Abstract
Pressure-induced unfolding of 23-kDa protein from spinach photosystem II has been systematically investigated at various experimental conditions. Thermodynamic equilibrium studies indicate that the protein is very sensitive to pressure. At 20°C and pH 5.5, 23-kDa protein shows a reversible two-state unfolding transition under pressure with a midpoint near 160 MPa, which is much lower than most natural proteins studied to date. The free energy (ΔGu) and volume change (ΔVu) for the unfolding are 5.9 kcal/mol and −160 ml/mol, respectively. It was found that NaCl and sucrose significantly stabilize the protein from unfolding and the stabilization is associated not only with an increase in ΔGu but also with a decrease in ΔVu. The pressure-jump studies of 23-kDa protein reveal a negative activation volume for unfolding (−66.2 ml/mol) and a positive activation volume for refolding (84.1 ml/mol), indicating that, in terms of system volume, the protein transition state lies between the folded and unfolded states. Examination of the temperature effect on the unfolding kinetics indicates that the thermal expansibility of the transition state and the unfolded state of 23-kDa protein are closer to each other and they are larger than that of the native state. The diverse pressure-refolding pathways of 23-kDa protein in some conditions were revealed in pressure-jump kinetics.
INTRODUCTION
Understanding protein folding mechanisms remains one of the major challenges in protein science. Various physical and chemical perturbations such as temperature, pH, or chemical denaturants have been widely used as experimental variables to explore protein unfolding. These studies are carried out mostly at atmospheric pressure, and have provided considerable information on the process (Buck et al., 1994; Creighton, 1993; Dobson et al., 1998; Kim and Baldwin, 1982, 1990). However, the knowledge gained so far is insufficient for gaining a full understanding how proteins fold into native state. According to Le Chatelier's principle, pressure is a fundamental thermodynamic variable in addition to temperature and chemical potential, and governs the chemical equilibrium of a state occupying smaller volume at higher pressure. Unfolding of proteins has been found to mostly accompany a reduction in the volume of a protein-solvent system (Heremans, 1982; Mozhaev et al., 1996; Silva and Weber, 1993; Weber and Drickamer, 1983; Zipp and Kauzmann, 1973). Using high pressure to explore protein-unfolding offers a number of advantages compared with other physical or chemical perturbations. It provides valuable information on the volume change upon unfolding, activation volumes, and thermal expansibility. More and more experimental data have been reported based on high pressure techniques, including Fourier transform infrared, nuclear magnetic resonance, small-angle x-ray scattering, high pressure densitometry, fluorescence, and fourth-derivative ultraviolet (UV) absorbance spectra (Desai et al., 1999; Jonas, 2002; Lange and Balny, 2002; Mohana-Borges et al., 1999; Ruan and Balny, 2002; Winter, 2002; Woenckhaus et al., 2001).
Photosystem II (PSII) is a multisubunit membrane protein complex performing light-induced electron transfer and water-splitting reactions, leading to the evolution of molecular oxygen. In green algae and higher plants, PSII contains three extrinsic proteins of 33-kDa, 23-kDa, and 17-kDa functioning to stabilize the Mn-cluster that directly catalyzes the water-splitting reaction. In our previous work, the pressure-unfolding of 33-kDa protein isolated from spinach photosystem II was studied by several approaches, including intrinsic (tryptophan) and extrinsic 8-anilino-naphthalino-sulfonic acid fluorescence, as well as fourth-derivative UV absorbance spectra (Ruan et al., 2001, 2003). These measurements revealed that the protein is very sensitive to pressure. A pressure of 180 MPa can totally and reversibly unfold the protein at 20°C and pH 6.0. This transition pressure is one of the lowest observed for natural (nonmutant) proteins so far. We also demonstrated that the pressure-unfolding of 33-kDa protein can be significantly modulated by physical or chemical perturbation. The unfolding free energy was found to decrease by increasing temperature, whereas the unfolding volume change (ΔVu) remained constant, suggesting that the change in thermal expansibility of the folded and unfolded state for 33-kDa protein is very small. This result differs significantly from that observed in Snase and trp repressor (Desai et al., 1999; Panick et al., 1999; Woenckhaus et al., 2001). Interestingly, through examining the pressure stability of the 33-kDa protein in the presence of sucrose and NaCl, it was found that the stabilization effect of these reagents was associated not only with an increase in free energy of unfolding, but also with a decrease in the absolute value of the unfolding volume change. The increase in unfolding free energy has been interpreted as arising from preferential hydration proposed by Timasheff (1993), whereas the decrease in the volume change has been suggested to result from several effects of these reagents on protein including osmotic stress (Ruan et al., 2003).
The unique behavior of 33-kDa protein under pressure motivated us to study another extrinsic protein from spinach PSII, i.e., 23-kDa protein (also intimately implicated in oxygen production), which modulates the Ca2+ and Cl− requirement for oxygen evolution and therefore has been widely studied (Berthold et al., 1981; Homann and Madabusi, 1993; Murata and Miyao, 1985; Ono et al., 1992; Seidler, 1994). The 23-kDa protein consists of 186 amino acid residues with two tryptophan residues located at residue numbers 34 and 167, respectively (Seidler, 1994). We found that, upon unfolding, the intrinsic tryptophan fluorescence of the protein exhibits both a large spectral shift and a substantial decrease in intensity. Furthermore, 23-kDa protein can be easily unfolded by pressure. A pressure of 200 MPa can completely unfold the protein at pH 5.5 and 20°C. These make it very convenient to follow the unfolding/refolding of the protein in both equilibrium and kinetics studies. The kinetics of protein unfolding/folding can provide direct information on the structural characteristics of the transition state relative to the native and unfolded states of the protein, revealing important aspects of the rate-limiting step in the process. Recently, with the development of the pressure-jump technique, a number of in-depth studies have been reported (Desai et al., 1999; Mohana-Borges et al., 1999; Panick et al., 1999; Panick and Winter, 2000; Vidugiris et al., 1995; Woenckhaus et al., 2001). However, the number of pressure-jump kinetics studies remains very limited compared with that of equilibrium studies. Here we present a systematic investigation of equilibrium and kinetics of the unfolding/refolding of 23-kDa protein induced by changes in pressure.
MATERIALS AND METHODS
Purification of the 23-kDa protein
The 23-kDa protein was purified from spinach photosystem II which was isolated from market spinach according to the method of Berthold et al. (1981) and then treated with 1 M NaCl at 1.0 mg chl/ml for 30 min in the dark to release the 23-kDa and 17-kDa extrinsic proteins. The NaCl-extract was diluted sixfold with 30 mM citric acid at pH 4.0, and immediately purified with a Bio-Scale S column (Bio-Rad, Hercules, CA) which has been equilibrated with 30 mM citric acid at pH 4.0. The 23-kDa protein was eluted with a NaCl gradient from 350 mM to 650 mM. All the treatment and purification procedures were carried out at 4°C or on ice. The purity of the eluted protein was confirmed by SDS-polyacrylamide gel analysis to be sufficient for this study (data not shown). The purified protein was dissolved in 0.1 M pH 5.5 4-morpholine-ethanesulfonic acid (MES) buffer, unless otherwise indicated.
Fluorescence measurements
Fluorescence measurements were carried out using either an Aminco Bowman Series 2 fluorescence-spectrophotometer (SLM Aminco, Foster City, CA) or an SLM 48000 fluorescence-spectrophotometer (SLM Aminco) in which the sample housings were modified at the INSERM laboratory and at the Shanghai laboratory, respectively, to measure fluorescence under pressure from 0.1 MPa to 600 MPa for the former and from 0.1 MPa to 300 MPa for the latter through thermostated pressure bombs. The fluorescence spectra were quantified by specifying the center of spectral mass 〈ν〉, which was defined and used by Silva et al. (1986),
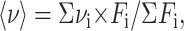 |
(1) |
where νi is the wavenumber and Fi the fluorescence intensity at νi. The excitation wavelength for the intrinsic tryptophan fluorescence was 295 nm.
The degree of unfolding or degree of transition (α) is related to 〈ν〉 by the formula
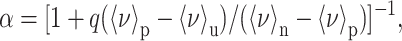 |
(2) |
where q is the ratio of the quantum yield of the unfolded state over the native state; 〈ν〉p is the center of spectral mass at pressure p; and 〈ν〉u and 〈ν〉n are the corresponding quantities for the unfolded and native states, respectively. The free energy and volume change upon unfolding, ΔGu and ΔVu, were calculated according to the method of Li et al. (1976). Equation 2 was also used to calculate α from the data obtained by using the fourth-derivative absorbance spectroscopy, in which the 〈ν〉 values were substituted with the corresponding accumulation amplitude for the fourth-derivative experiments.
Fourth-derivative UV absorbance spectra
Absorption spectra of the protein between 260 and 300 nm were recorded at 20°C using a modified Cary3 (Varian, Palo Alto, CA) absorption spectrophotometer described elsewhere allowing experiments in a pressure range from atmospheric pressure to 500 MPa through a thermostated high-pressure bomb. The fourth-derivative absorbance spectra were calculated from the corresponding absorption spectrum as described previously (Lange et al., 1996).
Measurement of thermal unfolding of 23-kDa protein
The measurement of the protein unfolding induced by temperature was carried out in the same way as in the fluorescence measurements mentioned above, except that the pressure was kept at atmospheric pressure. After a predefined temperature was reached, the protein was incubated at that temperature for 15 min, and then the fluorescence spectrum excited at 295 nm was recorded. This procedure was repeated with increasing temperature step-by-step until the maximum emission wavelength of the fluorescence spectrum shifted to 350 nm, indicating that the tryptophan residue of 23-kDa protein has been completely exposed to the aqueous solution caused by thermal denaturation.
Measurement of unfolding-folding kinetics induced by pressure-jump
The measurements of unfolding-folding kinetics induced by pressure-jump were carried out on an Aminco Bowman Series 2 fluorescence spectrophotometer connected with a pressure jump device made in the INSERM laboratory. Positive or negative pressure-jumps up to 100 MPa were performed in the pressure range from 0.1 to 600 MPa, with a dead-time of 5–10 ms. The tryptophan fluorescence intensity at 330 nm was used to monitor the unfolding-folding kinetics of the protein after positive or negative pressure jumps. The relaxation profiles of the unfolding/folding under given pressures were assumed to be single-exponential processes and were fit to get the relaxation time τ,
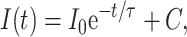 |
(3) |
where I(t) is the fluorescence intensity at time t, I0 is the intensity at time zero, and C is the asymptotic value at a given pressure. The inverse of the relaxation time is the apparent rate constant at the given pressure, kapp(p). For a two-state folding/unfolding process the apparent rate constant kapp(p) and the unfolding equilibrium constant Ku(p) have the following relations with the unfolding rate constant ku(p) and folding rate constant kf(p) at pressure p:
 |
(4) |
 |
(5) |
Equations 4 and 5 can be used to calculate ku(p) and kf(p) from kapp(p) and Ku(p) at the respective pressures, in which Ku(p), the unfolding equilibrium constant at pressure p, is calculated from Kou, the unfolding equilibrium constant at atmospheric pressure according to
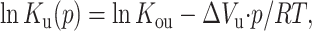 |
(6) |
where ΔVu is the volume change upon unfolding; and p, R, and T indicate the pressure, universal gas constant, and temperature, respectively. Meanwhile, Kou is obtained from the equilibrium measurement. The unfolding and folding rate constant at atmospheric pressure (kof and kou) and activated volume for unfolding and folding (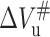 and
and 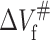 ) can be obtained from Eqs. 5 and 6, respectively, based on ku(p) and Δkf(p) at different pressures:
) can be obtained from Eqs. 5 and 6, respectively, based on ku(p) and Δkf(p) at different pressures:
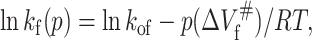 |
(7) |
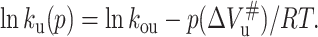 |
(8) |
Measurement of unfolding kinetics induced by guanidinium hydrochloride (Gdm-HCl)
Kinetics measurements of 23-kDa protein unfolding induced by guanidinium hydrochloride (Gdm-HCl) were performed on a mini stopped-flow device (SLM Aminco) by mixing native 23-kDa protein with Gdm-HCl at a ratio 1:1 to yield the desired Gdm-HCl concentration. The tryptophan emission spectra were recorded by a charge-coupled device spectrophotometer (Acton Research, Acton, MA) mounted on an SLM 48000. The excitation wavelength was 295 nm.
RESULTS
Pressure-unfolding of the 23-kDa protein
The tryptophan fluorescence spectra of the 23-kDa protein under various pressures were shown in Fig. 1. The maximum emission wavelength of the protein was at 325 nm, indicating that the average environment of two tryptophan residues in 23-kDa protein is fairly hydrophobic. Upon increasing the pressure from atmospheric step by step, the maximum emission wavelength gradually shifted from 325 to 350 nm, which is the typical spectrum of free tryptophan in aqueous solution. This indicates that the tryptophan residues in the protein are gradually exposed to solvent due to increased pressure. Further increases in pressure did not yield any further spectral shift, and a plateau in the change of center mass was achieved at 200 MPa (see Fig. 1 B), indicating that the tryptophan residues in 23-kDa protein were totally exposed to solvent as a result of pressure-induced unfolding. The decrease in fluorescence intensity with the pressure observed in Fig. 1 A also supports this conclusion. The profiles of the center mass of 23-kDa protein during the unfolding and refolding (Fig. 1 B) indicate that the unfolding is reversible and that the unfolding is a two-state transition. A small degree of hysteresis is observed in the equilibrium profile. Based on the data in Fig. 1, the free energy (ΔGu) and the volume change (ΔVu) upon unfolding were calculated as described in Materials and Methods; they were 5.60 kcal/mol and −150.3 ml/mol, respectively. The value of ΔVu of 23-kDa protein is much larger than that of 33-kDa protein (−120 ml/mol), considering that its molecular weight is 75% of the latter. The inset in Fig. 1 is the fourth-derivative absorption spectra of the protein under pressures from 0.1 to 200 MPa. The fourth-derivative absorption of protein (especially the spectral range from 280 to 286 nm and from 287 to 297 nm) can be used to study the local environment characteristics of tyrosine and tryptophan residues (Lange et al., 1996). The spectra in these two regions present both a distinct blue shift and decrease in amplitude upon increasing pressure, indicating that the tyrosine and tryptophan residues in 23-kDa protein become more and more exposed to the solvent upon pressure unfolding (Lange et al., 1996). As observed in fluorescence measurement, when the pressure was raised to 180 MPa and higher, no further spectral change in the inset can be observed. This observation indicates that all tyrosine and tryptophan residues in 23-kDa protein were completely exposed to solvent, leading to the same conclusion as above, i.e., that the protein was unfolded by a pressure ∼200 MPa. The calculated free energy and standard volume change from this measurement are 5.81 kcal/mol and 157.6 ml/mol, respectively, and these agree reasonably with the values obtained in fluorescence (see Table 1). The Gdm-HCl unfolding of 23-kDa protein was also examined. It was found that 23-kDa protein was readily unfolded by Gdm-HCl. The entire transition occurs between 1.0 and 1.5 M Gdm-HCl. The free energy was 6.85 kcal/mol, again in reasonable agreement with the values obtained from pressure-induced unfolding.
FIGURE 1.
Fluorescence emission spectra of 23-kDa protein under various pressures. (A) The fluorescence emission spectra of 23-kDa protein under pressures from 0.1 to 220 MPa (top to bottom). Protein concentration: 0.1 mg/ml in 0.1 M MES buffer, pH 5.5. Excitation wavelength: 295 nm. (Inset) The fourth-derivative UV spectra of 23-kDa protein under pressures 0.1–200 MPa (from right to left). Protein concentration: 0.6 mg/ml in 0.1 M MES buffer, pH 5.5. All measurements were carried out at 20°C. (B) The center of spectral mass in compression (▪) and decompression (•). The values were obtained from the spectra shown in A by the calculation described in Materials and Methods.
TABLE 1.
Thermodynamics parameters of 23-kDa protein at various temperatures
| Temperature (°C) | ΔGu (kcal/mol) | ΔVu (ml/mol) | p0.5 (MPa) |
|---|---|---|---|
| 3 | 5.36 | −202.6 | 118 |
| 10 | 5.77 | −174.2 | 138 |
| 20 | 5.60 | −150.3 | 156 |
| 35 | 4.43 | −120.0 | 152 |
| 45 | 3.88 | −129.7 | 128 |
Temperature effect on the pressure-unfolding
The pressure-induced unfolding profiles of 23-kDa protein at various temperatures were shown in Fig. 2. It can be seen that whatever the temperature is from 3 to 45°C, the protein were totally unfolded in pressure range of 160–200 MPa, and the maximum emission wavelengths of the protein tryptophan fluorescence were shifted to 350 nm (data not shown). The lines in Fig. 2 A represent the fitting of the data to a simple two-state unfolding transition, and the values calculated for the Gibbs free energy and volume change of 23-kDa protein upon unfolding, ΔGu and ΔVu, are listed in Table 1. The ΔGu at various temperatures shown in Table 1 and the nonlinear profile of lnKu versus temperature in Fig. 2 B indicate that the protein is more stable at ∼20°C. In other words, 23-kDa protein is destabilized at either higher or lower temperatures as compared to 20°C. The highest values of p1/2, the transition pressure midpoint at various temperatures listed in Table 1, was at 20°C (156 MPa), which also supports this conclusion. According to Fig. 2 A, the pressure required to unfold 23-kDa protein at various temperatures were estimated and plotted as a p/T phase diagram in Fig. 2 C. The temperature needed to denature the protein at atmospheric pressure was 85°C determined from the thermal-denaturation experiment. The phase diagram exhibits the well-known curvature for heat and cold denaturation of proteins due to the large decrease in heat capacity upon folding (Panick et al., 1999; Smeller, 2002). The similar temperature dependence of the stability was also observed in trp repressor, Snase, 33-kDa protein, and was attributed to the large increase in heat capacity upon unfolding (Desai et al., 1999; Panick et al., 1999). The temperature dependence of the unfolding ΔVu in Fig. 3 indicates that the ΔVu is roughly linearly decreased in absolute value with temperature increase. The slope of the linear fit of ΔVu versus temperature is ∼1.8 ml/mol deg.
FIGURE 2.
Temperature effect on 23-kDa protein unfolding induced by pressure. (A) The pressure-unfolding profile of 23-kDa protein at various temperatures. The degrees of unfolding were calculated from the tryptophan fluorescence spectra of the protein under pressures at temperatures 3°C (▪), 10°C (□), 20°C (•), 35°C (▵), and 45°C (▾). Protein concentration: 0.1 mg/ml in 0.1 M MES buffer, pH 5.5. (B) Natural logarithm of Ku, the equilibrium constant of 23-kDa protein unfolding versus the inverse of temperature. The values of Ku were calculated from the data in A as described in Materials and Methods. (C) The p/T-stability diagram of 23-kDa protein.
FIGURE 3.
The temperature dependence of the equilibrium volume changes for unfolding (ΔVu) and activation transition for unfolding 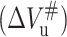 as well as folding
as well as folding 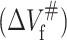 of 23-kDa protein.
of 23-kDa protein.
The stabilizing effect of NaCl and sucrose
In our previous study on 33-kDa protein, it was clearly found that protein-stabilizers, such as sucrose, glycerol, and NaCl, significantly protected the 33-kDa from pressure-unfolding and that the stabilization effect was associated not only with the increase in free energy, but also with the reduction in the volume change upon unfolding (Ruan et al., 2003). In Fig. 4, the unfolding curves of 23-kDa protein in the presence of various concentrations of NaCl (Fig. 4 A) or sucrose (Fig. 4 B) were shown. Clearly, the presence of NaCl or sucrose shifted the pressure required for unfolding 23-kDa protein to higher ones. For instance, the p1/2 of the protein in the absence of sucrose was 156 MPa, but it shifted to 173, 188, and 238 MPa in the presence of 0.2, 0.4, and 0.8 M sucrose. A similar situation was also found with NaCl. These results indicate that sucrose or NaCl can significantly protect 23-kDa protein from pressure-unfolding, very similar to the observation on the 33-kDa protein. The free energy and the volume change upon unfolding of the protein obtained from the data in Fig. 4 were listed in Table 2, which indicated that the free energy increases whereas the unfolding ΔVu exhibits a significant decrease with increasing concentration of sucrose or NaCl. This result gives strong support to the previous conclusion obtained in the 33-kDa protein study—i.e., that reagents such as sucrose or NaCl protect protein against pressure-induced unfolding not only because of the increase in the free energy caused by the increase of surface tension around the protein molecules, but also because of the reduction in absolute value of the volume change upon unfolding (Frye and Royer, 1997; Ruan et al., 2003; Timasheff, 1993).
FIGURE 4.
Stabilization effect of NaCl and sucrose on 23-kDa protein against pressure unfolding. (A) Stabilization effect of NaCl on 23-kDa protein against pressure unfolding. The degrees of unfolding were calculated from the fluorescence spectra under various pressures in the presence of 0.0 M (▴), 0.5 M (•), and 1.0 M (▪) NaCl at 20°C. The excitation wavelength was at 295 nm; the protein was dissolved at 0.1 mg/ml in 0.1 M MES buffer, pH 5.5. (B) Stabilization effect of sucrose on 23-kDa protein against pressure unfolding. The degrees of unfolding were calculated from the fluorescence spectra under various pressures in the presence of 0.0 M (▴), 0.2 M (•), 0.4 M (▪), and 0.8 M (▾) sucrose at 20°C. The excitation wavelength was at 295 nm; the protein was dissolved at 0.1 mg/ml in 0.1 M MES buffer, pH 5.5.
TABLE 2.
Effect of NaCl and sucrose on thermodynamics parameters of 23-kDa protein
| ΔGu (kcal/mol) | ΔVu (ml/mol) | p0.5 (MPa) | |
|---|---|---|---|
| NaCl (M) | |||
| 0.0 | 5.60 | −150.3 | 156 |
| 0.5 | 6.42 | −141.2 | 188 |
| 1.0 | 7.01 | −120.2 | 243 |
| Sucrose (M) | |||
| 0.0 | 5.60 | −150.3 | 156 |
| 0.2 | 5.91 | −150.1 | 173 |
| 0.4 | 6.45 | −140.4 | 188 |
| 0.8 | 6.49 | −114.7 | 238 |
The kinetics of 23-kDa protein unfolding and refolding
A series of tryptophan fluorescence relaxation profiles of 23-kDa protein unfolding and refolding induced by pressure jumps at 20°C were plotted in Fig. 5. Decrease or increase in fluorescence intensity was observed after rapid positive or negative pressure jumps, respectively (Fig. 5, A and B), showing good reversibility of the pressure-induced unfolding as observed in equilibrium experiments mentioned above. All fluorescence relaxation profiles for both unfolding and refolding were well fitted with a single exponential function, indicating the unfolding-folding transition is a two-state process. The apparent rate constants at pressure p, kapp(p) were plotted as a function of pressure in Fig. 5 C. It is obvious that the variation of kapp(p) upon the increasing pressure shows two phases. In the first phase (from 120 to 150 MPa) kapp(p) decreases with pressure, whereas in the second (>150 MPa), kapp(p) increases with pressure. From this, it is expected that for 23-kDa protein the pressure dependence of the folding rate constant, kf(p), and the unfolding rate constant, ku(p), should behave in a contrary manner according to Eqs. 4, 7, and 8. The data in Fig. 5 were used to calculate kf(p) and ku(p) under various pressures as described in Materials and Methods. The results indicate that the pressure dependence of kf(p) and ku(p) is just as expected above; i.e., kf(p) slows down and ku(p) speeds up with the increases of pressure. From the slopes of the linear fitting of kf(p) and ku(p) versus pressure, the folding activation volume  and unfolding activation volume
and unfolding activation volume 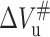 were respectively obtained. Meanwhile the rate constant for folding and unfolding at atmospheric pressure, kf and ku, were obtained by extrapolating the linear fitting curve to atmospheric pressure, respectively. Their values were listed in Table 3. As seen in the table, the folding activation volume of the protein,
were respectively obtained. Meanwhile the rate constant for folding and unfolding at atmospheric pressure, kf and ku, were obtained by extrapolating the linear fitting curve to atmospheric pressure, respectively. Their values were listed in Table 3. As seen in the table, the folding activation volume of the protein,  is positive (84.1 ml/mol), whereas
is positive (84.1 ml/mol), whereas 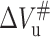 is negative (−66.2 ml/mol). As mentioned above, all the relaxation profiles for both unfolding and folding in Fig. 5 were well fit with a single exponential function. However, it was found that the relaxation profile of the 23-kDa protein refolding could only be fit with a two-exponential function in the event that the pressure span of the negative pressure-jump was larger (∼50 MPa or more) and the final pressure after jump was close to 100 MPa or lower. In the equilibrium and kinetics study described above, it was found that 100 MPa is the pressure for 23-kDa protein to begin unfolding significantly. Fig. 6, A–C, indicate that the relaxation profile of refolding after jump from 150 to 100 MPa can be well fit with a two-exponential function but not a single-exponential one. The relaxation times for the two phases are 3.4 and 77.5 s, respectively, whereby the fast phase dominates over the slower one. This observation indicates that the refolding of 23-kDa protein upon releasing pressure to pressures below the onset of the transition follows two paths. One is responsible for the slow relaxation time, which is on the same timescale with that in the unfolding and refolding at 100 MPa shown in Fig. 5 (80.0 s), suggesting that it would be the identical pathway with the unique unfolding observed above. The other pathway responsible for the fast relaxation time has not been observed other than in Fig. 6, implying that the populations of the corresponding intermediate become larger and detectable only under these conditions. The relaxation profile of 23-kDa protein refolding after the pressure jump from 200 to 150 MPa was also shown in Fig. 6 A. However, this relaxation profile could be fit quite well with a single-exponential function (Fig. 6 D). The same results were also obtained from the similar negative pressure jumps (not shown). This indicates that the two-phase phenomenon is not caused by the large-span pressure-jump. Furthermore, the unfolding relaxation after a positive jump from 100 MPa to 200 MPa was also fit well with a single-exponential function (Fig. 7), providing further evidence suggesting that the two-exponential relaxation for refolding observed above is reliable. More interestingly, the two-exponential relaxation for refolding seems to be related to temperature. The lower temperature renders it much more obvious (Fig. 7 C), whereas higher temperature has the opposite effect (Fig. 7 A). When the temperature was raised to 45°C, the relaxation profile of 23-kDa protein refolding after the similar big jump can be fit as a single-exponential function. All of these revealed that the pressure-refolding pathway of 23-kDa protein could be varied by the experimental conditions. The unfolding kinetics of 23-kDa protein induced by Gdm-HCl at 20°C was also examined by stopped-flow experiments. The final concentration of Gdm-HCl in the stopped-flow experiment was 1.5 M, which can completely denature the protein, as revealed in equilibrium study. The relaxation profile is well fit with a single-exponential function with a relaxation time of 8.5 s, significantly faster than that observed in pressure-unfolding kinetics. For the latter, the shortest (40 s) and the longest (80 s) relaxation time were observed at 200 and 100 MPa, respectively (Fig. 7 B and Fig. 5 A). The phenomenon that pressure slows down unfolding/folding kinetics is in agreement with the theoretical model presented by Hummer et al. (1998). These same phenomena have been observed in a number of proteins like Snase, Rnase, trp repressor, CI2, P13, and tendamistat (Blum et al., 1978; Desai et al., 1999; Holcomb and Van Holde, 1962; Kitahara et al., 2002; Mohana-Borges et al., 1999; Panick et al., 1998, 1999; Panick and Winter, 2000; Pappenberger, et al., 2000), leading to the conclusion that applying pressure does slow down protein folding/unfolding.
is negative (−66.2 ml/mol). As mentioned above, all the relaxation profiles for both unfolding and folding in Fig. 5 were well fit with a single exponential function. However, it was found that the relaxation profile of the 23-kDa protein refolding could only be fit with a two-exponential function in the event that the pressure span of the negative pressure-jump was larger (∼50 MPa or more) and the final pressure after jump was close to 100 MPa or lower. In the equilibrium and kinetics study described above, it was found that 100 MPa is the pressure for 23-kDa protein to begin unfolding significantly. Fig. 6, A–C, indicate that the relaxation profile of refolding after jump from 150 to 100 MPa can be well fit with a two-exponential function but not a single-exponential one. The relaxation times for the two phases are 3.4 and 77.5 s, respectively, whereby the fast phase dominates over the slower one. This observation indicates that the refolding of 23-kDa protein upon releasing pressure to pressures below the onset of the transition follows two paths. One is responsible for the slow relaxation time, which is on the same timescale with that in the unfolding and refolding at 100 MPa shown in Fig. 5 (80.0 s), suggesting that it would be the identical pathway with the unique unfolding observed above. The other pathway responsible for the fast relaxation time has not been observed other than in Fig. 6, implying that the populations of the corresponding intermediate become larger and detectable only under these conditions. The relaxation profile of 23-kDa protein refolding after the pressure jump from 200 to 150 MPa was also shown in Fig. 6 A. However, this relaxation profile could be fit quite well with a single-exponential function (Fig. 6 D). The same results were also obtained from the similar negative pressure jumps (not shown). This indicates that the two-phase phenomenon is not caused by the large-span pressure-jump. Furthermore, the unfolding relaxation after a positive jump from 100 MPa to 200 MPa was also fit well with a single-exponential function (Fig. 7), providing further evidence suggesting that the two-exponential relaxation for refolding observed above is reliable. More interestingly, the two-exponential relaxation for refolding seems to be related to temperature. The lower temperature renders it much more obvious (Fig. 7 C), whereas higher temperature has the opposite effect (Fig. 7 A). When the temperature was raised to 45°C, the relaxation profile of 23-kDa protein refolding after the similar big jump can be fit as a single-exponential function. All of these revealed that the pressure-refolding pathway of 23-kDa protein could be varied by the experimental conditions. The unfolding kinetics of 23-kDa protein induced by Gdm-HCl at 20°C was also examined by stopped-flow experiments. The final concentration of Gdm-HCl in the stopped-flow experiment was 1.5 M, which can completely denature the protein, as revealed in equilibrium study. The relaxation profile is well fit with a single-exponential function with a relaxation time of 8.5 s, significantly faster than that observed in pressure-unfolding kinetics. For the latter, the shortest (40 s) and the longest (80 s) relaxation time were observed at 200 and 100 MPa, respectively (Fig. 7 B and Fig. 5 A). The phenomenon that pressure slows down unfolding/folding kinetics is in agreement with the theoretical model presented by Hummer et al. (1998). These same phenomena have been observed in a number of proteins like Snase, Rnase, trp repressor, CI2, P13, and tendamistat (Blum et al., 1978; Desai et al., 1999; Holcomb and Van Holde, 1962; Kitahara et al., 2002; Mohana-Borges et al., 1999; Panick et al., 1998, 1999; Panick and Winter, 2000; Pappenberger, et al., 2000), leading to the conclusion that applying pressure does slow down protein folding/unfolding.
FIGURE 5.
Relaxation profiles of 23-kDa protein tryptophan fluorescence after pressure jumps at 20°C. (A) Relaxation profiles after a set of positive jumps. The first pressure jump was started from 100 MPa. The final pressures after each jump (from top to bottom) were 128.8, 144.3, 159.5, 174.9, and 199.4 MPa, respectively. (B) Relaxation profiles after a set of negative jumps. The first negative pressure was started from 199.4 MPa. The final pressures after each negative jump (from bottom to top) were 172.0, 157.2, 142.7, 127.9, and 104.6 MPa, respectively. The solid lines through the points in A and B represent the fits to a single-exponential function. The protein concentration in experiment was 0.1 mg/ml in 0.1 M MES buffer, pH 5.5, and the excitation and detective emission wavelength was at 295 nm and 330 nm, respectively. (C) The apparent unfolding rate constant at various pressures. The values were obtained from the relaxation profiles after positive pressure jumps, as partially shown in A.
TABLE 3.
The kinetics parameter of 23-kDa protein at various temperatures
| kou(s−1) |
 (ml/mol) (ml/mol) |
kof (s−1) |
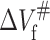 (ml/mol) (ml/mol) |
|
|---|---|---|---|---|
| Temperature (°C)* | ||||
| 3 | 3.9 × 10−6 | −99.6 | 0.012 | 98.7 |
| 10 | 3.2 × 10−5 | −71.9 | 0.860 | 101.6 |
| 20 | 1.3 × 10−4 | −66.2 | 1.870 | 84.1 |
| 35 | 1.4 × 10−3 | −56.6 | 1.990 | 63.4 |
| 45 | 2.7 × 10−2 | −44.6 | 12.570 | 85.3 |
| pH† | ||||
| 5.5 | 1.3 × 10−4 | −66.2 | 1.870 | 84.1 |
| 7.0 | 3.06 × 10−5 | −98.2 | 0.330 | 61.8 |
| 8.0 | 1.01 × 10−4 | −104.2 | 2.560 | 84.7 |
| 9.0 | 3.2 × 10−3 | −64.4 | 6.170 | 146.1 |
Protein at various temperatures was always at pH 5.5.
The temperature for experiments was at 20°C.
FIGURE 6.
Fluorescence intensity relaxation profiles of 23-kDa protein refolding after large negative jumps of pressure at 20°C. Shaded lines through the points of the two profiles in A represent the fits to a mono-exponential function; the solid line through the points of the profile for the jump from 150 to 100 MPa represents the fit to a double-exponential function. The protein concentration in experiment was 0.1 mg/ml in 0.1 M MES buffer, pH 5.5. The excitation and detective emission wavelength was at 295 nm and 330 nm, respectively. B–D show the residual of the fits at various conditions indicated on the figure.
FIGURE 7.
The fluorescence relaxation profiles of 23-kDa protein unfolding/refolding after positive pressure-jump from 100 to 200 MPa and negative jump from 200 to 100 MPa at 45°C (A), 20°C (B), and 10°C (C), respectively. Shaded lines through the points of each profile represent fits to a mono-exponential function and the solid lines represent fits to a double-exponential function. The protein concentration in experiment was 0.1 mg/ml in 0.1 M MES buffer, pH 5.5. The excitation and detective emission wavelength was at 295 nm and 330 nm, respectively.
The temperature effect on pressure-jump kinetics of 23-kDa protein unfolding and refolding
The kinetics of 23-kDa protein unfolding and folding induced by pressure-jump were examined at various temperatures tested in the equilibrium study above. The related kinetics parameters were obtained the same way as those at 20°C, and were listed in Table 3. As seen in Fig. 3 B, the unfolding activation volume change 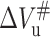 is roughly linearly decreased in absolute value with increasing temperature. The slope of the linear fitting is ∼1.3 ml/deg mol. Detailed examination of data in Fig. 3 indicates that the temperature dependence of
is roughly linearly decreased in absolute value with increasing temperature. The slope of the linear fitting is ∼1.3 ml/deg mol. Detailed examination of data in Fig. 3 indicates that the temperature dependence of 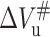 and ΔVu is more closely correlated, whereas, the temperature dependence of
and ΔVu is more closely correlated, whereas, the temperature dependence of  is different from that of
is different from that of 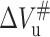 and ΔVu. The same similarity was also reported in trp repressor by Desai and co-workers, who suggested that the folding activation volume be responsible for the temperature dependence of ΔVu (Desai et al., 1999). As indicated above, the slope in Fig. 3 A is simply equal to the difference in the coefficient of thermal expansion between the unfolded and native states; consequently, the slope of Fig. 3 B should correspond to the difference in the coefficient of thermal expansion between the transition and native states. Thus, the thermal expansibility of 23-kDa protein transition state is larger than that of the native state by ∼1.3 ml/deg mol. Summarizing all the data above, a volume diagram of native, transition, and unfolded states of 23-kDa protein folding/unfolding was obtained according to Royer's schematic diagram (Desai et al., 1999). The diagram shown in Fig. 8 is very similar to that described by Desai et al. (1999) and Silva et al. (2001). The increases in thermal expansibility of the transition state relative to the native state indicates that the significant structural constraints are introduced between the transition and folded states. Furthermore, as already indicated above, the thermal expansibility of the unfolded state of 23-kDa protein is larger than that of its folded state by ∼1.8 ml/deg mol. Accordingly, it is clear that the thermal expansibility of the transition state and unfolded state are closer with each other (although the latter is a little larger than the former) and both of them are larger than that of the folded state. The decrease in thermal expansibility of the transition state relative to the unfolded state leads to a similar conclusion—namely, that the significant structural constraints also existed in the rate-limiting step for the folding. This conclusion was also reported in the study on staphylococcal nuclease reported by Woenckhaus et al. (2001).
and ΔVu. The same similarity was also reported in trp repressor by Desai and co-workers, who suggested that the folding activation volume be responsible for the temperature dependence of ΔVu (Desai et al., 1999). As indicated above, the slope in Fig. 3 A is simply equal to the difference in the coefficient of thermal expansion between the unfolded and native states; consequently, the slope of Fig. 3 B should correspond to the difference in the coefficient of thermal expansion between the transition and native states. Thus, the thermal expansibility of 23-kDa protein transition state is larger than that of the native state by ∼1.3 ml/deg mol. Summarizing all the data above, a volume diagram of native, transition, and unfolded states of 23-kDa protein folding/unfolding was obtained according to Royer's schematic diagram (Desai et al., 1999). The diagram shown in Fig. 8 is very similar to that described by Desai et al. (1999) and Silva et al. (2001). The increases in thermal expansibility of the transition state relative to the native state indicates that the significant structural constraints are introduced between the transition and folded states. Furthermore, as already indicated above, the thermal expansibility of the unfolded state of 23-kDa protein is larger than that of its folded state by ∼1.8 ml/deg mol. Accordingly, it is clear that the thermal expansibility of the transition state and unfolded state are closer with each other (although the latter is a little larger than the former) and both of them are larger than that of the folded state. The decrease in thermal expansibility of the transition state relative to the unfolded state leads to a similar conclusion—namely, that the significant structural constraints also existed in the rate-limiting step for the folding. This conclusion was also reported in the study on staphylococcal nuclease reported by Woenckhaus et al. (2001).
FIGURE 8.
Schematic diagram of the temperature effect on the volume for unfolding of 23-kDa protein. F, U, and T represent folded, unfolded, and transition-state of the protein, respectively. The thermal coefficients of expansion of the specific volumes of each state is indicated by their respective α-values.
The temperature dependences of the folding and unfolding rate constants of 23-kDa protein are different. The natural logarithms of the folding rate constant at atmospheric pressure, ln kof, under various temperatures were plotted versus the inverse of temperature 1/T in Fig. 9 A, in which the non-Arrhenius behavior of the folding rate constant can be found (Oliverberg et al., 1995). However, the natural logarithms of unfolding rate constant, ln kou, are linearly decreased with 1/T (Fig. 9 B). Therefore it is suggested that the nonlinear temperature dependence of 23-kDa protein stability observed in Fig. 2 B is mainly attributed to the non-Arrhenius behavior, which is easy to understand mathematically from Kou = kou/kof according to the expressions in Eq. 5. The non-Arrhenius behavior observed in the temperature dependence of the folding rate constant is typical for proteins and implies that the heat capacity of the protein has a large decrease upon the formation of the transition state from unfolded state (Oliverberg et al., 1995; Scalley and Baker, 1997).
FIGURE 9.
The temperature dependence of the folding rate constant (A) and unfolding rate constant (B) at atmospheric pressure. The experimental conditions were the same as in Fig. 5, except the temperatures were varied in the range of 3–45°C.
DISCUSSION
The present study used tryptophan fluorescence to explore the folding/unfolding processes of the 23-kDa extrinsic protein of photosystem II. Since the 23-kDa protein from spinach contained two tryptophan residues which are widely separated in the sequence (34 and 167), changes in the tryptophan fluorescence may reflect global changes in the protein structure rather than changes in some local domain in the protein. Our results showed that the 23-kDa protein is very sensitive to pressure- and chemical reagent-induced denaturation. The protein can be totally unfolded by a pressure of 180 MPa or 1.5–2 M Gdm-HCl at room temperature. This sensitivity is very similar to that of 33-kDa protein, another extrinsic protein of PSII. The pressure and concentration of Gdm-HCl required for unfolding of these two proteins are much lower than those required for unfolding of other proteins reported so far. For instance, the unfolding of ribonuclease A needs a pressure of 700 MPa (Panick and Winter, 2000). In our previous studies, it was also found that the pressure of 600 MPa could only transit bovine trypsin into a molten globular state (Ruan et al., 1997, 1999) and 650 MPa pressure could not totally unfold the phospholipase A2 yet (Ruan et al., 1998). This is remarkable, and naturally imposes a question as to why these two extrinsic proteins of PSII are so easy to unfold; in other words, why are they so unstable in their free form in solution? The answer to this question may be related to the functions of these proteins, as they are originally bound to the PSII membranes to stabilize the Mn-cluster required for water-splitting. Once they were dissociated from their binding sites, their structure may not be maintained in a rigid form, leading to an easy unfolding under very mild conditions. This may in turn suggest that these proteins undergo conformational changes upon binding to PSII. Indeed, the 33-kDa protein has been reported to be a naturally unfolded protein; upon binding to PSII, it undergoes some conformational changes to enable its proper function (Hutchison et al., 1998; Lydakis-Simantiris et al., 1999). Alternatively, the unfolding of proteins can be considered a kind of modulation to protein conformation and structure. The easy modulation of the 33-kDa and 23-kDa proteins by a change in physical or chemical conditions may provide a protection mechanism for the PSII of higher plants. When the environmental conditions are changed, the extrinsic proteins are modulated through unfolding, which may reduce the effect of the environmental change on other proteins of the system and enable them to perform the normal functions. It is also possible that the conformational changes that resulted from the easy modulation of these proteins act as a signal to induce corresponding adaptations of the system. In either event, all of these phenomena should be the result of biological evolution. It is thus natural to ask about the third extrinsic protein, the 17-kDa protein of higher plant PSII. Although we have not finished the pressure study on the 17-kDa protein yet, we have found that this protein is also easily denatured by a low pressure of ∼250 MPa pressure (data not shown). Thus, the easy unfolding seems to be a universal feature for the three extrinsic proteins of higher plant PSII. On the other hand, in cyanobacteria, only the 33-kDa protein is present, and the 23-kDa and 17-kDa proteins are replaced by cytochrome c-550 and a 12-kDa protein. It will be interesting to study the stability and unfolding of the cyanobacterial extrinsic proteins also. Whatever the origin and function of the easy modulation of these proteins may be, these proteins provide ideal models to explore the kinetic processes of pressure-induced unfolding-refolding because of their low stability and good performance of intrinsic fluorescence during unfolding.
As mentioned above, the slope of the linear fit of ΔVu versus temperature in Fig. 3 is ∼1.8 ml/mol deg. According to Zipp and Kauzman (1973), this slope is equal to the difference in the coefficient of thermal expansion of the protein in the unfolded, relative to the folded, state. Consequently, the thermal expansibility of the unfolded state of 23-kDa protein is larger than that of the folded state by ∼1.8 ml/deg mol; the value is very similar to that of Met-myoglobin (Zipp and Kauzmann, 1973). It was reported that this value for Snase and trp repressor was ∼1.0 ml/deg mol (Desai et al., 1999; Panick et al., 1999). Meanwhile, the unfolding ΔVu of 33-kDa protein was almost independent of temperature from 3 to 45°C (Ruan et al., 2001). These observations indicate that the overall change in the coefficient of the thermal expansion of protein between native and unfolded state seems to be in a range of 0∼2 ml/deg mol and is dependent on individual proteins.
Our results revealed that the 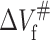 of the protein is positive, while the
of the protein is positive, while the 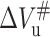 is negative. According to Eqs. 7 and 8, when pressure is increased, the positive
is negative. According to Eqs. 7 and 8, when pressure is increased, the positive 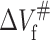 will slow down the folding rate constant whereas the negative
will slow down the folding rate constant whereas the negative 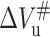 will increase the unfolding rate constants. These are inconsistent with the fact observed in this work that higher pressure speeds up the unfolding of 23-kDa protein. The positive activation volume for protein folding has been reported for Snase, CI2, and trp repressor (Desai et al., 1999; Mohana-Borges et al., 1999; Vidugiris et al., 1995; Woenckhaus et al., 2001). It has been considered that the physical basis for the volume change of protein unfolding is related to the reduction of free volume or voids that exist within the folded structure, resulting in more efficient packing of water molecules around the protein chain (Frye et al., 1996; Frye and Royer, 1998; Hummer et al., 1998). Accordingly, the rate-limiting step in 23-kDa protein folding involves the dehydration of a significant portion of the polypeptide chain and the formation of free volume in packing defects. The negative unfolding activation volume of 23-kDa protein means that the volume of the protein transition state is smaller than the native state, implying that some of the free volume and voids existing in the latter would be eliminated upon unfolding and the protein in the transition state is more solvated than in the folded state (Hummer et al., 1998). It is clear from the negative
will increase the unfolding rate constants. These are inconsistent with the fact observed in this work that higher pressure speeds up the unfolding of 23-kDa protein. The positive activation volume for protein folding has been reported for Snase, CI2, and trp repressor (Desai et al., 1999; Mohana-Borges et al., 1999; Vidugiris et al., 1995; Woenckhaus et al., 2001). It has been considered that the physical basis for the volume change of protein unfolding is related to the reduction of free volume or voids that exist within the folded structure, resulting in more efficient packing of water molecules around the protein chain (Frye et al., 1996; Frye and Royer, 1998; Hummer et al., 1998). Accordingly, the rate-limiting step in 23-kDa protein folding involves the dehydration of a significant portion of the polypeptide chain and the formation of free volume in packing defects. The negative unfolding activation volume of 23-kDa protein means that the volume of the protein transition state is smaller than the native state, implying that some of the free volume and voids existing in the latter would be eliminated upon unfolding and the protein in the transition state is more solvated than in the folded state (Hummer et al., 1998). It is clear from the negative 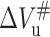 and positive
and positive 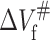 for 23-kDa protein that in terms of system volume, the transition state lies between the folded and unfolded states (see Fig. 8), very similar to the case of trp repressor, CI2, and P13 (Desai et al., 1999; Kitahara et al., 2002; Mohana-Borges et al., 1999). As seen in Table 3, the folding rate constant at atmospheric pressure, kof, is only ∼1.4 × 104 times faster than kou at 20°C, much smaller than that observed in other stable proteins such as trp repressor in which the difference between kof and kou is 5 × 108 (Desai et al., 1999). The smaller difference can help us understand why it is so easy for 23-kDa protein to be unfolded by pressure.
for 23-kDa protein that in terms of system volume, the transition state lies between the folded and unfolded states (see Fig. 8), very similar to the case of trp repressor, CI2, and P13 (Desai et al., 1999; Kitahara et al., 2002; Mohana-Borges et al., 1999). As seen in Table 3, the folding rate constant at atmospheric pressure, kof, is only ∼1.4 × 104 times faster than kou at 20°C, much smaller than that observed in other stable proteins such as trp repressor in which the difference between kof and kou is 5 × 108 (Desai et al., 1999). The smaller difference can help us understand why it is so easy for 23-kDa protein to be unfolded by pressure.
The kinetic behavior of 23-kDa protein unfolding is very similar to that of the trp repressor, except that the latter is more stable and needs the cooperation of GdmHCl to unfold at mild pressures. Nearly all the characteristics of unfolding kinetics reported for the trp repressor were observed in 23-kDa protein. This implies that these characteristics might be common features for protein unfolding induced by high pressure. This hypothesis, of course, needs to be examined with more proteins.
Acknowledgments
The authors thank Dr. Catherine A. Royer for a critical reading of the manuscript and fruitful discussion and comments.
This work was supported by a grant to K.R. from the National Natural Science Foundation of China (30370303), a grant from Institute National de la Santé et de la Recherche Médicale/Academia China (to C.B. and K.R.), and a grant from the State Key Basic Research of China to C.X. (G1998010100).
Abbreviations used: ΔGu, the free energy for the unfolding transition; ΔVu, the volume change for the unfolding transition;  the activation volume for unfolding;
the activation volume for unfolding;  the activation volume and folding.
the activation volume and folding.
References
- Berthold, D. A., G. T. Babcock, and C. F. Yocum. 1981. A highly resolved, oxygen-evolving photosystem II preparation from spinach thylakoid membranes: EPR and electron transport properties. FEBS Lett. 134:231–234. [Google Scholar]
- Blum, A. D., S. H. Smallcombe, and R. L. Baldwin. 1978. Nuclear magnetic resonance evidence for a structural intermediate at an early stage in the refolding of ribonuclease A. J. Mol. Biol. 118:305–316. [DOI] [PubMed] [Google Scholar]
- Buck, M., S. E. Radford, and C. M. Dobson. 1994. Amide hydrogen exchange in a highly denatured state. Hen egg-white lysozyme in urea. J. Mol. Biol. 237:247–254. [DOI] [PubMed] [Google Scholar]
- Creighton, T. E. 1993. Proteins. W.H. Freeman, New York.
- Desai, G., G. Panick, M. Zein, R. Winter, and C. A. Royer. 1999. Pressure-jump studies of the folding/unfolding of trp repressor. J. Mol. Biol. 288:461–475. [DOI] [PubMed] [Google Scholar]
- Dobson, C. M., A. Sali, and M. Karplus. 1998. Protein folding from theoretical and experimental point of view. Angew. Chem. 110:908–935. [Google Scholar]
- Frye, K. J., C. S. Perman, and C. A. Royer. 1996. Testing the correlation between ΔA and ΔV of protein unfolding using M-value mutants of staphylococcal nuclease. Biochemistry. 35:10234–10239. [DOI] [PubMed] [Google Scholar]
- Frye, K. J., and C. A. Royer. 1997. The kinetic basis for the stabilization of staphylococcal nuclease by xylose. Protein Sci. 6:789–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye, K. J., and C. A. Royer. 1998. Probing the contribution of internal cavities to the volume change of unfolding under pressure. Protein Sci. 7:2217–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heremans, K. 1982. High pressure effects on proteins and other biomolecules. Annu. Rev. Biophys. Bioeng. 11:1–21. [DOI] [PubMed] [Google Scholar]
- Holcomb, D. N., and K. E. Van Holde. 1962. Ultracentrifugal and viscometric studies of the reversible thermal denaturation of ribonuclease. J. Phys. Chem. 66:1999–2006. [Google Scholar]
- Homann, P. H., and L. V. Madabusi. 1993. Modifications of the thermoluminescence properties of Ca2+ depleted photosystem II membranes by the 23 kDa extrinsic polypeptide and by oligocarboxylic acids. Photosynth. Res. 35:29–39. [DOI] [PubMed] [Google Scholar]
- Hummer, G., S. Garde, A. E. García, M. E. Paulaitis, and L. R. Pratt. 1998. The pressure dependence of hydrophobic interactions is consistent with the observed pressure denaturation of proteins. Proc. Natl. Acad. Sci. USA. 95:1552–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison, R. S., S. D. Betts, C. F. Yocum, and B. A. Barry. 1998. Conformational changes in the extrinsic manganese stabilizing protein can occur upon binding to the photosystem II reaction center: an isotope editing and FT-IR study. Biochemistry. 37:5643–5653. [DOI] [PubMed] [Google Scholar]
- Jonas, J. 2002. High-resolution nuclear magnetic resonance studies of proteins. Biochim. Biophys. Acta. 1595:145–159. [DOI] [PubMed] [Google Scholar]
- Kim, P. S., and R. L. Baldwin. 1982. Specific intermediates in the folding reactions of small proteins and the mechanism of protein folding. Annu. Rev. Biochem. 51:459–489. [DOI] [PubMed] [Google Scholar]
- Kim, P. S., and R. L. Baldwin. 1990. Intermediates in the folding reactions of small proteins. Annu. Rev. Biochem. 59:631–660. [DOI] [PubMed] [Google Scholar]
- Kitahara, R., C. Royer, H. Yamada, M. Boyer, J. L. Saldana, K. Akasaka, and C. Roumestand. 2002. Equilibrium and pressure-jump relaxation studies of the conformational transitions of P13MTCP1. J. Mol. Biol. 320:609–628. [DOI] [PubMed] [Google Scholar]
- Lange, R., N. Bec, V. V. Mozhaev, and C. Balny. 1996. Fourth derivative UV-spectroscopy of proteins under high pressure. II. Application to reversible structural changes. Eur. Biophys. J. 24:284–292. [Google Scholar]
- Lange, R., J. Frank, J. L. Saldana, and C. Balny. 1996. Fourth derivative UV-spectroscopy of proteins under high pressure. I. Factors affecting the fourth derivative spectrum of the aromatic amino acids. Eur. Biophys. J. 24:277–283. [Google Scholar]
- Lange, R., and C. Balny. 2002. UV-visible derivative spectroscopy under high pressure. Biochim. Biophys. Acta. 1595:80–93. [DOI] [PubMed] [Google Scholar]
- Li, T., J. Hook, H. K. Drickamer, and G. Weber. 1976. Plurality of pressure-denatured forms in chymotrypsinogen and lysozyme. Biochemistry. 15:5571–5580. [DOI] [PubMed] [Google Scholar]
- Lydakis-Simantiris, N., R. S. Hutchison, S. D. Betts, B. A. Barry, and C. F. Yocum. 1999. Manganese stabilizing protein of photosystem II is a thermostable, natively unfolded polypeptide. Biochemistry. 38:404–414. [DOI] [PubMed] [Google Scholar]
- Mohana-Borges, R., J. L. Silva, J. Ruiz-Sanz, and G. de Prat-gay. 1999. Folding of a pressure-denatured model protein. Proc. Natl. Acad. Sci. USA. 96:7888–7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozhaev, V. V., K. Heremans, J. Frank, P. Masson, and C. Balny. 1996. High pressure effects on protein structure and function. Proteins. 24:81–91. [DOI] [PubMed] [Google Scholar]
- Murata, N., and M. Miyao. 1985. Extrinsic membrane proteins in the photosynthetic oxygen-evolving complex. Trends Biochem. Sci. 10:122–124. [Google Scholar]
- Oliverberg, M., Y. J. Tan, and A. R. Fersht. 1995. Negative activation enthalpies in kinetics of protein folding. Proc. Natl. Acad. Sci. USA. 92:8926–8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, T., S. Izawa, and Y. Inoue. 1992. Structural and functional modulation of the manganese cluster in calcium-depleted photosystem II induced by binding of the 24-kiloDalton extrinsic protein. Biochemistry. 31:7648–7655. [DOI] [PubMed] [Google Scholar]
- Panick, G., R. Malessa, R. Winter, G. Rapp, K. J. Frye, and C. A. Royer. 1998. Structural characterization of the pressure-denatured state and unfolding/refolding kinetics of staphylococcal nuclease by synchrotron small-angle x-ray scattering and Fourier-transform infrared spectroscopy. J. Mol. Biol. 275:389–402. [DOI] [PubMed] [Google Scholar]
- Panick, G., G. J. Vidugiris, R. Malessa, G. Rapp, R. Winter, and C. A. Royer. 1999. Exploring the temperature-pressure phase diagram of staphylococcal nuclease. Biochemistry. 38:4157–4164. [DOI] [PubMed] [Google Scholar]
- Panick, G., and R. Winter. 2000. Pressure-induced unfolding/refolding of ribonuclease A: static and kinetic Fourier transform infrared spectroscopy study. Biochemistry. 39:1862–1869. [DOI] [PubMed] [Google Scholar]
- Pappenberger, G., C. Saudan, M. Becker, A. Merbach, and T. Kiefhaber. 2000. Denaturant-induced movement of the transition state of protein folding revealed by high-pressure stopped-flow measurements. Proc. Natl. Acad. Sci. USA. 97:17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan, K., R. Lange, N. Bec, and C. Balny. 1997. A stable partly denatured state of trypsin induced by high hydrostatic pressure. Biochem. Biophys. Res. Commun. 239:150–154. [DOI] [PubMed] [Google Scholar]
- Ruan, K., R. Lange, Y. Zhou, and C. Balny. 1998. Unusual effect of high hydrostatic pressure on basic phospholipase A2 from venom of Agkistrodon halys Pallas. Biochem. Biophys. Res. Commun. 249:844–848. [DOI] [PubMed] [Google Scholar]
- Ruan, K., R. Lange, F. Meersman, K. Heremans, and C. Balny. 1999. Fluorescence and FTIR study of the pressure-induced denaturation of bovine pancreas trypsin. Eur. J. Biochem. 265:79–85. [DOI] [PubMed] [Google Scholar]
- Ruan, K., C. Xu, Y. Yu, J. Li, R. Lange, N. Bec, and C. Balny. 2001. Pressure-exploration of the 33-kDa protein from spinach photosystem II particle. Eur. J. Biochem. 268:2742–2750. [DOI] [PubMed] [Google Scholar]
- Ruan, K., and C. Balny. 2002. High pressure static fluorescence to study macromolecular structure-function. Biochim. Biophys. Acta. 1595:94–102. [DOI] [PubMed] [Google Scholar]
- Ruan, K., C. Xu, T. Li, J. Li, R. Lange, and C. Balny. 2003. The thermodynamic analysis of protein stabilization by sucrose and glycerol against pressure-induced unfolding—the typical example of the 33-kDa protein from spinach photosystem II. Eur. J. Biochem. 270:1654–1661. [DOI] [PubMed] [Google Scholar]
- Scalley, M. L., and D. Baker. 1997. Protein folding kinetics exhibit an Arrhenius temperature dependence when corrected for the temperature dependence of protein stability. Proc. Natl. Acad. Sci. USA. 94:10636–10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler, A. 1994. Expression of 23-kDa protein from the oxygen-evolving complex of higher plants in Escherichia coli. Biochim. Biophys. Acta. 1187:73–79. [DOI] [PubMed] [Google Scholar]
- Silva, J. L., E. W. Moles, and G. Weber. 1986. Pressure dissociation and conformational drift of the β-dimer of tryptophan synthase. Biochemistry. 25:5780–5786. [DOI] [PubMed] [Google Scholar]
- Silva, J. L., and G. Weber. 1993. Pressure stability of proteins. Annu. Rev. Phys. Chem. 44:89–113. [DOI] [PubMed] [Google Scholar]
- Silva, J. L., D. Fouguel, and C. A. Royer. 2001. Pressure provides new insights into protein folding dynamics and structure. Trends Biochem. Sci. 26:612–618. [DOI] [PubMed] [Google Scholar]
- Smeller, L. 2002. Pressure-temperature diagrams of biomolecules. Biochim. Biophys. Acta. 1595:11–29. [DOI] [PubMed] [Google Scholar]
- Timasheff, S. N. 1993. The control of protein stability and association by interactions with water: how do solvents affect these processes? Annu. Rev. Biophys. Biomol. Struct. 22:67–97. [DOI] [PubMed] [Google Scholar]
- Vidugiris, G. J., J. L. Markley, and C. A. Royer. 1995. Evidence for a molten globule-like transition state in protein folding from determination of activation volumes. Biochemistry. 34:4909–4912. [DOI] [PubMed] [Google Scholar]
- Weber, G., and H. G. Drickamer. 1983. The effect of high pressure upon proteins and other biomolecules. Q. Rev. Biophys. 16:89–112. [DOI] [PubMed] [Google Scholar]
- Winter, R. 2002. Synchrotron x-ray and neutron small–angle scattering of lyotropic lipid mesophases, model biomembranes and proteins in solution at high pressure. Biochim. Biophys. Acta. 1595:160–184. [DOI] [PubMed] [Google Scholar]
- Woenckhaus, J., R. Kohling, P. Thiyagarajan, K. C. Littrell, S. Seifert, C. A. Royer, and R. Winter. 2001. Pressure-jump small-angle x-ray scattering detected kinetics of staphylococcal nuclease folding. Biophys. J. 80:1518–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipp, A., and W. Kauzmann. 1973. Pressure denaturation of metmyoglobin. Biochemistry. 12:4217–4228. [DOI] [PubMed] [Google Scholar]











