Abstract
Here we report a 2.3-Å crystal structure of scallop myosin S1 complexed with ADP⋅BeFx, as well as three additional structures (at 2.8–3.8 Å resolution) for this S1 complexed with ATP analogs, some of which are cross-linked by para-phenyl dimaleimide, a short intramolecular cross-linker. In all cases, the complexes are characterized by an unwound SH1 helix first seen in an unusual 2.5-Å scallop myosin-MgADP structure and described as corresponding to a previously unrecognized actin-detached internally uncoupled state. The unwinding of the SH1 helix effectively uncouples the converter/lever arm module from the motor and allows cross-linking by para-phenyl dimaleimide, which has been shown to occur only in weak actin-binding states of the molecule. Mutations near the metastable SH1 helix that disable the motor can be accounted for by viewing this structural element as a clutch controlling the transmission of torque to the lever arm. We have also determined a 3.2-Å nucleotide-free structure of scallop myosin S1, which suggests that in the near-rigor state there are two conformations in the switch I loop, depending on whether nucleotide is present. Analysis of the subdomain motions in the weak actin-binding states revealed by x-ray crystallography, together with recent electron microscopic results, clarify the mechanical roles of the parts of the motor in the course of the contractile cycle and suggest how strong binding to actin triggers both the power stroke and product release.
The myosin head (subfragment one, or S1) consists of a motor domain (MD) and a lever arm that amplifies small conformational changes of the motor (Fig. 1). The MD comprises four subdomains (the 50-kDa upper and lower subdomains, the N-terminal subdomain, and the converter) linked by three flexible single-stranded joints (switch II, the relay, and the so-called SH1 helix). Motor function results from coupled rigid body rearrangements of the subdomains coordinated with conformational changes in the joints (1, 2). During the acto-myosin contractile cycle (3–5), the MD undergoes a number of conformational changes as the myosin head transduces ATP hydrolysis into mechanical work. The affinity of myosin for actin in the cycle is often described in terms of “strong” and “weak” actin-binding states. Myosin crystal structures have thus far been obtained only in the absence of actin, and three conformations have been identified, but their place in the contractile cycle has been controversial (6). The first or near-rigor conformation (7), was obtained in the absence of nucleotide, and, because the lever arm orientation was similar to that observed in electron microscopy (EM) images of actomyosin in the rigor state, this conformation was thought to be similar to a true rigor (i.e., strong actin-binding, nucleotide-free) state. More recent studies (8, 9) have shown, however, that this conformation corresponds to a weak actin-binding state occurring shortly after myosin detaches from actin, and that this state can bind to but cannot hydrolyze ATP. In this state, switch II is in an extended conformation that does not interact with the nucleotide-binding site. The second, or pre-power stroke conformation, has a primed lever arm (i.e., approximately perpendicular to the actin filament) (2, 10–12). In this conformation, switch II is bent and has moved in to interact with the nucleotide-binding pocket, leading to the enzymatic transition state (ADP⋅Pi) of the nucleotide (2, 10, 11, 13). Switch II also forms a specific salt bridge and hydrogen bond interactions with switch I that stabilize the pre-power stroke state (12). In both conformations, the so-called SH1 helix is intact and separates two reactive cysteine sulfhydryls, SH1 and SH2 (14), by more than 18 Å. These myosin crystal structures could not account, however, for the biochemical observations (15–17) that relatively short cross-linkers such as para-phenyl dimaleimide (p-PDM, ≈12 Å) interact with the two groups. Recently, an unusual conformation of scallop myosin S1 complexed with MgADP has been visualized (1) in which the SH1 helix is unwound, providing an explanation for how cross-linking can occur. In that work, it was shown that a small conformational change in the switch II loop (a catalytic loop of the nucleotide-binding pocket) produces rearrangements of the subdomains in the motor that destabilize the SH1 helix. The unwound SH1 helix, in turn, effectively uncouples the converter/lever arm module from the motor. Based on these and other findings, this third conformation has been interpreted as an “actin-detached internally uncoupled state” that binds ATP in the contractile cycle (note that this state has been called “detached,” refs. 1 and 2).
Figure 1.
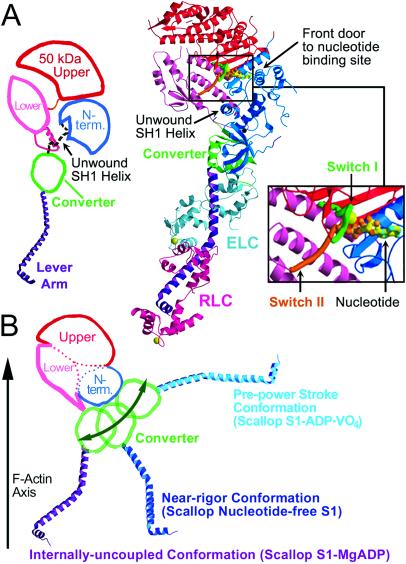
Overview of the internally uncoupled scallop S1 conformation. (A) (Center) The internally uncoupled S1-ADP⋅BeFx structure (50-kDa upper subdomain is shown in red, 50-kDa lower subdomain is shown in pink, N-terminal subdomain is shown in blue, converter is shown in green, lever arm heavy chain is shown in purple, essential light chain is shown in cyan, regulatory light chain is shown in magenta). Notable features of the structure include the unusual position of the lever arm and the unwound SH1 helix. Three other scallop structures (S1-AMPPNP, S1-ATP[γ-S]-p-PDM, and S1-ADP-p-PDM), as well as the previously reported scallop S1-MgADP (1), show the same conformation, although the orientations of the respective lever arms vary slightly. (Right Inset) Expanded view of the nucleotide binding size. (Left) Schematic diagram of the internally uncoupled conformation, showing the subdomains and the approximate location of the disordered SH1 helix (light chains not shown). (B) For comparison, a two-dimensional projection of the three weak actin-binding S1 conformations observed in scallop crystal structures (light chains not shown) (1, 2). The 50-kDa upper and N-terminal subdomains of the three structures are superimposed to show the large changes in the relative positions of the converter and lever arm (see also ref. 2). In each conformation, the 50-kDa lower subdomain adopts a slightly different position with respect to the 50-kDa upper and N-terminal subdomains, leading to a markedly different orientation of the converter. The converter, in turn, controls the position of the lever arm (1). Movement of the 50-kDa lower subdomain is not represented in this schematic diagram. This subdomain rotates to maintain contact with the converter as S1 adopts each of the three conformations. See Fig. 5 for a more detailed three-dimensional description of the subdomain motions.
Here we extend the previous S1-MgADP study by determining a 2.3-Å internally uncoupled state structure of scallop S1 complexed with ADP⋅BeFx, an ATP analog. This structure allows us to visualize ATP γ-phosphate interactions in the nucleotide-binding pocket. We have also determined three lower resolution (2.8–3.8 Å) structures of internally uncoupled scallop S1 complexed with ATP analogs [adenosine 5′-[β,γ-imido]triphosphate (AMPPNP) or adenosine 5′-[γ-thio]triphosphate (ATP[γ-S])] or ADP. The S1-ADP and S1-ATP[γ-S] structures contain an intramolecular cross-link by p-PDM. In addition, we have determined a 3.2-Å nucleotide-free structure of scallop S1 in the near-rigor state, which suggests a conformational change in switch I (a second catalytic loop of the nucleotide-binding pocket), clarifying the relationship between the near-rigor and internally uncoupled conformations. The results also include an analysis of certain mutations near the SH1 helix that, together with other crystallographic and EM evidence, provide information on the mechanical roles of various structural elements in the myosin head.
Methods
S1 from scallop (Argopecten irradians) striated muscle myosin was prepared as described (1) and was complexed with either ADP⋅BeFx, AMPPNP, ATP[γ-S], ADP, or made nucleotide free. S1-ADP and S1-ATP[γ-S] (3 mg/ml) each were cross-linked by a 2-min treatment with 0.5 mM nucleotide and 20 mM p-PDM at 20°C. Crystals were grown by vapor diffusion at 4°C. All structures were determined by molecular replacement techniques, followed by model building and refinement (see Table 1 for crystallographic statistics). Additional details of protein preparation and crystallographic methods are published as supporting information on the PNAS web site, www.pnas.org.
Table 1.
Data collection and refinement statistics
| ADP⋅BeFx | AMPPNP | ATP-γ-S-p-PDM | ADP-p-PDM | Nucleotide-free | |
|---|---|---|---|---|---|
| Data collection | |||||
| Resolution range, Å | 30.0–2.3 | 40.0–3.0 | 40.0–3.8 | 55.0–2.8 | 50–3.2 |
| Unique reflections/multiplicity | 49,703/4.4 | 31,019/3.5 | 14,451/3.0 | 30,488/2.0 | 20,298/4.2 |
| Average I/σ | 12.0 | 11.7 | 10.6 | 16.0 | 11.2 |
| Rsym, % (All/outer shell*) | 9.7/24.7 | 7.7/47.2 | 8.4/27.4 | 3.5/15.3 | 10.9/33.5 |
| Completeness (%): | |||||
| All data/outer shell* | 79.8/72.7 | 80.6/53.7 | 84.6/45.1 | 74.4/22.4 | 88.5/65.8 |
| 55–3.4 Å range | 90.7 | 93.1 | – | 95.0 | 93.0 |
| Refinement | |||||
| Sigma cutoff | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 |
| Completeness in range, % | 77.9 | 73.1 | 78.8 | 72.0 | 84.1 |
| R factor/Rfree, % | 23.0/26.9 | 27.7/31.7 | 23.8/32.8 | 28.0/32.7 | 25.8/31.3 |
| Mean B factor | 55.8 | 79.1 | 53.4 | 89.8 | 84.1 |
| rms bond lengths, Å/angles, °† | 0.010/1.46 | 0.015/2.51 | 0.011/1.77 | 0.010/1.93 | 0.008/1.54 |
| No. of protein/water atoms | 8,476/185 | 8,237/7 | 8,069/6 | 8,282/31 | 8,127/10 |
| No. of prosthetic atoms | 40 | 34 | 34 | 30 | 8 |
| Cross-validated coordinate error, Ň | 0.41 | 0.64 | 0.69 | 0.67 | 0.49 |
The cell parameters of these 5 crystal structures are ADP⋅BeFx (P1: a = 51.6, b = 58.5, c = 133.3 Å, α = 81.1°, β = 84.9°, γ = 67.2°); AMPPNP (P1: a = 52.4, b = 58.6, c = 148.9 Å, α = 81.6°, β = 82.4°, γ = 87.4°); ATP-γ-S-p-PDM (P1: a = 51.8, b = 57.0, c = 151.7 Å, α = 95.2°, β = 96.5°, γ = 100.9°); ADP-p-PDM (P1: a = 51.8, b = 57.0, c = 150.5 Å, α = 95.6°, β = 96.3°, γ = 101.5°); and nucleotide-free near-rigor (P21: a = 159.8, b = 51.2, c = 84.0 Å, α = 90.0°, β = 99.5°, γ = 90.0°).
Highest resolution shell (10% of the theoretical data).
See ref. 36.
Results and Discussion
Comparison of Structures.
All four nucleotide-containing structures (S1-ADP⋅BeFx, S1-AMPPNP, S1-ATP[γ-S]-p-PDM, S1-ADP-p-PDM) reveal the same overall internally uncoupled conformation as scallop S1-MgADP (1); these include an unwound SH1 helix, an extended switch II loop, a constrained switch I conformation (see below), and similar orientations of the lever arm. Within each subdomain of the MD, little or no change in conformation is detected. The positions of the N-terminal and 50-kDa upper subdomains (Fig. 1) are well conserved in all myosin S1 crystal structures. When these subdomains are superimposed, the four nucleotide-containing structures are very similar to the scallop S1-MgADP structure in the MD, producing α-carbon root mean square deviations of less than 1.3 Å. The lever arm α-carbons of these structures diverge, however, from those of S1-MgADP by as much as 3.3 Å, possibly as a result of crystal packing or because the unwinding of the SH1 helix reduces the constraints on the orientation of the converter/lever arm module. The converter/lever arm orientation is normally constrained chiefly by connection to the SH1 helix and contacts with the flexible relay joint (see Fig. 1 and ref. 1).
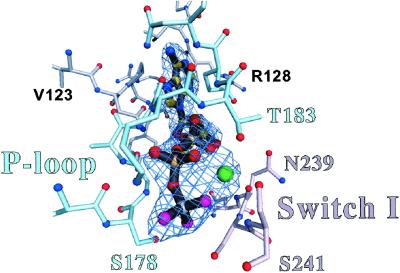
In the active site of all four internally uncoupled structures, electron density for the nucleotide is prominent (Fig. 2). In the case of S1-ADP⋅BeFx, the resolution is high enough to locate the phosphate oxygens and the fluorides (where BeFx is an analog for the γ-phosphate). In the other structures, the low resolution precludes this identification. Nevertheless, the γ-phosphate sulfur of ATP[γ-S] has strong density in the S1-ATP[γ-S]-p-PDM structure. Moreover, a strong electron density bulge in all four structures identifies the position of the Mg2+ atom, which is critical for activating ATP hydrolysis. In all of these weak actin-binding states, the Mg2+ cation is coordinated by several oxygen ligands, including one from the nucleotide β-phosphate, one from the γ-phosphate, as well as the hydroxyls of switch I residue S241 and P loop residue T183 (13). As in the near-rigor conformation (8, 9), ATP hydrolysis cannot occur in the internally uncoupled conformation because switch II is positioned away from the nucleotide pocket, in an extended conformation. The near-rigor and internally uncoupled conformations differ mainly as a result of small twists about three residues of the switch II loop in the internally uncoupled state (scallop residues I461 through G463; ref. 1). Nevertheless, the active sites of the scallop S1-ADP⋅BeFx structure and a Dictyostelium discoideum near-rigor myosin-ATP structure (8) are nearly identical. In particular, the interactions between the nucleotide phosphates, Mg2+ atom, the P loop and switch I loop are conserved. These results suggest that the near-rigor nucleotide-containing and internally uncoupled conformations are enzymatically very similar, despite large conformational differences in the positions of the 50-kDa lower subdomain and the converter/lever arm module.
Figure 2.
Nucleotide electron density. The nucleotide binding site is shown together with a simulated annealing Fo − Fc omit map of ADP⋅BeFx, contoured at the 5.0 σ level. The P loop (part of the N-terminal subdomain) is shown in sky-blue, and switch I (lower right, part of the 50-kDa upper subdomain) is shown in pink. Included in this view is the electron density attributed to the Mg2+ ion (green ball). The γ-phosphate position is occupied by BeFx (Be in gray and F in magenta). The other internally uncoupled structures show approximately the same nucleotide conformation, but at a lower resolution. The interactions of the Mg2+ nucleotide with the P loop and switch I are virtually identical to those previously reported at higher resolution in a near-rigor Dictyostelium truncated myosin-ATP structure (8).
The nucleotide-free S1 structure crystallizes in the near-rigor conformation previously reported in chicken skeletal S1 (7) and at lower resolution in scallop (2), as well as in D. discoideum truncated myosin MD structures (8, 9, 11–13). This structure is not expected to occur in the contractile cycle, but, as we show below, small differences between this and the nucleotide-containing near-rigor structures give insights into the conformation of acto-myosin in the true rigor state. This structure differs from the internally uncoupled state by an α-carbon root mean square difference of more than 6 Å in the MD and more than 60 Å in the lever arm, due for the most part to rotation of the converter/lever arm module and the 50-kDa lower subdomain. The motor domains of the scallop and all other near-rigor structures superimpose closely except for the exact position of the 50-kDa upper and lower subdomains with respect to the N-terminal subdomain (see next section). All of these near-rigor structures are characterized by an intact SH1 helix, and the lever arm, when present, assumes a very different orientation than that of the internally uncoupled state (1, 2, 7) (Fig. 1).
We have previously interpreted the internally uncoupled conformation as an ATP state, though the original structure contained ADP in the nucleotide-binding site (1), but this interpretation was controversial (18). The current visualization of ATP analogs in the nucleotide-binding pocket might be considered to confirm our original view. Moreover, because cross-linking by p-PDM identifies the internally uncoupled S1-ATP conformation as a weak actin-binding state (see below), it is likely that this conformation occurs in the contractile cycle as the second ATP state (M*-ATP, see Fig. 3B), which Bagshaw and Trentham (19) predicted from biochemical kinetic studies. It has been established, however, that the nucleotide-binding pocket is fairly promiscuous and can accommodate either ADP or ATP in both near-rigor and internally uncoupled states (as reported here and in refs. 1, 8, and 9). By contrast, note that in the absence of nucleotide, only the near-rigor conformation is obtained (2, 7, 8). These results suggest that the binding of ATP analogs or ADP establishes an equilibrium in the absence of actin in which switch II changes conformation to sample both the near-rigor and internally uncoupled states.
Figure 3.
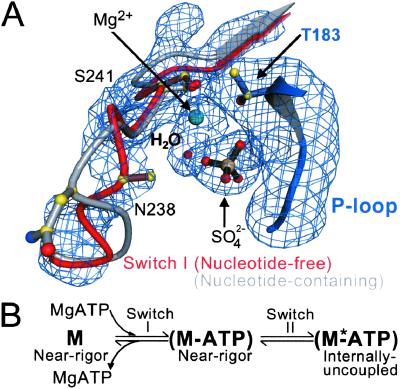
A conformational change in switch I on the binding of nucleotide. (A) Shown is a composite of four difference Fourier (Fo − Fc) electron density omit maps of switch I and the surrounding region of the nucleotide-binding site of the nucleotide-free near-rigor structure. Each map was generated by torsional simulated-annealing in which atoms were omitted either for a segment of switch I (residues N235 to R242), the Mg2+ ion and a water molecule, a sulfate ion, or part of the P loop (residues A180 to T183). The maps are contoured at the 3.5 σ level. Superimposed on the maps is the proposed conformational change of switch I in the transition from the nucleotide-free to the nucleotide-bound (ATP and its analogs, or ADP) states. The P loop of the scallop near-rigor and S1-ADP⋅BeFx structures are superimposed. The “empty” nucleotide-binding site of the near-rigor conformation is shown in color (switch I in red, P loop in blue, Mg2+ ion in green, with a sulfate ion occupying the β-phosphate position). In gray is the same site in the internally uncoupled scallop S1-ADP⋅BeFx structure (nucleotide not shown), which shows the conformation of switch I seen in all nucleotide-containing myosin crystal structures. When nucleotide is present in the binding site, it appears to displace part of switch I, most notably residue N238. The internally uncoupled state is initiated by small changes in the switch II loop (not shown, see ref. 1). (B) The equilibrium equation (in vitro) between the empty and nucleotide-containing near-rigor myosin structures, leading to the internally uncoupled state. Shown also are the steps at which switches I and II change. An asterisk is assigned to the internally uncoupled ATP state (right) to indicate that this conformation may correspond to the second ATP state predicted in biochemical kinetics studies (19). The near-rigor nucleotide-free state (left) is not likely to be part of the in vivo acto-myosin contractile cycle.
Switch I in the Near-Rigor Nucleotide-Free and Internally Uncoupled Conformations.
Although the nucleotide-free scallop S1 structure (in the near-rigor conformation) is determined at a relatively low resolution (3.2 Å), we have been able to identify a small nucleotide-dependent conformational change in the switch I loop that may bias this equilibrium (Fig. 3). The electron density map indicates that the switch I loop becomes somewhat disordered in this near-rigor nucleotide-free state, and that its most probable conformation appears to differ from that observed in any nucleotide-containing structure, including Dictyostelium (8, 9, 11–13), chicken (7, 10), and scallop (1, 2). We have modeled this change as a bending of part of the switch I loop by ≈2.5 Å. The bending could be driven by a ≈180° rotation (“flip”) of residue N238 made possible by the absence of nucleotide interactions with residue N237. However, we cannot see density beyond the β-carbon of N238, so that side chain interactions are uncertain. This conformational change of switch I in the scallop S1 is similar in magnitude to that observed in part of the switch I loop of Ras-p21 upon GTP hydrolysis (20). In Ras, however, this flip is centered about a residue equivalent to S241 (a Mg2+ ligand), not N238, which does not appear to be a Mg2+ ligand. Because of the low resolution of the structure, the position of the Mg2+ ion could only be tentatively determined on the basis of optimal coordination distances. Our observations on the structure of switch I in scallop S1 indicate that this loop is less constrained in the nucleotide-free than in the nucleotide-containing structures. In all nucleotide-containing structures, switch I is constrained by hydrogen bonds that N237 forms with the α- and (when present) γ-phosphates (1, 2, 7–13). The flexibility of switch I permitted in the nucleotide-free structure may be related to a small rigid-body rotation (≈5°) of the 50-kDa upper and lower subdomains (counterclockwise when the molecule is viewed as in Fig. 1). This movement is centered about switch I and the preceding β-hairpin and is best seen when the N-terminal subdomains of near-rigor structures are superimposed. The small rotation would increase the frequency with which transient stabilizing contacts are made between the SH1 helix and the helical portion of the relay. As a result, the equilibrium would favor the near-rigor conformation in the absence of nucleotide (Fig. 3B). On the basis of this argument, it appears that switch I senses the content of the nucleotide-binding pocket and indirectly influences the stability of the SH1 helix. This interpretation also suggests why SH1-SH2 cross-linking by p-PDM proceeds very slowly in the absence of nucleotide (17). The rigid-body rotation of the 50-kDa upper subdomain in the nucleotide-free near-rigor structure also widens the so-called “front door” (21) to the nucleotide-binding site by ≈2 Å. If this rotation were to continue, it would eventually close the actin-binding portion of the cleft, and there are indications from EM studies (ref. 22; see also K. Holmes, available at www.mpimf-heidelberg.mpg.de/∼holmes/muscle/weak_strong.html) and fluorescence studies (23) that this change occurs when myosin binds strongly to actin. We note that a similar rigid-body movement can be observed in the 2.8-Å chicken skeletal nucleotide-free near-rigor S1 structure (7), albeit without bending in switch I, but this movement is less pronounced than in scallop S1 and has not previously been reported. Nevertheless, this rotation would not be possible if switch I were constrained by specific interactions with a nucleotide.
The Cross-Linker.
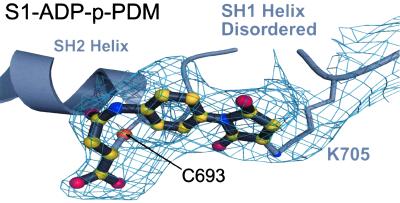
The crystal structures of scallop S1 with p-PDM provide the first visualization of the thiol cross-link in the myosin head. We find that the cross-link in scallop S1 is unusual: one of the rings of p-PDM opens and cross-links SH2 to the side chain of K705, not to SH1 as expected (Fig. 4). Early biochemical studies with skeletal muscle myosin have identified the cross-linked residues as SH1 and SH2 (24, 25). A more recent study indicates that the SH2 group in scallop S1 appears to be the site of the first reaction with p-PDM, in contrast to the cross-linking properties of skeletal muscle isoforms, where SH1 is more reactive than SH2 (Nitao and Reisler, personal communication). We find that in both the scallop S1-ADP-p-PDM and S1-ATP[γ-S]-p-PDM structures, SH1 does not participate in the cross-linking. It has been reported that in actin at high pH, p-PDM also cross-links a cysteine sulfhydryl to a lysine residue (26).
Figure 4.
Electron density for the cross-linker. Shown is the simulated annealing Fo − Fc omit map for the electron density of p-PDM together with the cross-linker modeled into the structure of S1-ADP-p-PDM. To generate this map, contoured at the 3.0 σ level, residues C693 and K705 were omitted along with the p-PDM model. The SH1 helix is disordered, including the SH1 sulfhydryl (C703). Biochemical studies with rabbit muscle myosin have indicated that this thiol can be cross-linked to the SH2 sulfhydryl (C693) by p-PDM (15, 24, 25). However, this map reveals that, in scallop S1, p-PDM cross-links the SH2 sulfhydryl to the side chain of K705, instead of SH1. The scallop S1-ATP[γ-S]-p-PDM structure gives the same result.
The ability to cross-link S1-ADP with p-PDM is also an indication of the physiological state of the myosin head. Although S1-ADP is reported as a strong actin-binding state in the acto-myosin cycle (3, 4), rabbit skeletal acto-S1 cannot be cross-linked by p-PDM (17). Moreover, rabbit skeletal S1-ADP-p-PDM, like S1-ATP, has a lower affinity for the actin filament (F actin) than unmodified S1-ADP (27, 28). The affinity of scallop S1 for actin is also significantly reduced by cross-linking with p-PDM (unpublished results). These findings indicate that this intramolecular cross-link occurs only in an actin-detached form of S1-ADP.
Mutations Near the SH1 Helix.
A number of mutations near the SH1 helix indicate the importance of this element in the functioning of the motor. Mutations R702C (R701 in scallop) and N93K (N95 in scallop) in human nonmuscle myosin IIA (NMIIA) substantially reduce both actin-activation of the MgATPase and the motility velocity (¶ and J. Sellers, personal communication). Crystal structures of various myosin isoforms (e.g., PDB ID codes 2MYS, 1BR1, and 1KK7) show that R702C breaks side chain hydrogen bonds that help stabilize the last turn of the SH1 helix, immediately before the G707 pivot (scallop G706, Fig. 5). (Note that in most isoforms, two glycine residues, one on each side of the SH1 helix, have been pictured as pivots for the rotation of the converter; refs. 10, 29, and 30.) Similarly, N93K breaks a hydrogen bond that helps stabilize the first turn of N-terminal subdomain helix C. The two residues flanking this asparagine form van der Waals contacts and hydrogen bonds that help to stabilize the SH1 helix and position it correctly about the G695 pivot (scallop numbering). Weakening these interactions may cause the SH1 helix to wobble on the G695 pivot, which, along with increased flexibility of the helix, would dislocate the G706 pivot as well. This mutation affects the entire SH1 helix and, correspondingly, it inhibits myosin activity more completely than R702C. (Note that the residue in chicken skeletal myosin equivalent to N93 is a histidine, which is more firmly anchored than the asparagine by extensive van der Waals and hydrogen bond contacts with nearby residues. It is therefore possible that the SH1 helix in chicken skeletal myosin is more stable than its counterpart in either human NMIIA, chicken smooth, or scallop myosins.) The mutations described above demonstrate that the stability of the SH1 helix and the glycine pivots is vital to the proper functioning of the motor.
Figure 5.
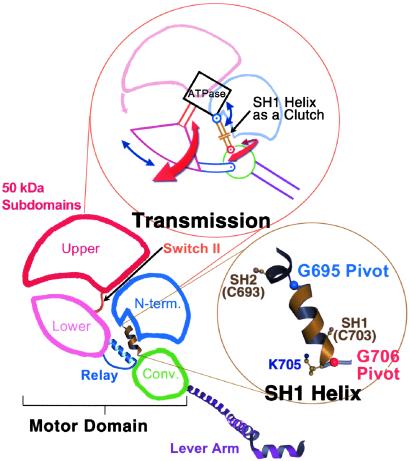
Mechanical components of the motor. Schematic diagram of the myosin head (shown here in the near-rigor conformation), depicting the four subdomains of the motor together with the lever arm (light chains not shown). (Upper Inset) a mechanical representation of the motor's “transmission,” illustrated to highlight the roles of various components. Switch II functions like a piston by delivering work from the enzyme (“ATPase”) to the 50-kDa lower subdomain, which converts this work to rotational motion, roughly comparable to the role of a crank shaft. During the transition from the near-rigor to the pre-power stroke conformation (with the lever arm moving from “down” to “up”), the rotation of the 50-kDa lower subdomain can be divided into two roughly perpendicular vector components (red and blue arrows). The red arrows represent rotation about axes in the plane of the paper, and the blue arrows represent rotation about axes perpendicular to the plane of the paper (described previously as twist and tilt motions, respectively; ref. 40). The converter rotates about the glycine pivots along these vector components as shown (1, 10) and, in addition moves about a hinge in the relay (not shown, see text and ref. 1). During this transition, the molecule passes through the internally uncoupled state, in which the SH1 helix unwinds. The converter/lever arm orientation then becomes less constrained, and torque cannot be delivered from switch II to the converter/lever arm module. Thus, the function of the SH1 helix can be compared with that of an automobile clutch, which can disengage the gear train and drive shaft from the motor. Based on EM studies of myosin cross-bridges bound to actin and the x-ray crystallographic nucleotide-free near-rigor structure, it appears that rotation of the 50-kDa upper subdomain (pink arrow) on strong binding to actin closes the front of the cleft and triggers both the power stroke and product release (see text). A related two-dimensional mechanical model of the myosin motor has been published (41), but revisions are needed to agree with recent experimental findings. (Lower Inset) The SH1 helix flanked by the glycine pivots.
Mechanical Implications.
This analysis prompts us to consider the possible mechanical roles played by different structural elements of the motor, based on a lever arm model and independent of the precise physiological pre- and post-power stroke conformations. Specific aspects of the motor's mechanism may be illustrated by different (albeit imperfect) mechanical analogues, one of which is the automobile engine. The 50-kDa lower subdomain has been described as functioning like a piston in the motor (31), but it seems more appropriate to assign that role to the switch II loop, which moves in and out of the nucleotide-binding pocket during enzymatic activity. Small conformational changes in switch II are converted into rotational motion by the 50-kDa lower subdomain. The relay (on one end of this subdomain; refs. 1 and 10), the converter, and the SH1 helix may be pictured as the transmission of the myosin motor (Fig. 5). The relay delivers torque from the switch II loop to the converter. In addition, this relay helix bends about a hinge near its center during the pre-power stroke conformation, allowing the 50 kDa subdomain to approach closely to the N-terminal subdomain. The converter moves about this hinge while simultaneously rotating about two pivots (G695 and G706), located on either side of the SH1 helix (10, 29, 30). The precise locations of the glycine pivots, which may differ in different isoforms, are important in determining both the range of motion of the converter and the conserved contacts it can make with the relay. In the final stage of torque transmission, the converter directs the rotational motion of the lever arm (1, 10).
The internally uncoupled conformation demonstrates a second role for the SH1 helix, in addition to controlling the location of the glycine pivots. When the SH1 helix unwinds, it effectively disengages the converter/lever arm module from the motor; i.e., although the module remains associated with the relay, torque cannot be transmitted from the motor to the converter because the relay is a flexible joint. At this stage of the contractile cycle, myosin-ATP is in equilibrium with nucleotide-free myosin (32). If the SH1 helix did not disengage the motor to produce the internally uncoupled state, rapid but transient reattachment of the head to actin might introduce an unproductive impedance to the sliding of the actin filament. Moreover, the greater freedom of motion for the converter/lever arm module resulting from the unwound SH1 helix may help facilitate the transition to the pre-power stroke conformation. The SH1 helix can therefore be described as a kind of clutch that controls coupling between the converter and the rest of the motor. According to this picture, the internally uncoupled state occurs during the contractile cycle as an ATP state, between the near-rigor and pre-power stroke conformations.
To consider the possible effects of strong binding of myosin to actin on the mechanics of the motor, we must rely on low-resolution (EM) data, because crystal structures of such states are not yet available. The results (ref. 22, see also www.mpimf-heidelberg.mpg.de/∼holmes/muscle/weak_strong.html) suggest that strong actin binding is accompanied by a rotation of the 50-kDa upper subdomain that closes the actin-binding portion of the cleft and simultaneously opens the “front door,” facilitating release of MgADP (Fig. 5). It is likely that this rotation triggers the power stroke by breaking a critical salt bridge and several hydrogen bonds between switches I and II that stabilize the pre-power stroke conformation (12). As a result, switch II would change from a bent to an extended conformation similar to that found in the near-rigor crystal structures, producing a relatively small rotation of the 50-kDa lower subdomain and a larger rotation of the converter/lever arm module in the power stroke. Although the 50-kDa upper subdomain closes the actin-binding side of the cleft, the conformational change of switch II would open the base of the cleft (near the nucleotide-binding pocket) and provide a “back door” for phosphate release. Moreover, the new orientation of the 50-kDa upper subdomain would constrain the 50-kDa lower subdomain after the power stroke and thus stabilize relay interactions with the SH1 helix (the same interactions that we characterized above as transient stabilizing contacts in the near-rigor nucleotide-free crystal structure). These interactions would prevent the SH1 helix from unwinding immediately after the power stroke. In terms of the automobile analogy, strong myosin binding to actin is the spark plug charge that triggers the switch II piston motion; this interaction may also lock the SH1 clutch to prevent uncoupling of the converter/lever arm module from the rest of the motor immediately after the power stroke. Unlike the automobile engine, however, here the chemical step (i.e., hydrolysis of ATP to form protein bound ADP⋅Pi) and the mechanical output of acto-myosin are not simultaneous; rather, on cleavage of the ATP, the energy is stored and then transduced to mechanical work on strong binding of myosin to actin (5, 33, 34).
Coda.
The significance of the internally uncoupled S1-ATP state discovered in 1999 (1) is only now becoming recognized (35). The contractile cycle is often described by referring simply to the pre- and postpower stroke states. But, as shown in this study (see also ref. 1), the internally uncoupled state is informative about the structural basis for certain mechanical properties of the myosin motor, including the key role played by the SH1 helix. Moreover, the rich biochemical literature on myosin cross-linking can be accounted for by this state. The nucleotide-free near-rigor S1 structure indicates that the switch I loop moves between two conformations—constrained and relaxed—in the presence and absence of nucleotide. This result suggests that, in addition to coordinating the Mg2+ cation during catalysis, switch I serves as one of the key sensors of the nucleotide in the binding pocket. The small changes we have detected in the conformation of switch I and the reorientation of the 50-kDa upper subdomain in the transition from the S1-MgATP near-rigor state to the nucleotide-free near-rigor state may presage the larger changes in these components expected when myosin binds strongly to actin in the true rigor state. These findings, together with results from EM studies, suggest key steps by which strong myosin binding to actin induces the power stroke. We picture that closure of the actin-binding cleft by rotation of the 50-kDa upper subdomain would break interactions between switches I and II, leading to relaxation of switch I and the piston-like motion of switch II.
Supplementary Material
Acknowledgments
We are grateful to S. Sprang, A. Houdusse, J. H. Brown, Y. Goldman, E. Reisler, and J. Sellers for helpful discussions, E. O'Neall Hennessey for her expert technical assistance, M. Love and the staffs of the Cornell High Energy Synchrotron Source and the Brookhaven National Laboratory for assistance with data collection, and C. Palmer and A. Brilliant for assistance with photography. This work was supported by National Institutes of Health Grant AR17346 and a grant from the Muscular Dystrophy Association (to C.C.), as well as National Institutes of Health Grant AR41808 (to C.C. and A.G.S.).
Abbreviations
- S1
subfragment 1
- AMPPNP
adenosine 5′-[β,γ-imido]triphosphate
- ATP[γ-S]
adenosine 5′-[γ-thio]triphosphate
- EM
electron microscopy
- MD
motor domain
- p-PDM
para-phenyl dimaleimide
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1KK7, 1KK8, 1KQM, 1L20, and 1KWO for near-rigor nucleotide-free S1, S1-ADP⋅BeFx, S1-AMPPNP, S1-ADP-p-PDM, and S1-ATPγS-p-PDM, respectively).
Hu, A., Wang, F. & Sellers, J. R. (2002) Biophys. J. 82, 407a (abstr.).
References
- 1.Houdusse A, Kalabokis V N, Himmel D, Szent-Györgyi A G, Cohen C. Cell. 1999;97:459–470. doi: 10.1016/s0092-8674(00)80756-4. [DOI] [PubMed] [Google Scholar]
- 2.Houdusse A, Szent-Györgyi A G, Cohen C. Proc Natl Acad Sci USA. 2000;97:11238–11243. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldman Y E. Cell. 1998;93:1–4. doi: 10.1016/s0092-8674(00)81137-x. [DOI] [PubMed] [Google Scholar]
- 4.Spudich J A. Nature (London) 1994;372:515–518. doi: 10.1038/372515a0. [DOI] [PubMed] [Google Scholar]
- 5.Lymn R W, Taylor E W. Biochemistry. 1971;10:4617–4624. doi: 10.1021/bi00801a004. [DOI] [PubMed] [Google Scholar]
- 6.Houdusse A, Sweeney H L. Curr Opin Struct Biol. 2001;11:182–194. doi: 10.1016/s0959-440x(00)00188-3. [DOI] [PubMed] [Google Scholar]
- 7.Rayment I, Rypniewski W R, Schmidt-Bäse K, Smith R, Tomchick D R, Benning M M, Winkelmann D A, Wesenberg G, Holden H M. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 8.Bauer C B, Holden H M, Thoden J B, Smith R, Rayment I. J Biol Chem. 2000;275:38494–38499. doi: 10.1074/jbc.M005585200. [DOI] [PubMed] [Google Scholar]
- 9.Gulick A M, Bauer C B, Thoden J B, Rayment I. Biochemistry. 1997;36:11619–11628. doi: 10.1021/bi9712596. [DOI] [PubMed] [Google Scholar]
- 10.Dominguez R, Freyzon Y, Trybus K M, Cohen C. Cell. 1998;94:559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 11.Smith C A, Rayment I. Biochemistry. 1996;35:5404–5417. doi: 10.1021/bi952633+. [DOI] [PubMed] [Google Scholar]
- 12.Fisher A J, Smith C A, Thoden J B, Smith R, Sutoh K, Holden H M, Rayment I. Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 13.Smith C A, Rayment I. Biochemistry. 1995;34:8973–8981. doi: 10.1021/bi00028a005. [DOI] [PubMed] [Google Scholar]
- 14.Wells J A, Yount R G. Methods Enzymol. 1982;85:93–116. doi: 10.1016/0076-6879(82)85013-1. [DOI] [PubMed] [Google Scholar]
- 15.Burke M, Reisler E. Biochemistry. 1977;16:5559–5563. doi: 10.1021/bi00644a026. [DOI] [PubMed] [Google Scholar]
- 16.Wells J A, Sheldon M, Yount R G. J Biol Chem. 1980;255:1598–1602. [PubMed] [Google Scholar]
- 17.Nitao L K, Reisler E. Biochemistry. 1998;37:16704–16710. doi: 10.1021/bi9817212. [DOI] [PubMed] [Google Scholar]
- 18.Cooke R. Curr Biol. 1999;9:R773–R775. doi: 10.1016/S0960-9822(00)80010-X. [DOI] [PubMed] [Google Scholar]
- 19.Bagshaw C R, Trentham D R. Biochem J. 1974;141:331–349. doi: 10.1042/bj1410331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milburn M V, Tong L, deVos A M, Brunger A, Yamaizumi Z, Nishimura S, Kim S H. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 21.Yount R G, Lawson J D, Rayment I. Biophys J. 1994;68:44s–49s. [PMC free article] [PubMed] [Google Scholar]
- 22.Volkmann N, Hanein D, Ouyang G, Trybus K M, DeRosier D J. Nat Struct Biol. 2000;7:1147–1155. doi: 10.1038/82008. [DOI] [PubMed] [Google Scholar]
- 23.Yengo C M, DeLaCruz E M, Chrin L R, Gaffney D P, II, Berger C L. J Biol Chem. 2002;277:24114–24119. doi: 10.1074/jbc.M111253200. [DOI] [PubMed] [Google Scholar]
- 24.Yamashita T, Soma Y, Kobayashi S, Sekine T. J Biochem (Tokyo) 1965;57:460–461. doi: 10.1093/oxfordjournals.jbchem.a128103. [DOI] [PubMed] [Google Scholar]
- 25.Bailin G, Bárány M. J Biol Chem. 1972;247:7815–7821. [PubMed] [Google Scholar]
- 26.Knight P, Offer G. Biochem J. 1978;175:1023–1032. doi: 10.1042/bj1751023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greene L E, Chalovich J M, Eisenberg E. Biochemistry. 1986;25:704–709. doi: 10.1021/bi00351a030. [DOI] [PubMed] [Google Scholar]
- 28.Chalovich J M, Greene L E, Eisenberg E. Proc Natl Acad Sci USA. 1983;80:4909–4913. doi: 10.1073/pnas.80.16.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinose F, Wang S X, Kidambi U S, Moncman C L, Winkelmann D A. J Cell Biol. 1996;134:895–909. doi: 10.1083/jcb.134.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geeves M A, Holmes K C. Annu Rev Biochem. 1999;68:687–728. doi: 10.1146/annurev.biochem.68.1.687. [DOI] [PubMed] [Google Scholar]
- 31.Vale R D, Milligan R A. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- 32.Trentham D R, Eccleston J F, Bagshaw C R. Q Rev Biophys. 1976;9:217–281. doi: 10.1017/s0033583500002419. [DOI] [PubMed] [Google Scholar]
- 33.Bagshaw C R, Trentham D R, Wolcott R G, Boyer P D. Proc Natl Acad Sci USA. 1975;72:2592–2596. doi: 10.1073/pnas.72.7.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldman Y E. Annu Rev Physiol. 1987;49:637–654. doi: 10.1146/annurev.ph.49.030187.003225. [DOI] [PubMed] [Google Scholar]
- 35.Homma K, Yoshimura M, Saito J, Ikebe R, Ikebe M. Nature (London) 2001;412:831–834. doi: 10.1038/35090597. [DOI] [PubMed] [Google Scholar]
- 36.Engh R A, Huber R. Acta Crystallogr A. 1991;47:392–400. [Google Scholar]
- 37.Kleywegt G J, Brünger A T. Structure (London) 1996;4:897–904. doi: 10.1016/s0969-2126(96)00097-4. [DOI] [PubMed] [Google Scholar]
- 38.Kleywegt G J, Bergfors T, Senn H, Le Motte P, Gsell B, Shudo K, Jones T A. Structure (London) 1994;2:1241–1258. doi: 10.1016/s0969-2126(94)00125-1. [DOI] [PubMed] [Google Scholar]
- 39.Luzzati V. Acta Crystallogr. 1952;5:802–810. [Google Scholar]
- 40.Corrie J E T, Brandmeier B D, Ferguson R E, Trentham D R, Kendrick-Jones J, Hopkins S C, van der Heide U A, Goldman Y E, Sabido-David C, Dale R E, et al. Nature (London) 1999;400:425–430. doi: 10.1038/22704. [DOI] [PubMed] [Google Scholar]
- 41.Becker E W. Proc Natl Acad Sci USA. 2000;97:157–161. doi: 10.1073/pnas.97.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.