Abstract
ATP-binding cassette (ABC) transporters are membrane-bound molecular pumps that form one of the largest of all protein families. Several of them are central to phenomena of biomedical interest, including cystic fibrosis and resistance to chemotherapeutic drugs. ABC transporters share a common architecture comprising two hydrophilic nucleotide-binding domains (NBDs) and two hydrophobic transmembrane domains (TMDs) that form the substrate pathway across the membrane. The conformational changes in the NBDs induced by ATP hydrolysis and the means by which they are transmitted to the TMDs to effect substrate translocation remain largely unknown. We have performed a molecular dynamics simulation of HisP, the well studied NBD of the bacterial histidine permease, to identify hinges and switches of the NBD conformational transitions and subunit–subunit interfaces. This analysis reveals that the TMDs regulate ATP hydrolysis by controlling conformational transitions of the NBD helical domains, and identifies the conformational changes and the crucial TMD:NBD interface, by which the energy of ATP hydrolysis is transmitted to the TMDs. We also define the conformational transitions of the Q-loop, a key element of the NBD mechanism, and identify pathways by which Q-loop switching is coordinated with TMD and NBD conformational changes. We propose a model for the catalytic cycle of ABC transporters that shows how substrate-binding and transport by the TMDs may be coordinated and coupled with ATP binding and hydrolysis in the NBDs.
ATP-binding cassette (ABC) transporters utilize the energy of ATP hydrolysis to translocate a wide variety of solutes across cellular membranes. These molecular pumps are found in all phyla and form one of the largest of all protein families (1). ABC transporters are central to many biomedical phenomena, including genetic diseases such as cystic fibrosis and multidrug resistance in cancer. These proteins share a common architectural organization comprising two hydrophilic nucleotide-binding domains (NBDs) located at the cytoplasmic surface of the membrane and two hydrophobic TM domains (TMDs) that form the translocation pathway. In prokaryotes, these domains are mostly expressed as separate protein subunits, whereas in eukaryotes they are usually fused into a single polypeptide (2). ABC transporter NBDs contain the Walker A and Walker B consensus sequences (3) characteristic of ATP-binding P-loop proteins (4), as well as a highly conserved “LSGGQ” signature sequence or C-motif (5). Although the NBDs of ABC transporters are generally closely homologous irrespective of the transporter's substrate specificity or phylogenetic origin (6), the TMDs exhibit relatively high sequence variability and are thought to contain the substrate-binding sites (7).
A number of x-ray crystal structures of ABC transporter NBDs have been reported (8–12), thus establishing the consensus fold of the cassette and serving as a basis for investigations into the mechanochemistry of the domain through comparative analysis. In all of these crystal structures, however, the catalytic site is unusually solvent exposed, exhibiting a deficiency in hydrogen bonding and electrostatic interactions with the nucleotide. To resolve this problem, we proposed that in the NBD dimer, the LSGG region completes the catalytic site of the opposite monomer (13). Subsequently, this configuration of the NBD dimer was observed in a crystal structure of the remotely related ABC-ATPase, the Rad50 DNA-repair enzyme. The Rad50 catalytic domain has a similar fold and an identical catalytic site to ABC transporter NBDs, containing the LSGG sequence as well as the D-loop and Q-loop structures (14). More recently, our proposed dimer configuration was found in crystal structures of two ABC transporter NBDs (15, 16). Thus, the interaction of the LSGG with the bound ATP has become a leading model for the dimerization of ABC transporter NBDs (17). Further progress in ABC transporter research was achieved recently with the first high-resolution structures of complete transporters from E. coli: MsbA, a putative lipid A transporter (18) and BtuCD, a vitamin B12 permease (15). These structures have provided insights into the interaction between the NBDs and the TMDs, which occurs through intimate contact between the NBDs and the intracytoplasmic domains (ICDs) that join the TM segments. Notwithstanding these advances, the conformational changes in the NBDs induced by ATP hydrolysis, and the means by which they are transmitted to the TMDs to effect substrate translocation, remain largely unknown.
The intrinsic flexibility of proteins empowers them to perform a wide spectrum of biochemical functions by means of spatial arrangement of structural elements. These conformational changes most often occur through hinge-bending movements, in which segments of preserved structure are connected by flexible joints that tether the domains and constrain their movement (19). Hinge-bending is believed to allow an induced fit of molecular surfaces in protein assembly and ligand docking. Classical molecular mechanics offers an extremely powerful tool for analyzing the dynamic, mechanistic properties of biomolecules, extending insights gained from static, single-conformation crystal structures. These techniques have been used to reveal switch regions and hinges of the conformation transition in G-proteins by identification of regions of high fluctuation of backbone torsion angles and Cα positions (20). In this study, we have performed an unconstrained molecular dynamics simulation of a well studied NBD with a shell of explicit water molecules to derive information about hinges and levers of the conformational transitions that occur in the NBDs during the catalytic cycle and about regions involved in subunit–subunit interactions.
Insights derived from our simulation, in conjunction with comparative analysis of available crystal structures, enable the delineation of key elements of the NBD mechanistic transitions, as well as TMD:NBD interfaces involved in the coordination of ATP hydrolysis and substrate translocation. We show how rotation of the NBD helical domain (α-domain) relative to the ATP-binding pocket is controlled by the TMDs to regulate ATP hydrolysis. We also outline how ATP hydrolysis generates conformational changes within the NBD and identify a highly variable loop within the α-domain as forming the crucial TMD:NBD interface through which conformational changes are transmitted to the TMDs to effect substrate translocation. We define the conformational transitions of the Q-loop and identify pathways by which this switching is coordinated with TMD and NBD conformational changes. Finally, we propose a mechanistic model for the catalytic cycle, which describes how substrate binding and transport by the TMDs may be coordinated and coupled with ATP binding and hydrolysis in the NBDs.
Materials and Methods
The crystal structure of the HisP subunit with bound ATP (1B0U; ref. 8) was used as the starting structure for the simulation. The coordinates for the catalytic magnesium were obtained by superimposing the HisP coordinates with those of Rad50 with bound MgAMP-PNP (1FTU; ref. 14). The resultant structure was then solvated to produce a 6-Å water shell that consisted of 1,826 TIP3P water molecules (21). The entire system consisted of 9,553 atoms. Molecular dynamics (MD) was performed by using X-PLOR V.3.851 (22) with the CHARMM22 all-atom force field (23). A 1.5 fs time-step was used in all MD calculations. The internal geometry of all water molecules and the bond lengths of all hydrogen atoms were constrained to equilibrium values by using the SHAKE algorithm (24). A cutoff of 14 Å was used for the nonbonded list generation, with shift and switching functions engaging at 12 Å. Initial velocities were assigned according to a Maxwellian distribution, and temperature was maintained by mimicking coupling to a water bath (25).
The solvated starting structure was minimized by using conjugate gradient minimization to a 0.5 kcal/mol⋅Å rms gradient with all protein heavy atoms fixed. Water molecules and protein hydrogens then were further minimized during a 10-ps molecular dynamics run at 300 K, in which all protein heavy atoms were again fixed. This starting model then was minimized with positional constraints on the NCαCO backbone. A 100 kcal/mol⋅Å2 force constant was used to minimize the system to a 0.5 kcal/mol⋅Å rms gradient. The constraints were gradually removed by subsequent minimizations to a 0.1 kcal/mol⋅Å rms gradient by using force constants of 50, 15, 2, and 0 kcal/mol⋅Å2. The minimized structure was heated from 75 K to 300 K during a 5-ps molecular dynamics run. The simulation then was run for 390 ps at a constant temperature of 300 K. For analysis, the coordinates were saved every 0.3 ps. Coordinate sets from the last 30 ps of the simulation were averaged to obtain the final calculated solution structure. Figures from crystal structures were prepared with SETOR and SETORPLOT (26).
Results and Discussion
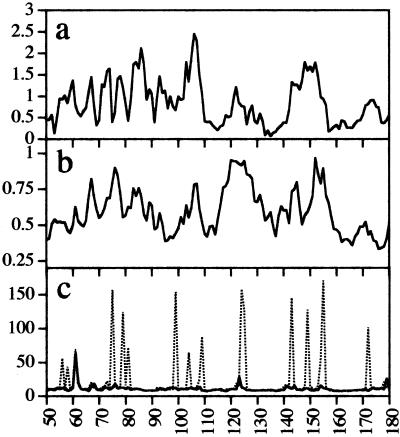
The conformational transitions essential to the activity of many proteins occur by hinge bending, in which relatively rigid parts of the protein pivot about flexible joints (19). This flexibility is utilized in recognition processes associated with protein assembly and ligand docking by allowing different conformational states to be accessed, thereby facilitating induced fit. We have performed MD calculations of the MgATP-bound form of HisP with a 6-Å shell of explicit solvent to determine areas of flexibility and, thus, to gain insight into possible hinges and levers of the conformational transitions that occur during the catalytic cycle and into regions involved in subunit–subunit interactions. The results of this simulation are presented in Fig. 1.
Figure 1.
Analysis of the coordinate trajectory of the MD simulation. The simulated structure equilibrates relatively rapidly with the potential energy of the system dropping to a plateau value of approximately −25,620 kcal/mol after 150 ps. The rms deviation from the starting crystal structure reaches a plateau value of 1.18 ± 0.09 Å between 150 ps and 390 ps. (a) rms deviation between the positions of the Cα atoms of the averaged calculated solution structure of MgATP-bound HisP with respect to the crystal structure. (b) rms fluctuations from the average structure for Cα atoms over the last 240 ps of the simulation. (c) rms fluctuations from the average of the φ (solid lines) and ψ (dotted lines) backbone dihedral angles during the last 240 ps of the simulation, plotted vs. the amino acid sequence number.
How Does the Q-Loop Mediate TMD–NBD Communication?
A highly conserved glutamine residue (Q100 in HisP) is located at the C terminus of β8 and is directly involved in the catalytic site (Figs. 2 and 3; HisP secondary structural elements are numbered as in ref. 8). The conserved glutamine is followed by an eight- or nine-residue segment known as the Q-loop (14), which joins the core domain to the α-domain (Fig. 2). The structural diversity and generally high crystallographic temperature factors for atoms within the Q-loop suggest that it is flexible and may undergo conformational changes during the catalytic cycle. Because the conserved glutamine interacts directly with the catalytic site, conformational transitions of the Q-loop are likely to be crucial to transporter function. However, the exact function and mechanism of the Q-loop is poorly understood.
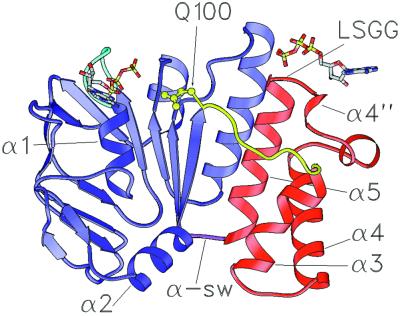
Figure 2.
Ribbon diagram of the HisP ATP-binding domain from Salmonella typhimurium (8). The ATP-binding core domain is shown in blue and the α-domain is shown in red. Secondary structure elements discussed in the text are indicated and numbered as in ref. 8. The Q-loop is yellow, the P-loop cyan, and the α-swivel (α-sw) magenta. Residue Q100 (yellow) is shown in ball and stick representation. Two ATP molecules are shown in stick representation with oxygen atoms shown in red, carbon in gray, nitrogen in blue, and phosphorous in yellow. The ATP molecule proximal to the LSGG represents the nucleotide bound in the opposite monomer of a Rad50-like HisP dimer and illustrates the interaction of the LSGG with the γ-phosphate. The coordinates of this ATP were determined by least squares alignment of the Walker A and LSGG transporter sequences of HisP with the equivalent segments of the MgAMP-PNP bound Rad50 homodimer.
Figure 3.
Structure-based sequence alignment of ABC transporter NBDs. S. typhymurium HisP crystallized with ATP (1B0U, ref. 8); Thermococcus litoralis MalK with pyrophosphate in the active site (1G29, ref. 9); Methanococcus jannaschii MJ1267 ABC MgADP bound (1G6H) and nucleotide-free (1GAJ, ref. 10); M. jannaschii MJ0796 ABC with MgADP bound (1F30, ref. 11); human cTAP1 with MgADP bound (1JJ7, ref. 12); and Escherichia coli MsbA complete ABC transporter for which only part of the NBD is resolved in the crystal structure (1JSQ, ref. 18). Secondary structural elements in the HisP stucture are numbered as in Hung et al. (8); α-helices are indicated by thick overbars and β-strands by thin overbars. Key regions discussed in the text are color coded as follows: Walker A, orange; Q-loop, yellow; ENI-motif, light green; C-motif, pale blue.
The five Q-loop residues immediately C terminal to the conserved glutamine are referred to as QL1–QL5. Our simulation shows that the peptide bond between QL4 and QL5 in HisP (L104-W105) undergoes high dihedral angle transitions (Fig. 1c), as does the ψ angle of F99, suggesting conformational hinges at these points. In the x-ray structures of HisP (8), MJ0796 (11), MalK, monomer A (9), and TAP1 (12), the Q-loop adopts an extended conformation in which the Cα atom of the conserved glutamine is proximal to the catalytic site. In contrast, the Q-loop in the two MJ1267 structures (10) is in a coiled conformation, and this is associated with partial unzipping of β-strands 8 and 9 at their C termini, resulting in withdrawal of the glutamine from the catalytic site. Examination of the Q-loop in the MJ1267 structures reveals that they diverge from the extended conformation principally because of a significant change in the backbone dihedral angle between QL4 and QL5 (P93-L94 in MJ1267). Moreover, the unzipping of β-strands 8 and 9 is associated with alterations in the dihedral angles of residues at the C terminus of β8, equivalent to F99 in HisP. Thus, our results indicate that the peptide bond QL4–QL5, and that equivalent to F99-Q100 in HisP, are hinge points in ABC transporters, mechanistically involved in the movement of the conserved glutamine in and out of the catalytic site, by mediating the switching of the Q-loop between an extended and a retracted or coiled conformation. The alteration in the backbone conformation of the Q-loop that occurs upon switching profoundly alters the spatial disposition of the side chains of QL1–QL4 (Fig. 4). The consequent alteration in the surface of the protein would affect interactions with the TMDs, which the MsbA and BtuCD structures reveal occurs between the ICDs and QL1–QL4 (Fig. 5). Residues QL1–QL3 are quite variable among NBDs (Fig. 3), which is consistent with the notion that they interact with the TMDs. Therefore, our analysis also shows that the conformational switching of the Q-loop may mediate communication between the TMDs/substrate-binding site(s) and the catalytic site(s) in ABC transporters.
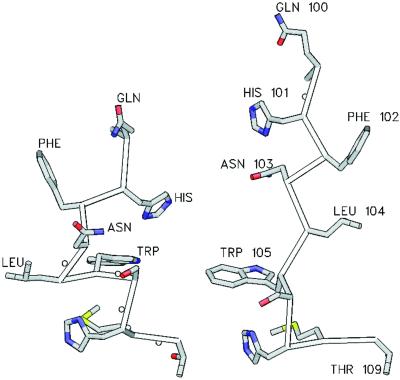
Figure 4.
View of the Q-loop in the retracted (Left) and extended (Right) conformations illustrating the alteration in the spatial dispositions of QL1–QL4. The backbone Cα trace is shown with side chains in ball and stick representation. The extended Q-loop is from HisP (1B0U), the retracted Q-loop is the backbone conformation of the Q-loop from MJ1267 (1G6H) with the side chains “mutated” to those of HisP using the application SWISS PDBVIEWER (27), with the lowest energy rotamer selected in each case.
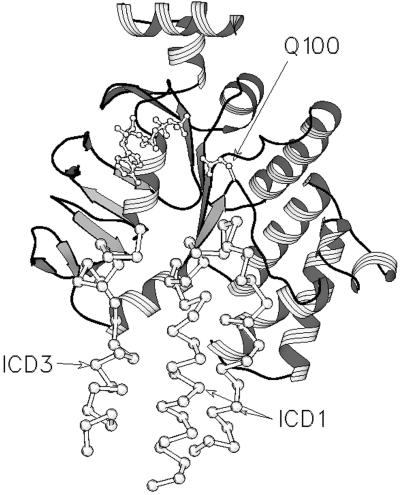
Figure 5.
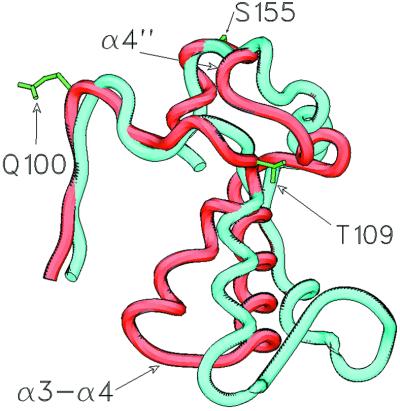
Structural alignment of the HisP structure (1B0U) with that of MsbA (1JSQ). The alignment was performed with SWISS PDBVIEWER by using the “best fit with structural alignment” option. HisP is depicted as a ribbon diagram. The bound ATP, the side chain of Q100 in HisP, and the Cα backbone of MsbA are drawn in ball and stick representation. Only the regions of ICD1 and ICD2 of MsbA that are proximal to the NBD are shown.
The α-Domain Interacts with the ICDs to Effect Substrate Transport.
At its C-terminal end, the Q-loop joins to an α-helix, one of three phylogenetically conserved α-helices (α3–α5 in HisP; Figs. 2 and 3) that comprise the α-domain. This domain has long been thought to be involved in transmitting the energy released by ATP hydrolysis to the TMDs to effect substrate translocation (28, 29). Although strong conservation in the packing core, but not the surface, of the α-domain is consistent with the idea that it contacts the cognate TMDs (11), the mechanism by which it transmits the energy of ATP hydrolysis to the TMDs is unknown. The results of our simulation strongly suggest that the loop between α3 and α4 within the α-domain is involved in subunit–subunit contacts, and that this interaction is the means by which conformational changes generated by ATP hydrolysis are transmitted to the TMDs to effect substrate translocation.
The region of the N-terminal half of the first helix of the α-domain (α3 in HisP) embodies the consensus motif S/T Φ X D/E N Φ, where X = any residue and Φ = hydrophobic residue (5). We shall refer to this region as the “ENI” motif. A structural alignment of the α-domains of six NBD crystal structures reveals a conserved spatial relationship between the LSGG and ENI motifs (Fig. 6). In contrast, residues in the C terminus of α3, within the highly variable loop joining α3 to α4 and within α4, all show significant positional variation among the NBD crystal structures (Fig. 6). This variability correlates with a peak in positional fluctuations and deviation from the starting structure observed in the simulation for residues 118–125 within the α3–α4 loop in HisP (Fig. 1 a and b). Together with the high torsion angle transitions observed for residues 123–125 during the simulation (Fig. 1c), these data suggest that the α3–α4 loop may be involved in subunit contacts. This notion is supported by the generally high backbone thermal B factors for atoms within this loop found in all of the NBD crystal structures and the high sequence and structural variability among ABC transporters in this region. Notably, in the MsbA structure, residues in the equivalent loop are proximal to ICD1 (Fig. 5). Thus, our dynamic analysis suggests that the residues within the ENI motif act as a fulcrum about which the loop at the C terminus of α3 pivots, and that this loop engages the TMDs.
Figure 6.
Structural alignment of the LSGGQQQ (C-motif) and S/T Φ X D/E N Φ (ENI-motif) of six different NBD crystal structures. The alignment was performed by using a least squares fit of the Cα backbone atoms of: HisP (1B0U); MalK (monomer A, 1G29); MalK (monomer B, 1G29); MJ1267 with ADP (1G6H); MJ1267 nucleotide-free (1GAJ); and MJ0796 (1F30). Cα atoms of residues in HisP are as indicated. The Liv-specific insertion between α3 and α4 in the α-domain in MJ1267 (10) has been omitted for clarity. TAP1, which has an insertion immediately upstream of the LSGG, has not been included in the alignment.
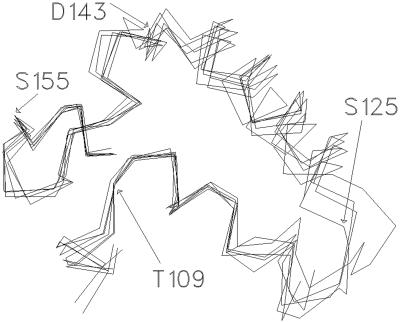
An identical structural alignment of the HisP and MsbA α-domains (Fig. 7) reveals that in the MsbA structure, α3 has pivoted about its N terminus, resulting in a large displacement of its C terminus and the adjoining α3–α4 loop, relative to α5. The qualitative impression is that, in MsbA, the peptide segment between the N terminus of α3 and the C terminus of α4 has pivoted about these two end-points and that this movement has resulted in a large displacement of residues at the C terminus of α3. Consistent with this idea, high backbone torsion angle transitions during the simulation at T109 (N terminus of α3) and D143 (C terminus of α4) indicate hinges at these points (Fig. 1c). Thus, our simulation suggests that the MsbA structure represents a distinct, possibly posthydrolysis, conformation of the α-domain in which the C terminus of α3 has undergone a conformation change relative to its position in all other NBD structures. Because this latter region is likely to engage the TMDs, we propose that this conformational change represents the power stroke of the catalytic cycle and transmits the energy of ATP hydrolysis to the TMDs.
Figure 7.
Structural alignment of the MsbA (1JSQ) and HisP (1B0U) α-domains using a least-squares fit of the Cα atoms of the LSGGQQQ (C-motif) and S/T Φ X D/E N Φ (ENI motif). The Cα trace is colored red for HisP and blue for MsbA. HisP residue side chains are colored green and are as indicated. Secondary structure elements discussed in the text also are indicated. The α-domain of the MsbA structure does not overlay as closely with that of HisP as do the α-domains of MJ1267, MJ0796, and MalK (Fig. 6). The positions of the Cα atoms of QL4–QL5, the first three residues of the ENI-motif (110–112 in HisP), and the LSGG residues (154–157 in HisP) are all within 1–2 Å of their equivalent residue in each structure. Although the spatial relationship between the N termini of α3 and α5 found in all of the NBD structures is approximately maintained in the MsbA structure, the relative orientation of the axes of these two helices is different, resulting in a significant difference in the position of the α3–α4 loop.
A Mechanistic Relay Role for α4".
N terminal to the LSGG sequence is a 10-residue segment that contains two small helices, α4′ and α4" in HisP (8). The backbone conformation of this segment is nearly identical between the six NBD crystal structures (Fig. 6). Two backbone–backbone hydrogen bonds formed by the carbonyl oxygen atoms of two residues N terminal to α4" (residues Q147 and K149 in HisP) to the amide nitrogen atoms of residues at the N terminus of α3 (ENI motif residues L111 and V110, respectively, in HisP) are observed in all structures, suggesting an important role for the interaction of these regions. Peaks in the positional fluctuations and deviation from the crystal structure of Cα atoms within α4" (Y150-H153 in HisP), together with high dihedral fluctuations for the ψ angles of K149 and L154 during the MD simulation (Fig. 1), indicate that α4" pivots about hinges at L154 and K149. This idea is consistent with the small but notable differences apparent in the backbone conformation of the α4" segment among the crystal structures (Fig. 6). Also, most significantly, in the MsbA structure, the α4" region seems to have pivoted about the Cα atoms of the residues at its N and C termini (I474 and L480 in MsbA; equivalent to Q147 and H153 in HisP), swinging away from α3, thus altering the conserved interactions with the N terminus of α3 (Fig. 7).
We have suggested (13) that, in ABC transporters, the free energy of ATP may be harnessed to produce conformational changes in the protein by means of direct interactions between the γ-phosphate of the bound nucleotide and the serine and second glycine of the LSGG sequence in a manner related to that in which this is achieved in other P-loop proteins, such as myosin and G-proteins. In addition to this, we propose that in the catalytic cycle of ABC transporters, upon formation of the pentacoordinate transition state of ATP hydrolysis, an alteration in the backbone conformation of either or both of these two signature sequence residues produces a conformational change that is propagated upstream to the α4" region, whereupon it becomes amplified by means of a fulcrum and lever mechanism involving rotation of α3 relative to α5 (the C-motif helix) to produce a large conformational change in the variable loop connecting α3 to α4 within the α-domain. This conformational change is thence transmitted to the TMDs to effect substrate transport. Thus the α4" region acts to relay the conformational change generated by ATP hydrolysis at the γ-phosphate, possibly by the alteration of the interaction of α4" with residues of the ENI motif. Because residues in α4" also contact QL4, which undergoes conformational changes upon switching of the Q-loop, it is likely that conformational changes in the α4" relay are involved in coordination of Q-loop switching with conformational changes induced in the α-domain by ATP hydrolysis.
What Is the TMD:NBD Interface?
The TMD:NBD interface is crucial for coordination of substrate binding and translocation with ATP binding and hydrolysis but until now has been largely unknown. The sequence and structural variability among ABC transporter NBDs of residues that span the C termini of α1 and α2 (residues 55–94 in HisP) suggest that they may be involved in direct interactions with the TMDs. The equivalent region has above-average temperature factors in all NBD crystal structures and shows some of the highest rms deviations from the crystal structure in our simulation (Fig. 1a), as would be expected if this region required interaction with other parts of the protein to adopt a stable conformation. Peaks in torsion angle fluctuations found in this region and the high positional fluctuations around residues 64–90 in our HisP simulation (Fig. 1 b and c) are also consistent with a role in intersubunit docking, and this conclusion is supported by both the MsbA and BtuCD structures, which reveal that sections of the ICDs are proximal to the NBD regions equivalent to residues 55–94 in HisP (Fig. 5). Further supporting this idea, mutations L90S, T96(+T) in HisP α2 (Fig. 2) have been found to disrupt signaling between the TMDs and the NBDs in the histidine permease, resulting in constitutive ATPase activity (8). Thus, we suggest that the positional and torsion angle fluctuations displayed by residues 56–90 in HisP during the simulation reflect, at least in part, a role for these regions in TMD–NBD interactions crucial to coupling of substrate binding and/or translocation to ATP hydrolysis.
The α-Swivel Mediates Coordination of Substrate Binding and Transport with ATP Binding and Hydrolysis.
Comparative analysis of ABC NBD crystal structures indicates that the α-domain moves as a rigid body and rotates relative to the ATP-binding pocket (10). This rotation occurs principally as a result of conformational changes in the short, solvent-exposed loop that joins the α-domain to β9. We refer to this loop as the α-swivel. In the Rad50 ABC-ATPase, the region containing the LSGG motif (equivalent to the α-domain in ABC transporters) is rotated 30° outward from the active site in the nucleotide-free structure, compared with its position in the MgAMP-PNP-bound structure, and this rotation is associated with a conformational change in the equivalent α-swivel (14). Thus, the α-domain undergoes important mechanistic conformational transitions relative to the core domain, and the conformation of the α-swivel is an important determinant of the orientation of the α-domain and, thus, the position of the LSGG. This idea accords with our simulation, which shows that the ψ angle of P172—on the HisP α-swivel—undergoes high torsion angle transitions, indicating a conformational hinge at this point (Fig. 1c). It is also clear from the positional fluctuations of Cα atoms during the simulation, that the α-swivel and Q100 are the relatively stable end-points to which the ABCα subdomain is flexibly attached.
HisP mutations P172L and P172T were found to release HisP from the regulatory control of the TM subunits, HisM and HisQ, resulting in uncoupled, constitutive ATPase activity (30). Nikaido and Ames (30) concluded that liganded substrate-binding protein triggers ATP hydrolysis by eliciting a conformational change in the TMDs, which alters the interaction with the NBD, thereby enabling ATP hydrolysis. We further articulate this idea by suggesting that, in the absence of bound substrate, the TMDs act by means of the α-swivel to block or stop conformational transitions of the α-domain, locking it in a hydrolysis-incapable conformation in which the LSGG cannot engage the opposite catalytic site. Substrate binding would alter this TMD:NBD interaction, allowing the α-domain to rotate and the LSGG to engage the opposite catalytic site, thus enabling ATP hydrolysis. In HisP, residues within the α-swivel are in contact with residues at the C terminus of α2 and may, therefore, be involved in a signal response transmitted from the TMDs through the α2 region (see above). This idea is supported by the fact that mutation of residues in HisP α2 have the same unregulated ATP hydrolysis as the P172 α-swivel mutations (8).
A Model for the Catalytic Cycle of an ABC Transporter Export Protein.
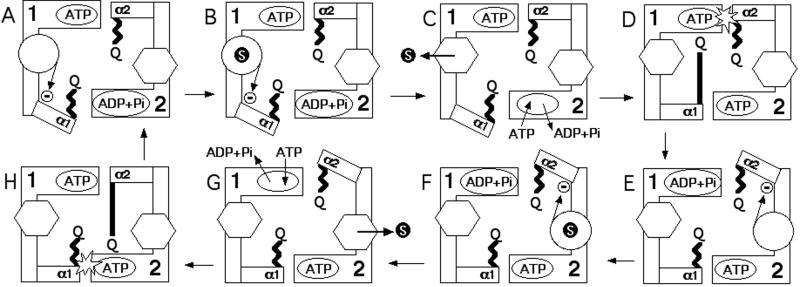
In Fig. 8, a model for the catalytic cycle of an ABC exporter is presented that incorporates the findings and conclusions of this and other studies. A number of assumptions as well as aspects of previous models are incorporated in this scheme, as follows. (i) ATP is hydrolysed alternately at each catalytic site (31, 32). (ii) There are two substrate-binding sites, and each substrate-binding site alternates between an outward-facing low-affinity conformation and an inward-facing high-affinity conformation (33). (iii) ATP hydrolysis leads to opening of the translocation pathway (30). To articulate this idea further we suggest that ATP hydrolysis at the catalytic site of one NBD changes the substrate-binding site associated with the opposite NBD from an outward- to an inward-facing conformation by means of conformational changes in the α-domain of the opposite NBD. (iv) Substrate-binding triggers a conformational change in the TMDs resulting in activation of the NBD (30). In this model, the conserved glutamine triggers ATP hydrolysis when it engages the ATP-bound catalytic site, which is not to imply that the glutamine is the catalytic base but simply that it completes the catalytic site in a manner that allows or facilitates hydrolysis. Because the regulatory and coordinating role of the TMDs in the model is effected through inhibition of NBD conformation changes, the model can account for the high ATPase activity often observed for isolated NBDs (31).
Figure 8.
Model for the catalytic cycle of an ABC exporter. The core domains of the NBDs are depicted as “L”-shaped blocks and named 1 and 2. The ATP-binding sites are depicted as oval shapes located in 1 and 2. The α-domains are depicted as rectangular boxes and are labeled α1 and α2, with the ends of these boxes closest to the opposite ATP-binding site representing the LSGG region. The Q-loop is represented by either a zigzag (retracted) or long, straight (extended) bar ending in the letter “Q.” A substrate molecule is represented by a black circle with the letter “S” inside it. The TMDs are depicted as a hexagon to indicate an outward-facing low-affinity substrate-binding site or a circle to indicate an inward-facing high-affinity substrate-binding site. The inward-facing conformation of the substrate-binding site is a high potential energy “spring-loaded” state, which changes to a low potential energy outward-facing conformation upon substrate binding. This substrate-triggered conformational change thereby effects substrate export. The inward-to-outward change in the conformation of the TMD removes the restraint on rotation of the α-domain mediated by the α-swivel, allowing the α-domain to move from an OFF conformation to an ON (hydrolysis-capable) conformation, in which its LSGG engages (binds) the ATP bound in the opposite NBD. The conserved glutamine triggers ATP hydrolysis (represented by the “flash” in steps D and H) when its Q-loop extends (long straight bar) and engages its own catalytic site (Q near ATP site) in which there is LSGG-bound ATP. (A) TMD1 starts empty in the inward-facing conformation, which locks α1 in the OFF orientation (indicated by the arrow with the circled dash). Substrate binds to TMD1 (B) and triggers the conversion of TMD1 to the outward-facing conformation, thus releasing the substrate (substrate export). (C) As a consequence of this conformational change, ADP and phosphate are exchanged for ATP in NBD2. Because of the removal of the block, (D) α1 can move into the ON conformation and its LSGG engages ATP bound at NBD2. Binding of ATP by α1 has converted the α1 Q-loop to the extended conformation. This conformational change enables its conserved glutamine to trigger hydrolysis at NBD1. (E) Hydrolysis at NBD1 converts TMD2 from the outward- to the inward-facing high-energy conformation and converts the Q-loop of α1 to the retracted conformation. It also locks α2 in the OFF orientation. Stages F–H are a symmetrical series of events as for B–D.
Our molecular dynamic analysis of HisP has identified some of the key interfaces involved in interdomain communication in ABC transporters, as well as hitherto unknown hinges of the NBD conformational transitions. In the context of recent crystal structures of other ABCs, as well as previous experimental data, our analysis allows a detailed delineation of the catalytic cycle of the transporter that, we believe, represents a significant leap forward in our understanding of these important and intriguing proteins.
Acknowledgments
We thank Giovanna Ames for insightful comments on manuscript drafts and Herbert Treutlein for helpful discussions.
Abbreviations
- ABC
ATP-binding cassette
- NBD
nucleotide-binding domains
- TMD
transmembrane domain
- ICD
intracytoplasmic domain
- MD
molecular dynamics
References
- 1.Higgins C F. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 2.Saurin W, Hofnung M, Dassa E. J Mol Evol. 1999;48:22–41. doi: 10.1007/pl00006442. [DOI] [PubMed] [Google Scholar]
- 3.Walker J E, Saraste M, Runswick M J, Gay N J. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith C A, Rayment I. Biophys J. 1996;70:1590–1602. doi: 10.1016/S0006-3495(96)79745-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bianchet M A, Ko Y H, Amzel M, Pedersen P L. J Bioenerg Biomembr. 1997;29:503–524. doi: 10.1023/a:1022443209010. [DOI] [PubMed] [Google Scholar]
- 6.Schneider E, Hunke S. FEMS Microbiol Rev. 1998;22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 7.Holland B, Blight M A. J Mol Biol. 1999;293:381–399. doi: 10.1006/jmbi.1999.2993. [DOI] [PubMed] [Google Scholar]
- 8.Hung L-W, Wang I X, Nikaido K, Liu P-Q, Ames G F-L, Kim S-H. Nature (London) 1998;396:703–707. doi: 10.1038/25393. [DOI] [PubMed] [Google Scholar]
- 9.Diederichs K, Diez J, Greller G, Müller C, Breed J, Schnell C, Vonrhein C, Boos W, Welte W. EMBO J. 2000;19:5951–5961. doi: 10.1093/emboj/19.22.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karpowich N, Martsinkevich O, Millen L, Yuan Y R, Dai P L, MacVey K, Thomas P J, Hunt J F. Structure (London) 2001;9:571–586. doi: 10.1016/s0969-2126(01)00617-7. [DOI] [PubMed] [Google Scholar]
- 11.Yuan Y R, Blecker S, Martsinkevich O, Millen L, Thomas P J, Hunt J F. J Biol Chem. 2001;276:32313–32321. doi: 10.1074/jbc.M100758200. [DOI] [PubMed] [Google Scholar]
- 12.Gaudet R, Wiley D C. EMBO J. 2001;20:4964–4972. doi: 10.1093/emboj/20.17.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones P M, George A M. FEMS Microbiol Lett. 1999;179:187–202. doi: 10.1111/j.1574-6968.1999.tb08727.x. [DOI] [PubMed] [Google Scholar]
- 14.Hopfner K-P, Karcher A, Shin D S, Craig L, Arthur L M, Carney J P, Tainer J A. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 15.Locher K P, Lee A T, Rees D C. Science. 2002;296:1091–1098. doi: 10.1126/science.1071142. [DOI] [PubMed] [Google Scholar]
- 16.Smith P C, Karpowich N, Millen L, Moody J E, Rosen J, Thomas P J, Hunt J F. Mol Cell. 2002;10:139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerr I D. Biochim Biophys Acta. 2002;1561:47–64. doi: 10.1016/s0304-4157(01)00008-9. [DOI] [PubMed] [Google Scholar]
- 18.Chang G, Roth C B. Science. 2001;293:1793–1800. doi: 10.1126/science.293.5536.1793. [DOI] [PubMed] [Google Scholar]
- 19.Gerstein M, Lesk A M, Chothia C. Biochemistry. 1994;33:6739–6749. doi: 10.1021/bi00188a001. [DOI] [PubMed] [Google Scholar]
- 20.Diaz J F, Wroblowski B, Engelborghs Y. Biochemistry. 1995;34:12038–12047. doi: 10.1021/bi00037a047. [DOI] [PubMed] [Google Scholar]
- 21.Jorgensen W, Chandrasekar J, Madura J, Impey R, Klein M. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 22.Brünger A T. X-PLOR. New Haven, CT: Yale Univ. Press; 1992. , Version 3.1. [Google Scholar]
- 23.Mackerell A D, Bashford D, Bellott M, Dunbrack R L, Evanseck J D, Field M J, Fischer S, Gao J, Guo H, Ha S, et al. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 24.Ryckaert J, Ciccotti G, Berendsen H. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 25.Berendsen H, Postma J, van Gunsteren W, DiNola A, Haak J. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 26.Evans S V. J Mol Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- 27.Guex N, Peitsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 28.Hyde S C, Emsley P, Hartshorn M J, Mimmack M M, Gileadi U, Pearce S R, Gallagher M P, Gill D R, Hubbard R E, Higgins C F. Nature (London) 1990;346:362–365. doi: 10.1038/346362a0. [DOI] [PubMed] [Google Scholar]
- 29.Mimura C S, Holbrook S R, Ames G F-L. Proc Natl Acad Sci USA. 1991;88:84–88. doi: 10.1073/pnas.88.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikaido K, Ames G F-L. J Biol Chem. 1999;274:26727–26735. doi: 10.1074/jbc.274.38.26727. [DOI] [PubMed] [Google Scholar]
- 31.Liu P-Q, Liu C E, Ames G F-L. J Biol Chem. 1999;274:18310–18318. doi: 10.1074/jbc.274.26.18310. [DOI] [PubMed] [Google Scholar]
- 32.Senior A E, Gadsby D C. Semin Cancer Biol. 1997;8:143–150. doi: 10.1006/scbi.1997.0065. [DOI] [PubMed] [Google Scholar]
- 33.van Veen H W, Margolles A, Muller M, Higgins C F, Konings W N. EMBO J. 2000;19:2503–2514. doi: 10.1093/emboj/19.11.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]