | −14 |
| 2‡ | 345 | 50.1 | −(1917–1992) | −8 | 15.5 | −3 | 15.1 | −3 |
| 3 | 335 | 61.2 | 2455–2797 | −24 | 14.8 | −1 | 12.6 | −13 |
| 4 | 205A§ | 50.7 | 5118–72 | −2 | 4.4 | −9 | 4.5 | −2 |
| 4 | 263 | 58.6 | 4986–5234 | −6 | 8.1 | −4 | 7.4 | −10 |
μ-decrement = 100 (μR − 1).
τ-decrement = 100 (τR − 1).
The 340-bp fragment with residues 1917–1992 replaced by 70-bp of normal DNA from pUC19.
So designated to distinguish this fragment from the 205-bp insertion derivative in Table 4.
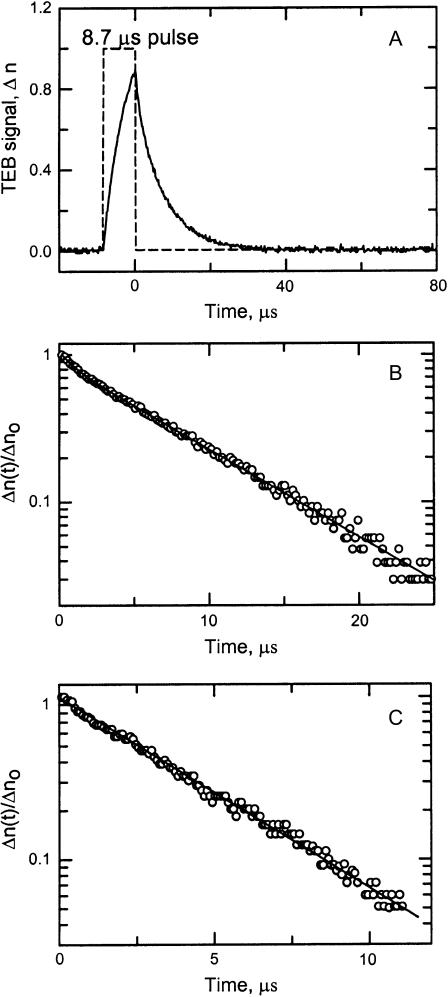
The curvature of restriction fragments containing sites 1–4 was also analyzed by TEB. A typical birefringence signal observed for the 263-bp fragment containing site 4 is illustrated in Fig. 3 A. A semilogarithmic plot of the decay of the birefringence as a function of time is shown in Fig. 3 B. Two relaxation times, 1.36 μs (12.9%) and 8.11 μs (87.1%), were required to describe the decay of the birefringence of this fragment (solid line through the data points). Previous studies have shown that DNA restriction fragments larger than ∼250 bp exhibit biexponential decay curves (Stellwagen, 1981; Lu et al., 2002), with the slower relaxation time corresponding to the end-over-end rotation of the macromolecule after removal of the electric field. Restriction fragments smaller than ∼250 bp exhibited monoexponential birefringence decays, as shown in Fig. 3 C for the 199-bp fragment containing site 2. This decay curve is well described by a single relaxation time of 3.74 μs.
FIGURE 3.
(A) Typical birefringence signal, Δn, observed for the 263-bp fragment in buffer B. The dashed line corresponds to the applied pulse (arbitrary scale); the somewhat noisy line is the birefringence signal. (B) Semilogarithmic plot of the decay of the birefringence of the 263-bp fragment in buffer B as a function of time. The fractional birefringence signal, Δn(t)/Δno, remaining at any time, t, after the pulse is removed is plotted as a function of time. The solid line is a two-exponential fit with relaxation times of 1.36 μs (12.9%) and 8.11 μs (87.1%). (C) Semilogarithmic plot of the decay of the birefringence of the 199-bp fragment in buffer B as a function of time. The solid line is a one-exponential fit with a relaxation time of 3.74 μs. The relaxation times obtained from 15 such measurements of each sample were averaged to give the relaxation times used to calculate the τ-ratios and τ-decrements.
The terminal relaxation times observed for restriction fragments containing sites 1–4 in buffers B and C, and the corresponding τ-decrements, are summarized in columns 6–9 of Table 1. The 357-bp restriction fragment containing site 1, near the start site of the VP3 gene, exhibited very small τ-decrements (from −2 to −3) in buffers B and C. Previous studies have shown that τ-decrements of this size fall within the range of values exhibited by normal DNA fragments and are not correlated with curvature (Lu et al., 2003b). Hence, the 357-bp fragment does not appear to be curved in free solution. The μ-decrement of −11 observed for this fragment is relatively small for a fragment of this size (compare with the μ-decrements of −24 and −39 observed for the 335- and 340-bp fragments, respectively, in Table 1). Hence, the curvature of the 357-bp fragment appears to be relatively small when measured either by polyacrylamide gel electrophoresis or TEB.
A 263-bp restriction fragment containing site 4, near the origin of replication and the start site of the genes for the large and small T antigens, exhibited τ-decrements ranging from −4 to −10 in buffers B and C, suggesting that this fragment is moderately curved in free solution. This fragment is also somewhat flexible, because of the difference in the τ-decrements observed in the two buffers. A 205-bp restriction fragment (called 205A) containing site 4, near the origin of replication, and a 335-bp fragment containing site 3, which includes the bidirectional terminus of transcription, exhibited large τ-decrements (from −9 to −13) in one of the buffers examined but very small τ-decrements (from −1 to −2) in the other buffer, suggesting that these fragments are anisotropically flexible. The mobility decrements observed for the 205- and 335-bp fragments were −2 and −24, respectively, reflecting in part the fact that μ-decrements are larger for larger DNA fragments because of sieving effects (Hagerman, 1985; Koo et al., 1986; Stellwagen and Stellwagen, 1990; Stellwagen, 1997). Further experiments are needed to verify that the 205-, 263-, and 335-bp fragments are anisotropically flexible.
The 199- and 340-bp fragments, both of which contain site 2 within the VP1 gene, exhibited large μ- and τ-decrements in all buffers examined (Table 1 and data not shown). The τ-decrements of the 199-bp fragment were exceptionally large (from −13 to −16) and relatively independent of the ionic content of the solution, suggesting that this fragment is intrinsically curved rather than anisotropically flexible. The τ-decrements of the 340-bp fragment ranged from −7 to −14 in buffers B and C, suggesting that this fragment is somewhat flexible as well as curved, possibly because of its greater size.
When 75 basepairs from the center of the 340-bp fragment were replaced by 70 basepairs of normal DNA from pUC19 plus 10 bp of linker DNA, creating a 345-bp construct, the μ-decrement was reduced from −39 to −8 and the τ-decrements were reduced from large negative values (−7 and −14) to the nonsignificant value of −3 (Table 1). Hence, the central 75 residues in the 340-bp fragment are necessary and sufficient to cause the curvature of this fragment. By inference, because these 75 basepairs are also present in the center of the 199-bp fragment, these 75 bp are also responsible for the curvature of the 199-bp fragment.
Temperature dependence of curvature in restriction fragments containing site 2
The τ-decrements observed for the 199- and 340-bp fragments in buffers B and C at various temperatures are summarized in Table 2. Large τ-decrements were observed for the 199-bp fragment in buffers B and C and for the 340-bp fragment in buffer C, suggesting that the intrinsic curvature of these fragments is relatively independent of temperature in these buffers. However, the τ-decrement of the 340-bp fragment decreased significantly with increasing temperature in buffer B, suggesting that the flexibility of this fragment increases with increasing temperature in solutions with higher ionic strengths. Because the 199-bp fragment is contained within the 340-bp fragment, the greater flexibility of the 340-bp fragment in buffer B must be due to sequences located on either side of the embedded 199-bp fragment. Further studies would be needed to attribute this increased flexibility to specific sequence motifs.
TABLE 2.
Temperature dependence of the τ-decrements of the 199- and 340-bp fragments
|
τ-Decrements
|
||||||
|---|---|---|---|---|---|---|
| Buffer B
|
Buffer C
|
|||||
| Fragment | 4° | 10° | 20° | 4° | 10° | 20° |
| 199 | −19 | −17 | −16 | −19 | – | −13 |
| 340 | −22 | −10 | −7 | −12 | −16 | −13 |
Quantitative analysis of curvature in the VP1 gene
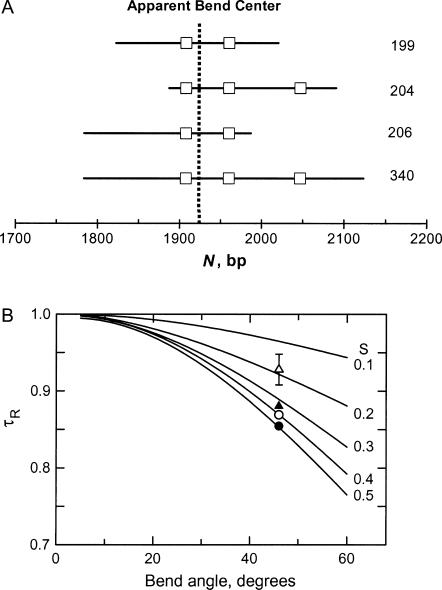
The magnitude of the apparent bend in the VP1 gene and its sequence location were determined by TEB, using the method of overlapping fragments (Lu et al., 2003b). Four overlapping restriction fragments containing 199, 204, 206, and 340 bp were created by suitable restriction enzyme digestion. The relationship of the four fragments is shown schematically in Fig. 4 A; for ease of reference, A- and T-tracts containing five or more residues are highlighted by square boxes. The relaxation times of the four overlapping fragments were measured in buffers B and C and compared with the relaxation times of normal fragments containing the same number of basepairs, taken from previously measured standard curves in the same two buffers (Lu et al., 2002). The τ-ratios calculated for the four fragments in buffers B and C were then averaged and compared with theoretical curves calculated as a function of bend angle and the fractional distance, S, of the bend from one end of the molecule (García Bernal and García de la Torre, 1980; Mellado and García de la Torre, 1982). Global fitting of the experimental τ-ratios to the theoretical curves, shown in Fig. 4 B, indicates that an apparent bend of 46 ± 2° is located at fractional distances of 0.50, 0.18, 0.33, and 0.41 from one end of the 199-, 204-, 206-, and 340-bp fragments, respectively. These fractional distances correspond to sequence position 1922 ± 2 bp, near the middle of the VP1 gene. The apparent bend angle of 46 ± 2° is similar to the apparent bend of 41 ± 6° observed in the M13 origin of replication, using TEB (Lu et al., 2003b) and atomic force microscopy (Lu et al., 2003a). The magnitude of the bend corresponds to the degree of curvature expected for 2–3 phased A-tracts (Levene et al., 1986; Koo et al., 1990; Ross et al., 1999; Podtelezhnikov et al., 2000; MacDonald et al., 2001; Barbič et al., 2003; Tchernaenko et al., 2003).
FIGURE 4.
(A) Schematic diagram of the relationship between the four overlapping restriction fragments used to analyze the curvature of the 199-bp fragment. For ease of reference, the square boxes indicate the positions of A- or T-tracts containing five or more residues; the vertical dashed line corresponds to the location of the apparent bend center at 1922 bp. The size of each restriction fragment is indicated at the right. (B) Comparison of average τ-ratios measured in buffers B and C at 20°C with theoretical curves calculated as a function of the apparent bend angle and relative position, S, with respect to the end of the fragment. From top to bottom, the symbols correspond to fragments containing: (▵) 204 bp; (▴) 206 bp; (○) 340 bp; and (•) 199 bp. The error bar bracketing the symbol for the 204-bp fragment corresponds to the average standard deviation of the τ-ratios measured for the various fragments in buffers B and C. The values of S obtained for the 204-, 206-, 340-, and 199-bp fragments are 0.18, 0.33, 0.41, and 0.50, respectively.
The sequence surrounding the apparent bend center in the 199-bp fragment includes four An- or Tn-tracts with n ≥ 4 and a mixed A3T4 element, as shown in Scheme 1. For easy reference, these sequence elements will be designated A6, A4T, A4, A3T4, and T7, respectively; the entire 59-bp sequence will be described as the curvature module.
SCHEME 1.
SV40 sequence from 1905 to 1964 bp. The A- and T-tracts are indicated by bold letters and underlining. The position of the apparent bend center at 1922 bp is marked by a vertical arrow.
Most of the A- and T-tracts in the curvature module do not occur in phase with the 10-bp helix repeat. A6, A4T, and A4 are each separated by 12 or 13 bp, counting the first A in each A-tract as position 1. Mixed A/T-tracts with no TA basepair steps have many of the properties of A-tracts (Hagerman, 1986; Leroy et al., 1988; Burkhoff and Tullius, 1988), and will be assumed to be equivalent to A-tracts in this discussion. The first A in A4 is separated from the first A in A3T4 by 18 bp. In addition, the first T in A3T4 is separated from the first T in T7 by 6 bp, suggesting that the bends caused by these two sequence elements might be in opposite directions and cancel each other.
Alternatively, because the A- and T-tracts in the curvature module are of different lengths, the separation between their 3′ ends may be more important than the separation of their 5′ ends, because A-tract bending increases in the 5′–3′ direction (Koo et al., 1986; Seela and Grein, 1992; Møllegaard et al., 1997; Maki et al., 2004). The terminal A residues in A6 and A4T are in phase and separated by 10 bp, but the terminal A residues in A4T, A4, and A3T4 are not in phase (separated by 13 and 17 bp, respectively). The terminal T residues in A3T4 and T7 are separated by 9 bp, suggesting that, in contrast to the above scenario, the bends caused by these two sequence elements might be additive. Hence, determination of the relative contribution of the various A- and T-tracts to the overall curvature of the 199-bp fragment should determine whether 5′- or 3′ phasing contributes more importantly to the curvature of the helix backbone.
Contribution of the A- and T-tracts near the apparent bend center to the curvature of the 199-bp fragment
To determine whether the intrinsic curvature of the 199-bp fragment is due to the five A- and T-tracts surrounding the apparent bend center, or to other proposed bending elements such as the G/C-rich regions between A-tracts (Goodsell et al., 1994) or the multiple CA/TG-bp steps (Bolshoy et al., 1991; Lyubchenko et al., 1993; Tonelli et al., 1998), the A- and T-tracts in the curvature module were modified by site-directed mutagenesis. All sequence mutants contained 198 or 199 bp, as indicated by their names. In one construct, called 199K, all five A- and T-tracts were mutated to other sequences. In other constructs, each of the A-tracts was modified individually, to give fragments 199F, 199B, 199C, 198A, and 199E. Because A3T4 and T7 occur very close together in the curvature module, a mutant (199A) was also constructed in which the CA dinucleotide between A3T4 and T7 was changed to TA, creating a 16-bp mixed A/T-tract. As a control for the latter construct, a mutant (199U) was constructed in which the CA dinucleotide was retained but A3T4 and T7 were changed to other sequences. The names of the various sequence mutants, the identities of the modified A-tract(s), and the μ- and τ-decrements observed for each fragment are compiled in Table 3. To avoid concentrating on the differences in the relaxation times observed in different buffers, the τ-decrements observed in buffers B, C, and E were averaged, giving the values reported in the last column of Table 3. Because the standard deviations of the average τ-decrements reflect the variation of the τ-values observed in different buffers, a relatively large standard deviation implies that the conformation of that particular sequence mutant is relatively flexible.
TABLE 3.
Characterization of sequence mutants with variable numbers of A-tracts
|
τ-Decrements
|
||||||
|---|---|---|---|---|---|---|
| Fragment Name | Mutated A-tract(s) | μ-Decrement | Buffer B | Buffer C | Buffer E | Average* |
| 199 | None | −24 | −16 | −13 | −21 | −17 ± 3 |
| 199K | A6, A4T, A4, A3T4, T7 | −2 | −1 | −4 | 0 | −1 ± 2 |
| 199F | A6 | −18 | −12 | −12 | −12 | −12 ± 1 |
| 199B | A4T | −17 | −9 | −8 | −12 | −10 ± 2 |
| 199C | A4 | −22 | −8 | −8 | −17 | −11 ± 4 |
| 198A | A3T4 | −14 | – | – | −14 | −14 |
| 199E | T7 | −18 | −15 | −15 | −12 | −14 ± 2 |
| 199A | A3T4, AT7† | −17 | −10 | −7 | −9 | −8 ± 1 |
| 199U | A3T4, AT7 | −13 | −9 | −4 | −8 | −7 ± 2 |
Average τ-decrement observed in buffers B, C, and E and the standard deviation.
Construct with T replacing C between A3T4 and T7, creating a 16-bp mixed A/T tract.
When all five A- and T-tracts surrounding the apparent bend center were mutated to other sequences, giving fragment 199K, the μ-decrement decreased from −24 to −2 and the average τ-decrement was reduced from −16 to −1, as shown in Table 3. Hence, the conformation of fragment 199K is that of normal, uncurved DNA. The conclusion must be drawn that the curvature of the 199-bp parent fragment is due primarily to the five unphased A- and T-tracts surrounding the apparent bend center.
The μ- and τ-decrements observed for fragments 199F, 199B, 199C, 198A, and 199E, each of which is missing one of the A- or T-tracts in the curvature module, were lower than observed for the parent 199-bp fragment (Table 3). Hence, all five A- and T-tracts contribute to the curvature of the parent fragment. The μ-decrements suggest that A4 contributes the least and A3T4 the most to the observed curvature, because modification of these sequence elements caused relatively small (199C) or relatively large (198A) changes in the μ-decrements, respectively. However, the average τ-decrements observed for fragments 199F, 199B, and 199C suggest that all three A-tracts, A6, A4T, and A4, contribute importantly to the observed curvature. The τ-decrements also suggest that curvature of the 199-bp fragment is very little affected by the loss of A3T4 or T7, as long as the other sequence element is present. The average τ-decrement was only reduced from −17, for the parent 199-bp fragment, to −14 for fragments 198A and 199E, missing A3T4 or T7, respectively. However, the average τ-decrement was reduced to −7 when both A3T4 and T7 were mutated to other sequences (fragment 199U). The relatively large μ- and τ-decrements observed for fragments 198A and 199E also indicate that the bends due to A3T4 and T7 are not out-of-phase. If they were, the μ- and τ-decrements observed for these fragments would have been larger than observed for the parent 199-bp fragment. Hence, the phasing of the terminal residues of A- and T-tracts of different lengths determines the additivity of the bends produced by each of these sequence elements. Similar results have been obtained with DNA oligomer ladders (Koo et al., 1986).
When the CA dinucleotide between A3T4 and T7 was mutated to TA, creating a 16-bp mixed A/T-tract with the sequence …AAATTTTTATTTTTTT… (fragment 199A), the μ-decrement and the average τ-decrement were reduced significantly (Table 3). Hence, the curvature of the parent 199-bp fragment was significantly reduced when two closely spaced A- or T-tracts were replaced by a long mixed A/T-tract. This result was not due to the loss of the CA dinucleotide between A3T4 and T7. If A3T4 and T7 were both modified by site-directed mutagenesis while retaining the CA dinucleotide in its original position, giving the sequence …AACGTTTCATCTAGAT… (fragment 199U), the μ-decrement and average τ-decrement were similar to those observed for fragment 199A with the long mixed A/T-tract. Hence, a long mixed A/T-tract appears to act as normal, non-A-tract DNA and does not contribute to the curvature of the helix backbone.
Derivatives containing inserts in or near the curvature module
The importance of the spacing between the unphased A- and T-tracts in the curvature module was investigated by inserting spacers of various lengths at position 1915, between A6 and A4T, position 1950, in the center of A3T4, or position 1988, outside the curvature module. Most of the inserted sequences were derived from the 10-bp ClaI linker (CCATCGATGG). However, due to base pairs added or removed during cloning procedures, the actual inserts ranged from 10 to 37 bp in size, as determined by sequencing. Two 204-bp constructs (204B and 204C) were also prepared, in which A6 and T7 were replaced by mixed-sequence DNA and a 5-bp spacer was inserted between A4T and A4 or between A4 and A3T4. To test the effect of the sequence of the added spacer on the observed results, a 231-bp construct containing three tandem copies of the ClaI linker was compared with a 236-bp construct containing three nested ClaI linkers inserted at the same site. Nested ClaI linkers can form a long inverted repeat (palindrome). The names of the various fragments, the insertion sites, and the sequences and sizes of the various inserts are given in the first four columns of Table 4. Various control fragments are listed in Table 5. The controls include 139- and 278-bp fragments that do not contain the curvature module, 204-, 206-, and 219-bp fragments that contain only part of the curvature module, so that the apparent bend center is located very close to one end of the fragment, and a 199-bp fragment (199G) with A6 and T7 mutated to random-sequence DNA. The latter fragment serves as a control for fragments 204B and 204C, in which A6 and T7 are mutated to the same sequences.
TABLE 4.
Characterization of insertion derivatives
| Fragment Name | Insertion site | Inserted sequence | Insertion size, bp | μ-Decrement | Average τ-decrement* |
|---|---|---|---|---|---|
| 204B† | 1926 | AGCTA | 5 | −3 | −4 ± 5 |
| 204C† | 1942 | AGCTA | 5 | −3 | −4 ± 3 |
| 205 | 1988 | CCATCGATGG | 10 | −24 | −17 ± 2 |
| 212 | 1913 | TGCCATCGATGG | 13 | −13 | −5 ± 1 |
| 213 | 1950 | TTCCATCGATGGAA | 14 | −12 | −6 ± 2 |
| 217 | 1988 | Nested ClaI linker dimers‡ | 22 | −24 | −8 ± 1 |
| 224 | 1913 | Nested ClaI linker dimers‡ | 25 | −13 | −2 ± 2 |
| 225 | 1950 | Nested ClaI linker dimers‡ | 26 | −14 | +1 ± 3 |
| 227 | 1913, 1950 | Two insertions§ | 14 | −7 | +1 ± 2 |
| 231 | 1913 | Tandem ClaI linker trimers¶ | 32 | −17 | −6 ± 1 |
| 236 | 1913 | Nested ClaI linker trimers‖ | 37 | −13 | −2 ± 2 |
Average τ-decrement observed in buffers B, C, and E and standard deviation.
Also, A6 and T7 replaced by TCTAGA.
Inserted sequence, CCATCGCCATCGATGGCGATGG.
Inserted sequences, CCATCGATGGCATG at 1913 and TTCCATCGATGGAA at 1950.
Inserted sequence, GCCCATCGATGGCCATCGATGGCCATCGATGG.
Inserted sequence, ATGCCATCGCCATCGCCATCGATGGCGATGGCGATGG.
TABLE 5.
Control fragments for the insertion derivatives
| Fragment Name | Sequence location or modification | μ-Decrement | Average τ-decrement* |
|---|---|---|---|
| 139 | 1993–2123 | −1 | +1 ± 2 |
| 199G | A4T, A4, A3T4 only | −11 | −11 ± 2 |
| 204 | 1894–2094 | −12 | −6 ± 1 |
| 206 | 1783–1993 | −18 | −12 ± 2 |
| 219 | 1913–2107 | −4 | +1 ± 1 |
| 278 | 1993–2262 | −3 | −1 ± 3 |
Average τ-decrement observed in buffers B, C, and E and standard deviation.
The insertion derivatives have the following properties. The 212-, 224-, 231-, and 236-bp fragments have 13-, 25-, 32-, and 37-bp spacers, respectively, added between A6 and A4T. The terminal A residues in these two A-tracts, which were in phase in the parent 199-bp fragment, are approximately out-of-phase in the 212- and 224-bp fragments and unphased in the 231- and 236-bp fragments. The 213- and 225-bp fragments have spacers of 14 and 26 bp, respectively, inserted between A3 and T4 in A3T4, creating a truncated A3T2 sequence element followed by a T4-tract. In these constructs, the terminal A in A4 is unphased with respect to the terminal A in A3T2, but is approximately in phase with the terminal T; the terminal T in A3T2 is out-of phase with the terminal T residues in T4 and T7. The 227-bp fragment has 14-bp spacers between A6 and A4T and between A4T and the truncated A3T2, making both pairs of A-tracts approximately out-of-phase with each other. In fragment 204B, the terminal A residues in A4T and A4 are separated by 17 bp. In fragment 204C, the terminal residues in A4 and A3T4 are approximately out-of-phase, separated by 26 bp. Finally, the 205- and 217-bp fragments have spacers of 10 and 18 bp, respectively, inserted outside the curvature module.
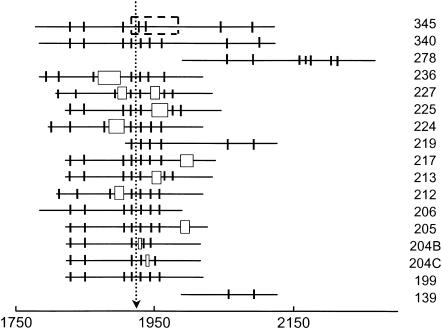
Schematic diagrams of the parent 199-bp fragment, constructs with added spacers and various controls are illustrated in Fig. 5. A- and T-tracts containing four or more residues are indicated by vertical lines; the inserted spacers are indicated by open rectangles with widths approximately proportional to the length of the spacer, and the location of the pUC19 sequence replacing the curvature module in the 345-bp fragment is indicated by a dashed rectangular box. The various constructs are aligned at the location of the apparent bend center in the parent 199-bp fragment. The size of each fragment is indicated in the key to the right.
FIGURE 5.
Schematic diagram of the 199-bp fragment, constructs with inserts at various positions, control fragments with sequences offset from the apparent bend center, and a 345-bp fragment with 75 bp of unbent DNA from pUC19 replacing 70 bp containing the curvature module. A- and T-tracts containing four or more residues are indicated by short vertical lines; the inserted spacers are indicated by open rectangles with widths approximately proportional to the length of the spacer, and the pUC19 substitution is indicated by a dashed rectangle. The sequences are aligned at the apparent bend center at 1922 bp, as indicated by the dotted line. Each fragment is identified in the key to the right by the total number of basepairs in the construct.
Electrophoretic mobilities observed for the insertion derivatives
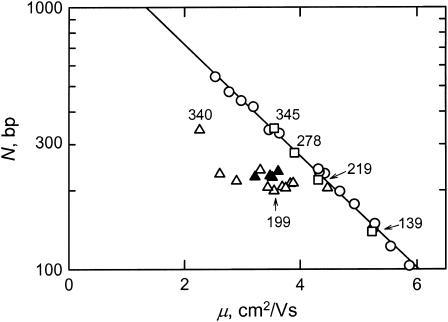
The electrophoretic mobilities observed for the various insertion derivatives in polyacrylamide gels are illustrated in Fig. 6. All constructs containing the curvature module (triangles) migrated anomalously slowly in polyacrylamide gels, whereas fragments without the curvature module (the 139-, 278-, and 345-bp fragments), or with the apparent bend center located close to one end (the 219-bp fragment), migrated with mobilities (open squares) equal to those observed for the fragments in a standard molecular weight ladder (open circles). The solid triangles correspond to those fragments that have normal birefringence relaxation times (see below); the open triangles correspond to fragments that have anomalously fast birefringence relaxation times and hence are curved in free solution.
FIGURE 6.
Semilogarithmic plot of DNA molecular weight, N, in basepairs, as a function of the electrophoretic mobility, μ, observed in a 6.9% T, 3% C polyacrylamide gel. (○) Normal fragments in the 50-bp ladder; (□) SV40 fragments without the curvature module or with the apparent bend center located very close to one end; (▵) insertion derivatives that are significantly curved in free solution; and (▴) insertion derivatives that are not curved, or only slightly curved, in free solution (see text). The drawn line corresponds to the fitting function describing the migration of the fragments in the 50-bp ladder. Selected fragments are identified for ease of reference.
The mobility decrements calculated for the various insertion derivatives and controls are given in column 5 of Table 4 and column 3 of Table 5. The μ-decrements can be grouped roughly into three categories: normal (μ-decrements ranging from −1 to −4), moderately anomalous (μ-decrements ranging from −7 to −14), and highly anomalous (μ-decrements ≥ −17). The normally migrating fragments include the 139- and 278-bp fragments that do not contain the curvature module, the 219-bp fragment with the apparent bend center located close to one end, and fragments 204B and 204C, in which A6 and T7 have been mutated to other sequences and 5-bp spacers have been inserted between A4T and A4 or A4 and A3T4, respectively. Because the μ-decrement observed for the control fragment 199G was moderately anomalous (−11), the original spacing between A4T, A4, and A3T4 contributes importantly to the curvature of the parent 199-bp fragment, even though these three A-tracts do not occur in phase with the helix repeat.
The fragments with the highest mobility decrements are the parent 199-bp fragment, the 205-and 217-bp fragments that have inserts outside the curvature module, and the 231-bp fragment that contains three tandem ClaI linkers in the curvature module. All other insertion derivatives exhibited moderate mobility decrements.
Birefringence relaxation times and τ-decrements observed for the insertion derivatives
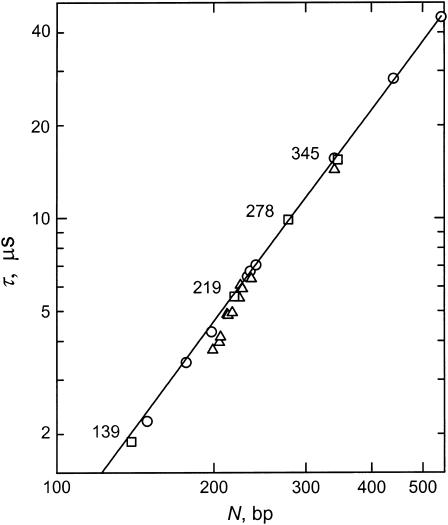
TEB relaxation times were measured for the insertion derivatives and various controls in buffers B, C, and E. A log-log plot of the terminal relaxation times (τ-values) observed in buffer B as a function of molecular weight is illustrated in Fig. 7, together with the τ-values observed for normal fragments of similar sizes measured in the same buffer (Lu et al., 2003b). The relaxation times of the normal fragments (open circles) exhibit a power-law dependence on molecular weight over the indicated size range. The relaxation times of the 139-, 219-, 278-, and 345-bp fragments (open squares), which do not contain the curvature module, have the apparent bend center located close to one end, or have had the curvature module replaced by a stretch of normal DNA from pUC19, fall on the same straight line as the normal fragments, indicating that these four fragments are not curved in free solution. Most, but not all, of the insertion derivatives (open triangles) exhibit terminal relaxation times that fall below the line describing the relaxation times of the normal fragments, indicating that these fragments are curved in free solution.
FIGURE 7.
Log-log plot of the dependence of the terminal relaxation times, τ, on DNA molecular weight, N, in bp, in buffer B at 20°C. (○) Normal control fragments; (▵) the parent 199-bp fragment, sequence mutants, and insertion derivatives; and (□) restriction fragments that do not contain the curvature module, have the apparent bend center located very close to one end, or have had the curvature module replaced by an equal-sized insert of normal DNA from pUC19. The solid line corresponds to the fitting function describing the τ-values of the normal control fragments.
The average τ-decrements observed for the insertion derivatives and controls in buffers B, C, and E are given in Table 4; for brevity, the individual τ-decrements are not given. The various insertion derivatives can be grouped into three categories based on their average τ-decrements: normal (average τ-decrements of −4 or less, particularly if the standard deviation is large), moderately anomalous (average τ-decrements ranging from −5 to −8), and highly anomalous (average τ-decrements ≥ −10). Fragments with normal τ-decrements, and hence normal conformations in free solution, include the 139- and 278-bp fragments missing the curvature module, the 204- and 219-bp fragments with the apparent bend center near one end, fragments 204B and 204C with 5-bp spacers between the three interior A-tracts in the curvature module, and the 224-, 225-, 227-, and 236-bp fragments with large nested ClaI linkers inserted into the curvature module.
The most highly anomalous insertion derivative is the 205-bp fragment, which contains a 10-bp insert outside the curvature module (compare with the average τ-decrement observed for the parent 199-bp fragment in Table 3). Hence, the 205-bp fragment is highly curved in free solution. The remaining fragments exhibited moderate τ-decrements and therefore appear to be moderately curved in free solution.
Comparison of solution and gel results
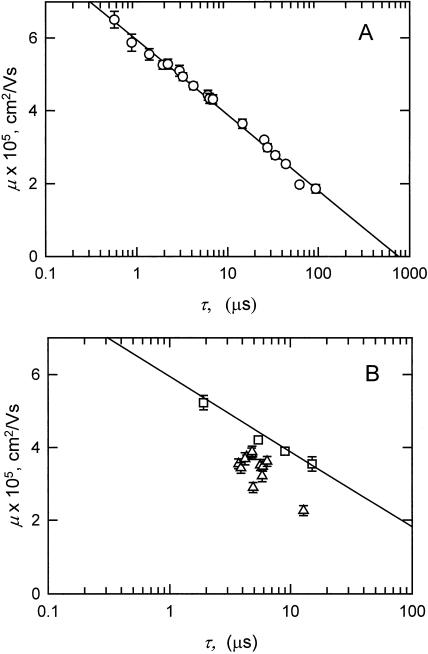
Two independent measures of DNA curvature have been used to characterize the various DNA fragments: anomalous electrophoretic mobilities in polyacrylamide gels and TEB relaxation times in free solution. For normal DNA fragments, there is a very good correlation between the μ-values measured in gels and the logarithm of the τ-values observed in free solution, as shown in Fig. 8 A. This relationship can be explained as follows. For the normal fragments, the dependence of electrophoretic mobility on molecular weight has the form: μ = a log(b/N), where N is the number of basepairs and a and b are constants (see Fig. 6). The dependence of birefringence relaxation times on molecular weight has the form: τ = c Nd, where c, and d are constants (Fig. 7). Expressing N in terms of τ, N = (τ/c)1/d; therefore, μ = a log[b/(τ/c)1/d] = a′ log(b′/τ), where a′ and b′ are constants. Hence, μ decreases with increasing log (τ), as shown in Fig. 8 A. However, this correlation breaks down for curved fragments, as shown in Fig. 8 B, where the random scatter of the points below the regression line calculated for the normal fragments (in Fig. 8 A) reflects the lack of correspondence between the solution and gel results.
FIGURE 8.
Dependence of the polyacrylamide gel mobilities (μ) observed for various DNA fragments on the logarithm of the birefringence relaxation times (τ) measured in buffer C. (A) Normal DNA fragments from Figs. 6 and 7, with the calculated linear regression line. (B) The 199-bp fragment, sequence mutants, and insertion derivatives. (▵) Fragments containing inserts of various sizes; and (□) fragments that do not contain the curvature module, have the apparent bend center located very close to one end, or have had the curvature module replaced by an equal-sized insert of normal DNA from pUC19. The solid line is the regression line for the normal fragments, taken from panel A, with the data points omitted for clarity. The error bars in panels A and B represent the standard deviation of duplicate or triplicate measurements of the mobilities and relaxation times; symbols without error bars had standard deviations smaller than the size of the symbol.
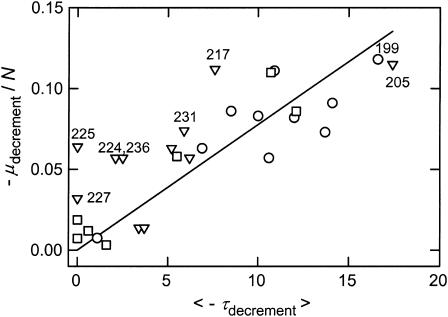
To better understand the discordant solution and gel results in Fig. 8 B, it is useful to compare the μ- and τ-decrements of the various fragments. However, direct comparison of the μ- and τ-decrements of molecules of different sizes is not warranted, because reptation effects cause electrophoretic mobilities to decrease approximately inversely with increasing size (Lerman and Frisch, 1982; Lumpkin and Zimm, 1982; Slater and Noolandi, 1989; Viovy, 2000). To compensate for this effect on the anomalous mobilities, the μ-decrements observed for each of the members of the 199-bp family were normalized by dividing by N, the number of basepairs in each fragment. The normalized μ-decrements and average τ-decrements observed for each fragment are compared in Fig. 9. In general, the normalized mobility decrements and average τ-decrements exhibited the same trends. Most fragments with large normalized μ-decrements had large average τ-decrements, and vice versa. However, the scatter of the data points about the linear regression line indicates that a quantitative relationship does not exist between the anomalous electrophoretic mobilities observed in polyacrylamide gels and the magnitude of the curvature observed in free solution.
FIGURE 9.
Comparison of the normalized μ-decrements (−μdecrement/N) with the average τ-decrements (<−τdecrement>) observed for various members of the 199-bp family, using values taken from Tables 3–5 and data not shown. (○) 199-bp fragment and sequence mutants; (▿) insertion derivatives; and (□) fragments that do not contain the curvature module, have the apparent bend center located very close to one end, or have had the curvature module replaced by an equal-sized insert of normal DNA from pUC19. The parent 199-bp fragment and other fragments discussed specifically in the text are identified.
The 224-, 225-, 227-, and 236-bp fragments fall in a class by themselves, because they exhibited moderate or large normalized μ-decrements but had very small or average τ-decrements with high standard deviations (Table 4). Hence, these four fragments appear to be curved when analyzed by gel electrophoresis, but have normal, although relatively flexible, conformations in free solution. These four fragments are indicated by the closed triangles in Fig. 6.
The 224-, 225-, and 236-bp fragments contain 25–37-bp inserts of nested ClaI linkers within the curvature module. Nested ClaI linkers, as shown in the footnotes to Table 4, are large inverted repeats, which could form hairpins or cruciform-like structures containing one or more unpaired bases at the ends of the loop(s). Cruciform formation is usually observed in supercoiled DNAs (reviewed in Sinden, 1994). However, it is possible that the stable curvature of the helix backbone in the curvature module can be relaxed by cruciform extrusion, making the formation of a branched structure energetically feasible. Restriction fragments containing bulged bases (Bhattacharyya and Lilley, 1989; Hsieh and Griffith, 1989; Rice and Crothers, 1989; Mills et al., 1994) or branched structures (Bell and Byers, 1983; Gough and Lilley, 1985; Murchie and Lilley, 1987) are known to migrate more slowly in polyacrylamide gels than their linear counterparts. Hence, the relatively high μ-decrements observed for the 224-, 225-, and 236-bp fragments suggest that they exist as an equilibrium mixture of duplex and hairpin/cruciform structures, which migrate as a single retarded band in polyacrylamide gels. High performance liquid chromatography has shown that the Dickerson dodecamer, which is an inverted repeat (CGCGAATTGCGC), exists as a mixture of linear and hairpin structures in solution (Braunlin et al., 2004). Conformational isomers have also been observed for multimers of a curved DNA fragment electrophoresed in polyacrylamide gels (Stellwagen, 2000). Conformational isomerization may be relatively slow in the polyacrylamide gel matrix because of “caging” effects (Fried and Liu, 1994; Fried and Bromberg, 1997). In free solution, conformational exchange would be relatively rapid (Braunlin et al., 2004), leading to a mixture of structures exhibiting, on average, normal conformations with very small τ-decrements. The 227-bp fragment, with two short inverted repeats inserted into the curvature module instead of a single long inverted repeat, exhibited a smaller μ-decrement, consistent with the formation of branched structure(s) with shorter arms. The 227-bp fragment is also conformationally flexible, because it exhibited very small τ-decrements with relatively large variations from one buffer to another.
The 231-bp fragment contains three tandem ClaI linkers at position 1913, instead of the nested ClaI linkers inserted at the same site in the 224- and 236-bp fragments. The normalized μ- and τ-decrements observed for the 231-bp fragment (Fig. 9) indicate that this fragment has moderate curvature in gels and free solution. Hence, long sequences inserted into the curvature module do not necessarily lead to discordant solution and gel results, only when the insert contains a long inverted repeat.
The 205- and 217-bp fragments, which have a single ClaI linker or two nested ClaI linkers, respectively, inserted at position 1988, outside the curvature module, exhibited very large normalized μ- and τ-decrements, as shown in Fig. 9. Hence, these two fragments appear to be highly curved by both methods of measurement. The results also suggest that the addition of a long inverted repeat to a DNA fragment does not lead to hairpin/cruciform formation and conformational isomerization unless the inverted repeat is located within an intrinsically curved region of the helix backbone.
Concluding remarks
Two major results have been obtained in this work. The first is that a sequence in the middle of the SV40 gene for the major capsid protein VP1 is intrinsically curved, with an apparent bend angle of 46 ± 2° centered at sequence position 1922 ± 2 bp. The apparent bend is not a sharp kink but a delocalized region of curvature distributed over ∼60 bp, called here the curvature module for brevity. Each of the five unphased A- and T-tracts in the curvature module contributes to the curvature of the helix backbone, because the μ- and τ-decrements are reduced when any of the A- or T-tracts are modified by site-directed mutagenesis. Curvature of the 199-bp fragment is eliminated when all of the A- and T-tracts are replaced by random-sequence DNA. Hence, A-tract bending is the primary cause of curvature in this region of the VP1 gene. Because the A- and T-tracts in the curvature module are not phased with the helix repeat, it is likely that the DNA backbone has a spiral shape and is not curved in a plane. The spacing of the three A- and T-tracts closest to the apparent bend center contributes importantly to the observed curvature; changing this spacing causes the modified construct to adopt a normal conformation in gels and free solution. In addition, a very long mixed A/T sequence, spanning more than one turn of the double helix, appears to act as normal DNA and does not contribute to the curvature of the helix axis.
The biological function of curvature in the middle of the VP1 gene is not clear. The apparent bend center is located at one of several sites that are sensitive to staphylococcal nuclease digestion at low temperatures (Sundin and Varshavsky, 1979). The apparent bend center also forms the boundary of a strong nucleosome positioning signal in vitro (Stein, 1987), suggesting that wrapping DNA around a histone core might be facilitated by a stably curved DNA backbone. However, in vivo, nucleosomes appear to be preferentially excluded from this site, possibly because it may be involved in the organization of minichromosomes into higher order structures (Ambrose et al., 1990).
The second major result of the current work is the observation that there is a very poor correlation between the anomalously slow electrophoretic mobilities observed in polyacrylamide gels and the curvature observed by TEB in free solution. The most discordant solution and gel results were observed when a long inverted repeat was inserted into the curvature module. Discordant solution and gel results were not observed when a long, noninverted repeat was inserted into the curvature module, or when a long inverted repeat was inserted into the parent 199-bp fragment outside the curvature module. It seems likely that the discordant solution and gel results were due to hairpin/cruciform extrusion when long inverted repeats were placed in an intrinsically curved region of the DNA backbone. The resulting branched structures would migrate anomalously slowly in polyacrylamide gels, but would have normal, somewhat flexible conformations in free solution because of branch point migration and rapid conformational exchange. Hence, restriction fragments that exhibit anomalously slow electrophoretic mobilities in polyacrylamide gels cannot automatically be assumed to be stably curved in free solution.
Acknowledgments
We gratefully acknowledge the expert technical assistance of Kurt Strutz in carrying out the circular permutation studies.
We also acknowledge financial support for this work from the National Institute of General Medical Sciences (grant GM29690).
Brock D. Weers' present address is Dept. of Biochemistry and Biophysics, Texas A&M University, College Station, TX 77843.
Abbreviations used: A, adenine; T, thymine; bp, base pair; TEB, transient electric birefringence; TAE, Tris-acetate-EDTA buffer; TBE, Tris-borate-EDTA buffer; τ, birefringence relaxation time; τ-ratio, τR = τcurved/τnormal; τ-decrement, 100 (τR − 1); mobility ratio, μR = μcurved/μnormal; μ-decrement, 100 (μR − 1).
References
- Allemann, R. K., and M. Egli. 1997. DNA recognition and bending. Chem. Biol. 4:643–650. [DOI] [PubMed] [Google Scholar]
- Ambrose, C., H. Lowman, A. Rajadhyaksha, V. Blasquez, and M. Bina. 1990. Location of nucleosomes in simian virus 40 chromatin. J. Mol. Biol. 214:875–884. [DOI] [PubMed] [Google Scholar]
- Anderson, J. N. 1986. Detection, sequence patterns and function of unusual DNA structures. Nucleic Acids Res. 14:8513–8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbič, A., D. P. Zimmer, and D. M. Crothers. 2003. Structural origins of adenine-tract bending. Proc. Natl. Acad. Sci. USA. 100:2369–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, L., and B. Byers. 1983. Separation of branched from linear DNA by two-dimensional gel electrophoresis. Anal. Biochem. 130:527–535. [DOI] [PubMed] [Google Scholar]
- Bevington, P. R. 1969. Data Reduction and Error Analyses for the Physical Sciences. McGraw-Hill, New York, NY.
- Bhattacharyya, A., and D. M. J. Lilley. 1989. The contrasting structures of mismatched DNA sequences containing looped-out bases (bulges) and multiple mismatches (bubbles). Nucleic Acids Res. 17:6821–6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolshoy, A., P. McNamara, R. E. Harrington, and E. N. Trifonov. 1991. Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles. Proc. Natl. Acad. Sci. USA. 88:2312–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunlin, W. H., I. Giri, L. Beadling, and K. J. Breslauer. 2004. Conformational screening of oligonucleotides by variable-temperature high performance liquid chromatography: dissecting the duplex-hairpin-coil equilibria of d(CGCGAATTCGCG). Biopolymers. 74:221–231. [DOI] [PubMed] [Google Scholar]
- Brukner, I., M. Dlakić, A. Savic, S. Susic, S. Pongor, and D. Suck. 1993. Evidence for opposite groove-directed curvature of GGGCCC and AAAAA sequence elements. Nucleic Acids Res. 21:1025–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brukner, I., S. Susic, M. Dlakić, A. Savic, and S. Pongor. 1994. Physiological concentration of magnesium ions induces a strong macroscopic curvature in GGGCCC-containing DNA. J. Mol. Biol. 236:26–32. [DOI] [PubMed] [Google Scholar]
- Burkhoff, A. M., and T. D. Tullius. 1988. Structural details of an adenine tract that does not cause DNA to bend. Nature. 331:455–457. [DOI] [PubMed] [Google Scholar]
- Crothers, D. M., T. E. Haran, and J. G. Nadeau. 1990. Intrinsically bent DNA. J. Biol. Chem. 265:7093–7096. [PubMed] [Google Scholar]
- Davis, N. A., S. S. Majee, and J. D. Kahn. 1999. TATA box DNA deformation with and without the TATA box-binding protein. J. Mol. Biol. 291:249–265. [DOI] [PubMed] [Google Scholar]
- Dlakić, M., and R. E. Harrington. 1995. Bending and torsional flexibility of G/C-rich sequences as determined by cyclization assays. J. Biol. Chem. 270:29945–29952. [DOI] [PubMed] [Google Scholar]
- Dlakić, M., and R. E. Harrington. 1996. The effects of sequence context on DNA curvature. Proc. Natl. Acad. Sci. USA. 93:3847–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew, H. R., and A. A. Travers. 1985. DNA bending and its relation to nucleosome positioning. J. Mol. Biol. 186:773–790. [DOI] [PubMed] [Google Scholar]
- Eckdahl, T. T., and J. N. Anderson. 1990. Conserved DNA structures in origins of replication. Nucleic Acids Res. 18:1609–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald, D. J., G. L. Dryden, E. C. Bronson, J. S. Williams, and J. N. Anderson. 1994. Conserved patterns of bending in satellite and nucleosome positioning RNA. J. Biol. Chem. 269:21303–21314. [PubMed] [Google Scholar]
- Fredericq, E., and C. Houssier. 1973. Electric Dichroism and Electric Birefringence. Clarendon Press, Oxford, UK.
- Fried, M. G., and J. L. Bromberg. 1997. Factors that affect the stability of protein-DNA complexes during gel electrophoresis. Electrophoresis. 18:6–11. [DOI] [PubMed] [Google Scholar]
- Fried, M. G., and G. Liu. 1994. Molecular sequestration stabilizes CAP-DNA complexes during polyacrylamide gel electrophoresis. Nucleic Acids Res. 22:5054–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Bernal, J. M., and J. García de la Torre. 1980. Transport properties and hydrodynamic centers of rigid macromolecules with arbitrary shapes. Biopolymers. 19:751–766. [Google Scholar]
- Goodsell, D. S., M. Kaczor-Grzeskowiak, and R. E. Dickerson. 1994. The crystal structure of C-C-A-T-T-A-A-T-G-G. Implications for bending of B-DNA at T-A steps. J. Mol. Biol. 239:79–96. [DOI] [PubMed] [Google Scholar]
- Gough, G. W., and D. M. J. Lilley. 1985. DNA bending induced by cruciform formation. Nature. 313:154–156. [DOI] [PubMed] [Google Scholar]
- Griffith, J. D., M. Bleyman, C. A. Rauch, P. A. Kitchin, and P. T. Englund. 1986. Visualization of the bent helix in kinetoplast DNA by electron microscopy. Cell. 46:717–724. [DOI] [PubMed] [Google Scholar]
- Hagerman, P. J. 1984. Evidence for the existence of stable curvature of DNA in solution. Proc. Natl. Acad. Sci. USA. 81:4632–4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman, P. J. 1985. Sequence dependence of the curvature of DNA: a test of the phasing hypothesis. Biochemistry. 24:7033–7037. [DOI] [PubMed] [Google Scholar]
- Hagerman, P. J. 1986. Sequence-directed curvature of DNA. Nature. 321:449–450. [DOI] [PubMed] [Google Scholar]
- Hagerman, P. J. 1990. Sequence-directed curvature of DNA. Annu. Rev. Biochem. 59:755–781. [DOI] [PubMed] [Google Scholar]
- Hizver, J., H. Rozenberg, F. Frolow, D. Rabinovich, and Z. Shakked. 2001. DNA bending by an adenine-thymine tract and its role in gene regulation. Proc. Natl. Acad. Sci. USA. 98:8490–8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, D. L., and N. C. Stellwagen. 1991. Estimation of polyacrylamide gel pore size from Ferguson plots of normal and anomalously migrating DNA fragments. I. Gels containing 3% N,N′-methylenebisacrylamide. Electrophoresis. 12:253–263. [DOI] [PubMed] [Google Scholar]
- Hsieh, C.-H., and J. D. Griffith. 1988. The terminus of SV40 DNA replication and transcription contains a sharp, sequence-directed curve. Cell. 52:535–544. [DOI] [PubMed] [Google Scholar]
- Hsieh, C.-H., and J. Griffith. 1989. Deletions of bases in one strand of duplex DNA, in contrast to single-base mismatches, produce highly kinked molecules: possible relevance to the folding of single-stranded nucleic acids. Proc. Natl. Acad. Sci. USA. 86:4833–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo, H.-S., J. Drak, J. A. Rice, and D. M. Crothers. 1990. Determination of the extent of DNA bending by an adenine-thymine tract. Biochemistry. 29:4227–4234. [DOI] [PubMed] [Google Scholar]
- Koo, H.-S., H.-M. Wu, and D. M. Crothers. 1986. DNA bending at adenine-thymine tracts. Nature. 320:501–506. [DOI] [PubMed] [Google Scholar]
- Lerman, L. S., and H. L. Frisch. 1982. Why does the electrophoretic mobility of DNA in gels vary with the length of the molecule? Biopolymers. 21:995–997. [DOI] [PubMed] [Google Scholar]
- Leroy, J.-L., E. Charretier, M. Kochoyan, and M. Gueron. 1988. Evidence from base-pair kinetics for two types of adenine tract structures in solution: their relation to DNA curvature. Biochemistry. 27:8894–8898. [DOI] [PubMed] [Google Scholar]
- Levene, S. D., H.-M. Wu, and D. M. Crothers. 1986. Bending and flexibility of kinetoplast DNA. Biochemistry. 25:3988–3995. [DOI] [PubMed] [Google Scholar]
- Lu, Y. J., B. Weers, and N. C. Stellwagen. 2002. DNA persistence length revisited. Biopolymers. 61:261–275. [DOI] [PubMed] [Google Scholar]
- Lu, Y. J., B. D. Weers, and N. C. Stellwagen. 2003a. Analysis of the intrinsic DNA bend in the M13 origin of replication by atomic force microscopy. Biophys. J. 85:409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y. J., B. D. Weers, and N. C. Stellwagen. 2003b. Analysis of DNA bending by transient electric birefringence. Biopolymers. 70:270–288. [DOI] [PubMed] [Google Scholar]
- Lumpkin, O. J., and B. H. Zimm. 1982. Mobility of DNA in gel electrophoresis. Biopolymers. 21:2315–2316. [DOI] [PubMed] [Google Scholar]
- Lyubchenko, Y. L., L. S. Shlyakhtenko, E. Appella, and R. E. Harrington. 1993. CA runs increase DNA flexibility in the complex of λ Cro protein with the OR3 site. Biochemistry. 32:4121–4127. [DOI] [PubMed] [Google Scholar]
- MacDonald, D., K. Herbert, X. L. Zhang, T. Polgruto, and P. Lu. 2001. Solution structure of an A-tract DNA bend. J. Mol. Biol. 306:1081–1098. [DOI] [PubMed] [Google Scholar]
- Maki, A. S., T. W. Kim, and E. T. Kool. 2004. Direct comparison of A- and T-strand minor groove interactions in DNA curvature at A tracts. Biochemistry. 43:1102–1110. [DOI] [PubMed] [Google Scholar]
- Marilley, M., and P. Pasero. 1996. Common DNA structural features exhibited by eukaryotic ribosomal gene promoters. Nucleic Acids Res. 24:2204–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini, J. C., S. D. Levene, D. M. Crothers, and P. T. Englund. 1982. Bent helical structure in kinetoplast DNA. Proc. Natl. Acad. Sci. USA. 79:7664–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill, G., and D. E. Fisher. 1998. DNA bending and the curious case of Fos/Jun. Chem. Biol. 5:R29–R38. [DOI] [PubMed] [Google Scholar]
- McNamara, P. T., and R. E. Harrington. 1991. Characterization of inherent curvature in DNA lacking polyadenine runs. J. Biol. Chem. 266:12548–12554. [PubMed] [Google Scholar]
- Mellado, P., and J. García de la Torre. 1982. Steady-state and transient electric birefringence of solutions of bent-rod molecules. Biopolymers 21:1857–1871. [Google Scholar]
- Merling, A., N. Sagaydakova, and T. E. Haran. 2003. A-tract polarity dominate the curvature in flanking sequences. Biochemistry. 42:4978–4984. [DOI] [PubMed] [Google Scholar]
- Mertz, J. E., and P. Berg. 1974. Viable deletion mutants of Simian Virus 40: selective isolation by means of a restriction endonuclease from Hemophilus parainfluenzae. Proc. Natl. Acad. Sci. USA. 71:4879–4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills, J. B., J. P. Cooper, and P. J. Hagerman. 1994. Electrophoretic evidence that single-stranded regions of one or more nucleotides dramatically increase the flexibility of DNA. Biochemistry. 33:1797–1803. [DOI] [PubMed] [Google Scholar]
- Milton, D. L., M. L. Casper, and R. F. Gesteland. 1990. Saturation mutagenesis of a DNA region of bend. Base steps other than ApA influence the bend. J. Mol. Biol. 213:135–140. [DOI] [PubMed] [Google Scholar]
- Milton, D. L., and R. F. Gesteland. 1988. Bends in SV40 DNA: use of mutagenesis to identify the critical bases involved. Nucleic Acids Res. 16:3931–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møllegaard, N. E., C. Bailly, M. J. Waring, and P. E. Nielsen. 1997. Effects of diaminopurine and inosine substitutions on A-tract induced DNA curvature. Importance of the 3′-A-tract junction. Nucleic Acids Res. 25:3497–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie, A. I. H., and D. M. J. Lilley. 1987. The mechanism of cruciform formation in supercoiled DNA: initial opening of central basepairs in salt-dependent extrusion. Nucleic Acids Res. 15:9641–9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickol, J., and D. C. Rau. 1992. Zinc induces a bend within the transcription III-A-binding region of the 5 S RNA gene. J. Mol. Biol. 228:1115–1123. [DOI] [PubMed] [Google Scholar]
- Ohki, R., M. Hirota, M. Oishi, and R. Kiyama. 1998. Conservation and continuity of periodic bent DNA in genomic rearrangements between the c-myc and immunoglobulin heavy chain μ loci. Nucleic Acids Res. 26:3026–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, W. K., and V. B. Zhurkin. 1996. Twenty years of DNA bending. In Biological Structure and Dynamics. R. H. Sarma and M. H. Sarma, editors. Adenine Press, Schenectady, NY. 341–370.
- Pauly, M., M. Treger, E. Westhof, and P. Chambon. 1992. The initiation accuracy of the SV40 early transcription is determined by the functional domains of 2 TATA elements. Nucleic Acids Res. 20:975–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Martin, J., and V. de Lorenzo. 1997. Clues and consequences of DNA bending in transcription. Annu. Rev. Microbiol. 51:593–628. [DOI] [PubMed] [Google Scholar]
- Podtelezhnikov, A. A., C. Mao, N. C. Seeman, and A. Vologodskii. 2000. Multimerization-cyclization of DNA fragments as a method of conformational analysis. Biophys. J. 79:2692–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poljak, L. G., and J. D. Gralla. 1987. The SV40 termination region exhibits an altered helical DNA conformation. Nucleic Acids Res. 15:5433–5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pörschke, D. 1991. Persistence length and bending dynamics of DNA from electrooptical measurements at high salt concentrations. Biophys. Chem. 40:169–179. [DOI] [PubMed] [Google Scholar]
- Pytkowicz, R., and C. T. O'Konski. 1959. Characterization of Helix Pomatia hemocyanin by transient electric birefringence. Biochim. Biophys. Acta. 36:466–470. [DOI] [PubMed] [Google Scholar]
- Rice, J. A., and D. M. Crothers. 1989. DNA bending by the bulge defect. Biochemistry. 28:4512–4516. [DOI] [PubMed] [Google Scholar]
- Ross, E. D., R. B. Den, P. R. Hardwidge, and L. J. Maher, III. 1999. Improved quantitation of DNA curvature using ligation ladders. Nucleic Acids Res. 27:4135–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder, K., S. Silver, L. DeLucia, A. E. Fanning, and P. Tegtmeyer. 1986. An altered DNA conformation in origin region I is a determinant for the binding of SV40 large T antigen. Cell. 44:719–725. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell. 2001. Molecular Cloning. 3rd Ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Satchwell, S. C., H. R. Drew, and A. A. Travers. 1986. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 191:659–675. [DOI] [PubMed] [Google Scholar]
- Schatzky-Schwartz, M., N. D. Arbuckle, M. Eisenstein, D. Rabinovich, A. Bareket-Samish, T. E. Haran, B. F. Luisi, and Z. Shakked. 1997. X-ray and solution studies of DNA oligomers and implications for the structural basis of A-tract-dependent curvature. J. Mol. Biol. 267:595–623. [DOI] [PubMed] [Google Scholar]
- Seela, F., and T. Grein. 1992. 7-Deaza-2′-deoxyadenosine and 3-deaza-2′-deoxyadenosine replacing dA within d(A6)-tracts: differential bending at 3′- and 5′-junctions of d(A6)·d(T6) and B-DNA. Nucleic Acids Res. 20:2297–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrader, T. E., and D. M. Crothers. 1990. Effects of DNA sequence and histone-histone interactions on nucleosome placement. J. Mol. Biol. 216:69–84. [DOI] [PubMed] [Google Scholar]
- Sinden, R. R. 1994. DNA Structure and Function. Academic Press, San Diego, CA.
- Sivolob, A. V., and S. N. Khrapunov. 1995. Translational positioning of nucleosomes on DNA: the role of sequence-dependent isotropic DNA bending stiffness. J. Mol. Biol. 247:918–931. [DOI] [PubMed] [Google Scholar]
- Slater, G. W., and J. Noolandi. 1989. The biased reptation model of DNA gel electrophoresis: mobility vs molecular size and gel concentration. Biopolymers. 28:1781–1791. [DOI] [PubMed] [Google Scholar]
- Stein, A. 1987. Unique positioning of reconstituted nucleosomes occurs in one region of simian virus 40 DNA. J. Biol. Chem. 262:3872–3879. [PubMed] [Google Scholar]
- Stellwagen, A., and N. C. Stellwagen. 1990. Anomalously slow electrophoretic mobilities of DNA restriction fragments in polyacrylamide gels are not eliminated by increasing the gel pore size. Biopolymers. 30:309–324. [DOI] [PubMed] [Google Scholar]
- Stellwagen, N. C. 1981. Electric birefringence of restriction enzyme fragments of DNA: optical factor and electric polarizability as a function of molecular weight. Biopolymers. 20:399–434. [DOI] [PubMed] [Google Scholar]
- Stellwagen, N. C. 1983. Anomalous electrophoresis of DNA restriction fragments on polyacrylamide gels. Biochemistry. 22:6186–6193. [DOI] [PubMed] [Google Scholar]
- Stellwagen, N. C. 1991. Electric birefringence of two small DNA fragments of the same molecular weight. Biopolymers. 31:1651–1667. [DOI] [PubMed] [Google Scholar]
- Stellwagen, N. C. 1995. Use of polyacrylamide gel electrophoresis to detect structural variations in kilobase-sized DNA. Electrophoresis. 16:691–699. [DOI] [PubMed] [Google Scholar]
- Stellwagen, N. C. 1997. DNA mobility anomalies are determined primarily by polyacrylamide gel concentration, not gel pore size. Electrophoresis. 18:34–44. [DOI] [PubMed] [Google Scholar]
- Stellwagen, N. C. 2000. Conformational isomers of curved DNA molecules can be observed by polyacrylamide gel electrophoresis. Electrophoresis. 21:2327–2334. [DOI] [PubMed] [Google Scholar]
- Stellwagen, N. C. 2003. Sequence-dependent bending in plasmid pUC19. Electrophoresis. 24:3467–3475. [DOI] [PubMed] [Google Scholar]
- Strutz, K., and N. C. Stellwagen. 1996. Intrinsic curvature of plasmid DNAs analyzed by polyacrylamide gel electrophoresis. Electrophoresis. 17:989–995. [DOI] [PubMed] [Google Scholar]
- Sundin, O., and A. Varshavsky. 1979. Staphylococcal nuclease makes a single non-random cut in the simian virus 40 viral minichromosome. J. Mol. Biol. 132:535–546. [DOI] [PubMed] [Google Scholar]
- Tchernaenko, V., H. R. Halvorson, and L. C. Lutter. 2003. Topological measurement of an A-tract bend angle: variation of duplex winding. J. Mol. Biol. 326:751–760. [DOI] [PubMed] [Google Scholar]
- Tirado, M. M., and J. García de la Torre. 1980. Rotational dynamics of rigid, symmetric top macromolecules. Application to circular cylinders. J. Chem. Phys. 73:1986–1993. [Google Scholar]
- Tirado, M. M., C. López Martínez, and J. García de la Torre. 1984. Comparison of theories for the translational and rotational diffusion coefficients of rod-like macromolecules. Application to short DNA fragments. J. Chem. Phys. 81:2047–2052. [Google Scholar]
- Tonelli, M., E. Ragg, A. M. Bianucci, K. Lesiak, and T. L. James. 1998. Nuclear magnetic resonance structure of d(GCATATGATAG)·d(CTATCATATGC): a consensus sequence for promoters recognized by σK RNA polymerase. Biochemistry. 37:11745–11761. [DOI] [PubMed] [Google Scholar]
- Wu, H.-M., and D. M. Crothers. 1984. The locus of sequence-directed and protein-induced DNA bending. Nature. 308:509–513. [DOI] [PubMed] [Google Scholar]
- Vacano, E., and P. J. Hagerman. 1997. Analysis of birefringence decay profiles for nucleic acid helices possessing bends: the τ-ratio approach. Biophys. J. 73:306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viovy, J.-L. 2000. Electrophoresis of DNA and other polyelectrolytes: physical mechanisms. Rev. Mod. Phys. 72:813–872. [Google Scholar]
- Zahn, K., and F. R. Blattner. 1987. Direct evidence for DNA bending at the lambda replication origin. Science. 236:416–422. [DOI] [PubMed] [Google Scholar]