Abstract
In Escherichia coli chemotaxis, signaling depends on modulation of the level of phosphorylation of CheY, a small protein that couples receptors and flagellar motors. Working in vivo, we used fluorescence resonance energy transfer (FRET) to measure the interaction of CheY∼P with its target, FliM. Binding of CheY∼P to FliM was found to be much less cooperative than motor switching; however, under the conditions of our experiment, most of the FliM appeared to be in the cytoplasm. We studied signal processing times in the chemotaxis pathway by measuring the changes in CheY∼P binding to FliM on flash release of caged chemoeffectors. Following sudden addition of attractant, the amount of CheY∼P bound to FliM decayed exponentially with a rate constant of about 2 s−1. Following sudden addition of repellent, FliM occupancy increased with a rate constant of about 20 s−1. Using these data, we were able to construct a simple model for the chemotactic pathway and to estimate values of rate constants for several key reactions.
Keywords: bacteria‖signal transduction‖cyan fluorescent protein‖yellow fluorescent protein
In Escherichia coli, information about changes in chemoeffector concentration is transmitted from specific receptors at the cell surface to the flagellar motors via a labile phosphorylated intermediate, CheY∼P. CheY is phosphorylated by the receptor-coupled kinase CheA and dephosphorylated by the phosphatase CheZ. Attractant binding to receptors lowers CheA activity, thus decreasing the level of CheY∼P. Attractant removal or repellent addition has the opposite effect. Cells adapt to constant levels of attractants or repellents by receptor methylation or demethylation, effected by the enzymes CheR and CheB, respectively. CheY∼P binds to FliM, a component of the switch complex at the cytoplasmic face of the flagellar motor, stabilizing the clockwise rotational state and destabilizing the counterclockwise rotational state. Most of the chemotaxis proteins, including CheA, CheY, and CheZ, are localized to receptor clusters at the cell poles (1, 2), although the reasons for such localization, especially of CheZ, remain unclear. The flagellar motors, on the other hand, are distributed at random on the sides of the cell. Thus, CheY∼P is produced at the cell pole and has to diffuse through the cytoplasm to reach the flagellar motors. For reviews of the chemotaxis pathway, see refs. 3 and 4.
The kinetics of changes in CheY∼P binding to FliM on attractant or repellent stimulation provide important information about the signal-processing pathway. Signal-processing times have been inferred previously from responses of tethered cells stimulated iontophoretically (5) or of swimming cells stimulated by release of caged attractants or repellents (6, 7). However, these methods require that one wait for motors to respond, and the interpretation of results is complicated by the nonlinear (ultrasensitive) dependence of motor bias on CheY∼P concentration (8).
To learn more about the binding of CheY∼P to FliM and about the kinetics of the chemotactic response, we extended our recent analysis of phosphorylation-dependent interactions of CheY with CheZ (9) and measured fluorescence resonance energy transfer (FRET) between cyan fluorescent protein (CFP) fused to the N terminus of FliM (CFP-FliM) and yellow fluorescent protein (YFP) fused to the C terminus of CheY (CheY-YFP). The FRET technique relies on the distance-dependent transfer of energy from an excited donor fluorophore (CFP) to an acceptor fluorophore (YFP) and allows one to monitor changes in protein interactions in real time in vivo (10, 11). We found that CheY∼P binds to FliM in vivo with a dissociation constant of about 3.7 μM, close to the concentration of CheY∼P required for a half-maximal motor response (8). This binding was much less cooperative than motor switching. The kinetics of CheY∼P binding to FliM was studied following flash release of caged attractant (aspartate) or repellent (protons), and the observed responses were modeled. Rates of the various reactions appear to be optimized for the time scale set by diffusion of CheY∼P through the cytoplasm.
Materials and Methods
Plasmids and Strains.
Fusions to CheY and FliM of yellow and cyan fluorescent protein (YFP and CFP, CLONTECH) were made as described (2, 9). The cheY-eyfp fusion was cloned into pTrc99A (AmpR, Pharmacia) under an isopropyl β-d-thiogalactoside (IPTG)-inducible promoter, yielding pVS18. The ecfp-fliM fusion was cloned into pBAD33 (CmR) under an arabinose-inducible promoter (12), yielding pVS31. The strains used for the FRET experiments were derived from strain DFB228 (ΔfliM), the gift of David Blair (University of Utah, Salt Lake City). VS120 (ΔcheY ΔfliM) and VS121 [Δ(cheY-cheZ) ΔfliM] were made by in-frame deletions of the corresponding chemotaxis genes (2). Strain VS125 [Δ2206(tap-cheZ) ΔfliM] was made by P1-transduction of the ΔfliM strain from RP2893 [Δ2206(tap-cheZ)], the gift of Sandy Parkinson (University of Utah, Salt Lake City). Growth with 34 μg/ml chloramphenicol and 100 μg/ml ampicillin was used for selection. Because the CheY/FliM interaction always was tested in a ΔcheY ΔfliM background, strain VS120 is referred to as wild type and only additional background mutations are indicated in the text. Strain VS125 is referred to as cheR cheB cheZ. YFP and CFP proteins were expressed as C-terminal fusions to 6× His tag from pVS47 and pVS48 plasmids, respectively, and purified using Ni-NTA spin kit (Qiagen, Valencia, CA).
Data Acquisition and Analysis.
Cells were grown and prepared for FRET as described (9). The levels of arabinose and IPTG induction were 100 μg⋅ml−1 and 0.05 mM, respectively, unless specified otherwise. We monitored fluorescence from a field of 300–500 cells in each experiment, and repeated each experiment at least three times by using different cell cultures. Fluorescence signals in cyan and yellow channels were detected using two photon-counting photomultipliers (H7421-40, Hamamatsu, Bridgewater, NJ) whose outputs were converted to analog signals by ratemeters (RIS-375, Rowland Institute). For the flow experiments the signals were filtered by 8-pole low-pass Bessel filters (3384, Krohn-Hite, Avon, MA) with 2 Hz cutoff frequency and sampled at 5 Hz by a computer data-acquisition system [National Instruments (Austin, TX) LABVIEW 5.1 and Macintosh G3]. For the flash experiments using wild-type cells the signals were filtered with 200 Hz cutoff frequency and sampled at 500 Hz, whereas for the flash experiments using cheZ cells the signals were filtered with 10 Hz cutoff frequency and sampled at 20 Hz. FRET was calculated as fractional change in CFP fluorescence, as described in the text. Alternatively, changes in cyan emission could be computed from changes in the ratios of yellow and cyan emission, as described (9). The two methods gave essentially the same results (data not shown).
Curve Fitting.
Fits were made to a multisite Hill model of the form FmaxYH/(YH + K1/2H), where Fmax is the value of FRET at saturating CheY-YFP concentrations, Y is the concentration of CheY-YFP, H is the Hill coefficient, and K1/2 is the apparent dissociation constant (Fig. 2A), or of the form M0 + [ΔMmax AH/(AH + K1/2H)], where M0 is the initial FliM occupancy, ΔMmax is the change in FliM occupancy observed at saturating stimulation, A is the step in attractant concentration, H is the Hill coefficient, and K1/2 is the value for the half-maximal response (Fig. 3). Decays following release of caged aspartate were fit to exponentials, [M0 − Mf] exp[−k(t − τ)] + Mf, where M0 is the initial FliM occupancy or initial CheY∼P level, Mf is the final FliM occupancy or final CheY∼P level, k is the decay rate, t is the time after the flash, and τ is a delay before the onset of the response.
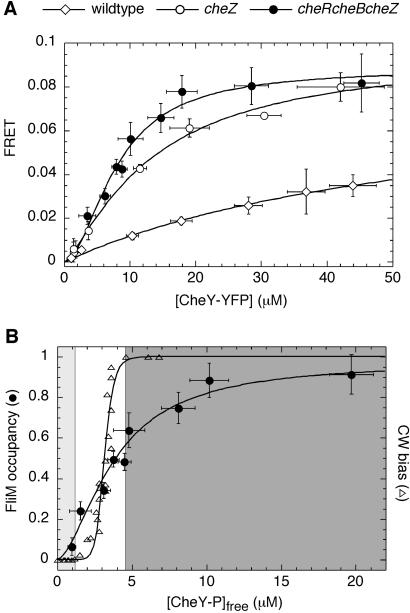
Figure 2.
Binding curves for CheY-YFP/CFP-FliM measured in vivo. (A) Dependence of FRET signals from unstimulated cell populations of wild-type (◊), cheZ (○), and cheRcheBcheZ (●) cells on the expression level of CheY-YFP, varied by induction with IPTG (0–0.3 mM). The CheY-YFP and CFP-FliM concentrations were determined as described in Materials and Methods. (B) Comparison of dependence of motor bias (▵) and FliM occupancy (●) on concentration of free cytoplasmic CheY∼P, [CheY∼P]free. Data for the motor bias and parameters for the Hill fit (dashed curve, H = 10.3, K1/2 = 3.1 μM) are taken from ref. 8. Data for FliM occupancy are recalculated from the data for the cheR cheB cheZ cells in (A) assuming that a FRET value of 0.088 corresponds to a FliM occupancy of 1, as explained in the text. Only part of the data are shown. The light-shaded area (Left) indicates the range of CheY∼P concentrations over which the rotation of the motor is exclusively counterclockwise, the unshaded area (Center) the range of concentrations over which the motor switches, and the dark-shaded area (Right) the range of concentrations over which the rotation is exclusively clockwise (CW). Fits by the Hill model are shown by solid lines. Error bars represent standard deviations of multiple experiments.
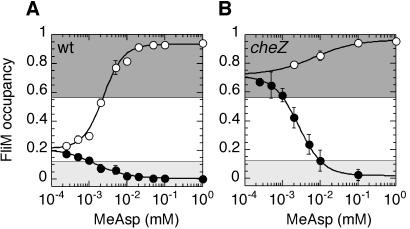
Figure 3.
Changes in FliM occupancy (as in Fig. 2B) shown as a function of changes in attractant concentration, for wild-type (A) and cheZ (B) cells. MeAsp was added (●) and then removed (○) in a sequence of steps of increasing size (as in Fig. 1). Curve fitting is described in Materials and Methods. Best-fit values for the wild type: initial FliM occupancy for a saturating stimulus, M0 = 0.193 ± 0.01; change in FliM occupancy for a saturating stimulus, ΔMmax = −0.193 ± 0.01 (attractant addition) and 0.67 ± 0.02 (attractant removal); K1/2 = 1.5 ± 0.2 μM (attractant addition) and 2.5 ± 0.2 μM (attractant removal), H = 1.0 ± 0.1 (attractant addition) and 1.6 ± 0.2 (attractant removal). Best-fit values for cheZ: M0 = 0.66 ± 0.01; ΔMmax = −0.66 ± 0.02 (attractant addition) and 0.23 ± 0.02 (attractant removal); K1/2 = 2.7 ± 0.2 μM (attractant addition) and 6.8 ± 2 μM (attractant removal), H = 1.3 ± 0.1 (attractant addition) and 0.8 ± 0.2 (attractant removal). Shaded areas and error bars are the same as in Fig. 2B.
Concentration Measurements.
The cellular concentrations of CheY-YFP and CFP-FliM were determined by sonicating a suspension of cells of known density and measuring the fluorescence of the suspension with the same microscope setup that was used for the FRET measurements, by flowing the suspension through the flow cell. Fluorescence standards were determined by adding known amounts of purified YFP and CFP proteins to sonicated suspensions of nonfluorescent cells of similar density. Cell number per milliliter of culture was determined by plating on nonselective LB plates, incubating overnight, and counting colonies. The average volume of the cell cytoplasm was determined as described (13), and was found to be 1.4 × 10−15 liters. The average intracellular protein concentration was then calculated by dividing the amount of CheY-YFP or CFP-FliM fusion protein per milliliter cell culture by the number of cells per milliliter of cell culture and the cytoplasmic volume. In a standard experiment, with cells grown in the presence of 100 μg⋅ml−1 arabinose and 0.05 mM IPTG, the concentration of CFP-FliM was 5.8 ± 0.6 μM, and the concentration of CheY-YFP was 17.9 ± 1.5 μM. The concentration of CheY-YFP at other induction levels was determined by comparing the levels of YFP fluorescence. The level of expression of CFP-FliM in our standard experiment, necessary for optimum complementation of motility, was about three times higher than the level of FliM in the wild type (1.7–2.0 μM; refs. 14 and 15). Applying fractionation by ultracentrifugation, Zhao et al. (15) found that in wild-type cells, most of FliM (≈75%) is present in either functional or incomplete flagellar motors. Using a similar approach, we estimated that at the expression level used in our experiments about 20–30% of CFP-FliM is present in such motors. The balance appears to be in the cytoplasm (data not shown).
Flash Release of Caged Chemoeffectors.
NPE-caged l-aspartate {N-[1-(2-nitrophenyl)ethyloxycarbonyl]aspartic acid, Calbiochem}, nitroindolyl-caged l-aspartate (the gift of John Corrie, National Institute for Medical Research, Mill Hill, London), and NPE-caged proton (2-hydroxyphenyl-1-(2-nitrophenyl)ethyl phosphate, Molecular Probes) were photolyzed by use of a xenon flash lamp [Chadwick–Helmuth (El Monte, CA) 35S driven by model 238 power supply, 200 W⋅s, 50 μs]. The flash lamp was mounted in an Oriel (Stamford, CT) housing (66055) equipped with a fused silica condensing lens (60076) and a Schott UG-5 filter (330 nm peak). The light was fed to the microscope with a liquid light guide (Oriel 77554) and focused onto the flow cell with a 25-mm fused silica lens (Oriel 41220). The light was reflected by a UV-enhanced aluminum mirror (Edmund Scientific, Barrington, NJ) and entered the flow cell from below, through a fused quartz bottom window. The focused spot was about 4 mm wide, much larger than the field of view. A UV-blocking filter (Chroma Technology, Brattleboro, VT) was inserted into the light path above the lower dichroic mirror to prevent UV light from reaching the photomultipliers. The UV flash generated an artifact present in the absence of cells or caged chemoeffectors (a pulse sensed by the photomultipliers that decayed in about 10 ms). This artifact was measured separately before each experiment and subtracted from the data. The efficiency of release of caged l-aspartate was calibrated using the amplitude of the FRET response in cheR cheB cells (9). In a typical experiment, 200 μM caged l-aspartate in tethering buffer supplemented with 5 mM DTT was drawn into the flow cell, and the cells were allowed to equilibrate for 5 min under slow flow (50 μl⋅min−1). The flow was stopped, and ≈10 μM of aspartate was released by a single flash. After data collection, the solution was exchanged by flow, and the procedure was repeated 10–20 times with the same cell culture. The experiment was repeated at least three times with different cell cultures. Release of NPE-caged l-aspartate produced a FRET response identical to that produced by nitroindolyl-caged l-aspartate, which is released much more rapidly; therefore, the rate of release is not limiting at these concentrations. Stimulation with 2.5-mM caged proton was performed similarly, except that the concentration of phosphate in the tethering buffer was reduced to 0.1 mM. The resulting pH change was calibrated by using the pH-sensitivity of the fluorescence of 0.05 μM fluorescein.
Modeling Response Kinetics.
A system of linear differential equations was used describing CheA autophosphorylation, phosphotransfer from phosphorylated CheA to CheY, binding of CheY∼P to FliM, binding of CheY∼P to CheZ, dephosphorylation of CheY∼P by CheZ, and autodephosphorylation of CheY∼P, but excluding adaptation reactions. The activity of chemotaxis pathway was expressed as the fraction of active CheA (16). The system of equations was solved using the NDSolve function of MATHEMATICA 3.0 (Wolfram Research, Champaign, IL). The complete description of the model including rate constants and protein concentrations is published as supporting information on the PNAS web site, www.pnas.org.
Results
CheY∼P Interaction with FliM Can Be Studied in Vivo by Using FRET.
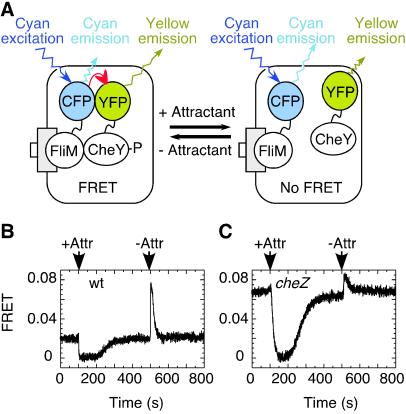
The experimental scenario is shown in Fig. 1. Here, cells are attached to the coverslip of a flow cell and attractant is added or removed. The addition of attractant lowers the level of CheY phosphorylation and leads to a decrease in energy transfer, and the removal of attractant has the opposite effect (Fig. 1A). Addition of attractant to wild-type cells (Fig. 1B) resulted in a rapid decrease in FRET followed by gradual recovery due to adaptation (receptor methylation). The subsequent removal of attractant resulted in a rapid increase in FRET followed by a relatively rapid recovery due to adaptation (receptor demethylation). As expected, the phosphatase-deficient cheZ mutant (Fig. 1C) showed a higher initial FRET level because of a higher steady-state CheY∼P level and a slower response to addition of attractant because of a longer CheY∼P lifetime.
Figure 1.
Changes in interactions between labeled proteins observed by FRET on chemotactic stimulation of E. coli cells. (A) Addition or removal of attractant changes the level of CheY phosphorylation, and thus the interaction between CheY and FliM. As a result, there is a change in energy transfer from CFP-FliM (donor) to CheY-YFP (acceptor), and a corresponding change in cyan and yellow emission levels. (B and C) Stepwise addition and subsequent removal of attractant to the cells attached to a coverslip resulted in changes of energy transfer, expressed here as the fractional decrement in CFP fluorescence (FRET), in (B) wild-type (wt) and (C) cheZ cells. Arrows indicate the time of addition (+Attr) or removal (-Attr) of attractant [30 μM α-methylaspartate (MeAsp)].
In our earlier work, FRET was defined in terms of the YFP/CFP fluorescence ratio, which is immune to variations in the intensity of the excitation light. In practice, our 442-nm laser proved sufficiently stable that we could work simply with the cyan emission. The emission background was measured with cells not containing CFP or YFP and subtracted from all other readings. The cyan emission at zero FRET, C0—this is the value observed in the absence of energy transfer—was measured after adding a saturating amount of attractant or by bleaching YFP, as described (9). In the figures, FRET means the fractional decrement in cyan fluorescence, (C0 − C)/C0, where C is the cyan emission measured during the experiment. The maximum value of this decrement, 0.088 ± 0.005, was obtained in unstimulated cheR cheB cheZ cells expressing large amounts of CheY-YFP (see below). These cells lack the methylesterase and phosphatase that compete with CheY for phosphorylation. Using the results of our model of response kinetics (see Materials and Methods and Discussion), we estimated, for the CheY-YFP concentration used in this work, that more than 99.5% of CheY-YFP is phosphorylated in this strain, in agreement with previous estimates (8, 17). Then, at high CheY-YFP levels, we expect all of FliM, whether built into motors or free, to be bound to phosphorylated CheY-YFP. So 0.088 is the fraction of the fluorescence of a CFP-FliM molecule quenched by binding of phosphorylated CheY-YFP—i.e., the FRET efficiency. This fraction is expected to depend on the 6th power of the separation, R, as R06/(R06 + R6), where R0 is the Förster radius, about 4.9 nm (18, 19). Therefore, the fraction 0.088 ± 0.005 implies a separation of the CFP and YFP fluorophores, in the complex of phosphorylated CheY-YFP with CFP-FliM, of 7.2 ± 0.1 nm.
Adding a saturating amount of attractant only reveals phosphorylation-dependent interaction between CheY and FliM. Bleaching YFP with a 532-nm laser (9) also can reveal any non-phosphorylation-dependent interactions. The two methods gave the same result as addition of a saturating amount of attractant, except at high concentrations of CheY-YFP, where a fraction of the CFP-FliM fluorescence was quenched in absence of CheY-YFP phosphorylation. At the highest CheY-YFP concentration we have used, 45 μM, about 3% of CFP-FliM fluorescence was quenched. This quenching probably was due to the formation of heterodimers of CFP and YFP, observed recently at high protein concentrations (20). The FRET efficiency predicted for the CFP/YFP heterodimer is ≈90% (19). Thus, in our experiment, <4% of CFP-FliM is expected to be bound by CheY-YFP in a phosphorylation-independent manner.
Binding of CheY∼P to FliM in Vivo Is Less Cooperative than the Motor Response.
To obtain CheY∼P/FliM binding curves—i.e., to measure the dependence of FliM occupancy on CheY∼P concentration—we first measured the binding of CheY-YFP (phosphorylated or not) to CFP-FliM, as shown in Fig. 2A. The intracellular concentrations of CFP-FliM and CheY-YFP in populations of cells with different genetic backgrounds expressing different levels of CheY-YFP were measured, as described in Materials and Methods. The abscissa in Fig. 2A is the total CheY-YFP concentration. The FRET level, shown on the ordinate, varies from strain to strain, reflecting various degrees of CheY phosphorylation. As argued above, the cheR cheB cheZ strain, showing the highest FRET levels, gives us the CheY∼P/FliM binding curve. A simple Hill fit gave a Hill coefficient of 1.8 ± 0.3 and an apparent dissociation constant of 8.0 ± 0.7 μM. For cheZ and wild-type cells, the Hill coefficients were 1.0 ± 0.2, but the apparent dissociation constants were substantially higher (13 ± 3 and 59 ± 13 μM, respectively), as expected, given that smaller fractions of CheY are phosphorylated. Here we assume that for CFP-FliM assembled into the motor, there is a linear increase in FRET with the number of bound CheY-YFP molecules.
To obtain the true dissociation constant of CheY∼P binding to FliM, the binding data for the cheR cheB cheZ cells are replotted in Fig. 2B as the fraction of FliM occupied versus the concentration of free cytoplasmic CheY∼P. This concentration was computed by subtracting from the total CheY∼P concentration the amount of CheY∼P bound to FliM and CheA. To calculate the amount of CheY∼P bound to FliM, we took the product of FliM occupancy and FliM concentration, 5.8 μM. To calculate the amount of CheY∼P bound to CheA, we assumed a dissociation constant of 7 μM (21) and a CheA concentration of 5 μM (22). A Hill fit to the data gave a Hill coefficient of 1.7 ± 0.3 and dissociation constant Kd = 3.7 ± 0.4 μM. Also shown are the data of Cluzel et al. (8) on motor bias. The motor transition is much steeper than the change in FliM occupancy, with a fit to the data giving a Hill coefficient of 10.3. Thus, the motor switches in a narrow window of FliM occupancies, and the range of CheY∼P concentrations accessible through observation of motor behavior (Fig. 2B, unshaded area) is much smaller than in our binding assay.
CheZ Does Not Play a Role in Signal Amplification.
Fig. 3 shows initial changes in FliM occupancy deduced from initial changes in FRET following addition and removal of α-methylaspartate (MeAsp) by flow, as defined in ref. 9, in a sequence of steps of increasing size, for wild-type cells (Fig. 3A) and cheZ cells (Fig. 3B). The values for K1/2 and the Hill coefficient obtained for the wild type (see Fig. 3 legend) indicate that the interaction of CheY∼P with FliM does not show a significantly higher cooperativity than the phosphorylation-dependent interaction of CheY with CheZ, observed earlier (9), which gave K1/2 of 2.6 ± 0.5 μM for attractant addition and 7.4 ± 0.4 μM for attractant removal, with H = 1.3 ± 0.1. Because the K1/2 values for the interaction of CheY with FliM are only 2-fold higher in the absence of CheZ, our results confirm the previous observations (9, 23) that CheZ does not play an important role in signal amplification in chemotaxis.
Fig. 3 also illustrates the advantage of using FRET to study chemotactic response. As in Fig. 2B, the unshaded areas indicate the range of FliM occupancies over which one would expect to see a motor response. Whereas the FRET assay is able to distinguish responses to the stimuli of up to 100 μM MeAsp, the motor would saturate at concentrations above 2 μM for wild-type cells and above 10 μM for cheZ cells. Moreover, with the motor assay, the response of cheZ cells to addition of attractant would appear less sensitive because of the high initial FliM occupancy, and the response to removal of attractant would not be detected at all.
Time-Resolved Study of Kinetics of the Chemotactic Response.
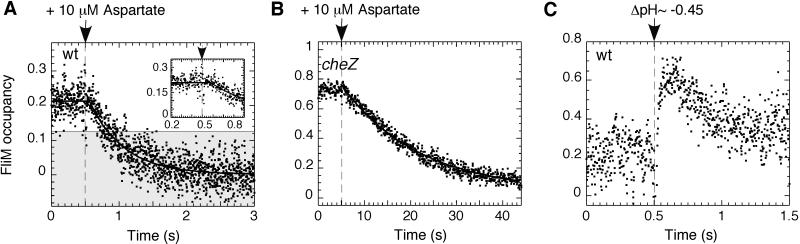
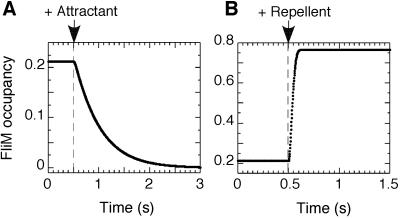
By using flash-stimulated release of caged attractants or repellents, we were able to make measurements of signal processing times in E. coli chemotaxis, as shown in Fig. 4. In wild-type cells stimulated with a saturating amount of the attractant, aspartate (Fig. 4A), there was an initial delay (65 ± 9 ms), followed by a rapid decay (with a first order rate constant, k = 2.0 ± 0.1 s−1). The delay was determined by the intersection of the exponential fit to the decay with the prestimulus baseline (Fig. 4A Inset). The rate constant is smaller than that measured in similar experiments following changes in behavior of free-swimming cells (7). As argued above, this might be due to the nonlinear nature of the motor response, because a saturating counterclockwise motor bias (shaded area) is reached sooner than zero FliM occupancy, which gives a lower value of the half-time of the response, and thus a higher value of apparent rate constant. The decrease of FliM occupancy in cheZ cells after stimulation was much slower (Fig. 4B), with k = 0.059 ± 0.001 s−1, as expected, given the absence of the phosphatase. The delay was not detected, because the filter used to smooth the data were set to a longer time span.
Figure 4.
Changes in FliM occupancy after flash-release of caged chemoeffectors. Attractant was released by UV flash at the times indicated by the arrows and thin vertical dashed lines. The smooth curves are exponential fits to the data; see Materials and Methods. FliM occupancy was calculated from the FRET signals as in Fig. 2B. (A) Response of wild-type cells to release of 10 μM caged l-aspartate. The horizontal solid line indicates the average prestimulus FliM occupancy. The data points just before and after the flash are shown with greater time resolution in the Inset. (B) Response of cheZ cells to release of 10 μM caged l-aspartate. (C) Changes in FliM occupancy after flash release of caged proton, resulting in a pH decrement of about 0.45.
Given the relationship between FliM occupancy and the total concentration of CheY∼P (Fig. 2A) we also could compute the corresponding CheY∼P decay curves (not shown). This gave us slightly larger rate constants, k = 2.2 ± 0.1 s−1 and 0.085 ± 0.001 s−1, corresponding to decay half times of t1/2 = 0.32 s and 8 s, respectively. Assuming that the off rate of CheY∼P from the FliM complex is fast relative to rate of dephosphorylation, these would be the half times for CheZ-catalyzed and spontaneous dephosphorylation, respectively.
Stimulation of wild-type cells by flash photolysis of a caged proton, a repellent, Fig. 4C, revealed no measurable delay (<20 ms). The maximum FliM occupancy was reached within 70–100 ms. This is in a good agreement with the value of ≈50 ms obtained for free-swimming cells (6). From the time required to reach ≈65% [1 − (1/e)] of the maximal repellent response (≈45 ms), the first-order rate constant of the repellent response was estimated as ≈22 s−1.
Discussion
Physiological responses of chemotactic E. coli cells have been studied extensively before, and many biochemical assays have been carried out on the reactions involved in the pathway. The use of FRET has allowed us to take these studies one step further to investigate responses at the level of the response regulator, CheY∼P. In our previous study (9), we used FRET between CheZ-CFP and CheY-YFP to infer the activity of the receptor kinase complex. Here, we used FRET between CFP-FliM and CheY-YFP to study their binding and its changes on chemotactic stimulation. We found that the binding of phosphorylated CheY-YFP to CFP-FliM in the cell is much less cooperative than the motor response. This finding agrees with allosteric transition models, where the binding function is generally less cooperative than the state function (24, 25). However, given that only a fraction of CFP-FliM in our experiment is incorporated into functional flagellar motors, the agreement with the allosteric models could be fortuitous. To make a precise comparison between binding of CheY∼P to the motor and motor switching, a better test would be to measure the binding affinities of phosphorylated CheY-YFP to CFP-FliM localized on a single motor. We hope that such a measurement will be possible using the FRET technique.
We combined FRET with flash release of caged chemoeffectors to determine signal processing times in the pathway. In wild-type cells, response times for changes in FliM occupancy are determined by several sets of rates: (i) rates of receptor-linked CheA autophosphorylation, CheY phosphorylation by CheA∼P, and CheY∼P dephosphorylation by CheZ; (ii) on- and off-rates of CheY∼P binding to FliM; and (iii) CheY∼P diffusion from receptor-kinase clusters on the cell pole across the cytoplasm to the flagellar motors or to cytoplasmic FliM. The phosphotransfer from CheA∼P to CheY is very fast (26), so the response to repellent (CheA-activating) stimuli is presumably limited by the rate of CheA autophosphorylation. From the kinetics of the repellent response, the rate constant for autophosphorylation of activated CheA should be >22 s−1, in good agreement with in vitro measurements (27) of 20–30 s−1. The response time to the sudden addition of attractant is determined in wild-type cells by CheZ-dependent dephosphorylation of CheY, giving the rate constant of ≈2 s−1. The 65-ms delay between stimulation and the onset of the response might be due, in part, to the ≈50-ms delay between CheY∼P binding to CheZ and CheZ activation observed in vitro (28). Our data also allowed us to estimate the on- and off-rates of CheY∼P binding to FliM. The time course of the repellent response implied an on-rate constant of >4 × 106 M−1⋅s−1 and, given the average Kd of CheY∼P binding to FliM of 3.7 μM, an off-rate of >15 s−1. This is close to the values for the on-rate constant, 3 × 106 M−1⋅s−1 (diffusion-limited), and the off-rate constant, 10 s−1, assumed in a recent model for motor switching (25).
More detailed modeling was performed using a set of linear differential equations describing phosphotransfer and binding reactions in the chemotaxis pathway. Using the rate estimates made above and values from the literature (see Materials and Methods and supporting information), we were able to reproduce the responses observed to addition of attractant or repellent shown in Figs. 4 A and C. The corresponding model values appear in Fig. 5 A and B. Note that adaptation was not included in our model; hence, the difference between Figs. 4C and 5B. The model allowed us to further refine the estimates of reaction rates required to match the observed response kinetics. To match the rate of increase in FliM occupancy observed on addition of repellent (Fig. 5B), the rate constant for autophosphorylation of activated CheA had to be set to 50 s−1, higher than the value estimated above. The delay predicted for this response was ≈5 ms, below the time resolution of our experiment. To match the rate constant of decay in FliM occupancy observed on addition of attractant (≈2 s−1; Fig. 5B), we had to set the concentration of CheZ in the cytoplasm to 1.1 μM. The total concentration of CheZ in the cell is 4–12 μM, depending on strain and growth conditions (13), but most of the CheZ is localized to receptor clusters (2), which might justify the lower value for the cytoplasmic concentration. Our simulation predicted a short (≈20 ms) delay in response to attractant because of a lag in dissociation of CheY∼P from FliM. Given the additional 50-ms delay in CheZ activation mentioned above, this might explain the observed delay of ≈65 ms.
Figure 5.
Simulation of excitation kinetics in chemotaxis. The system of linear differential equations (see Materials and Methods) was solved first for the steady state, which yielded an estimate for the fraction of CheA in the active state, 0.02. Addition of attractant reduced this fraction and addition of repellent increased it. The responses shown represent the relaxation of the system to the new steady state. Events linking changes in receptor occupancy to changes in kinase activity were assumed to be fast compared with the changes in protein phosphorylation levels. (A) Step-addition of attractant, simulated by changing the fraction of active CheA from 0.02 to 0. (B) Step-addition of repellent, simulated by changing the fraction of active CheA from 0.02–1. Arrows and thin vertical dashed lines indicate the time of stimulation.
The reactions in the chemotaxis pathway appear to be as fast as needed to work within the diffusion limit. The diffusion coefficient for CheY-GFP has been measured (8) (4.6 μm2⋅s−1), and the diffusion coefficient for CheY was estimated to have a similar value (ref. 29; of order 10 μm2⋅s−1). Thus, the time required for CheY∼P to diffuse ≈1 μm from a receptor cluster near one pole to the flagellar motors, distributed at random along the sides of the cell, is of order x2/2D ≈ 50 ms. This is the same order of magnitude as the time required for the repellent response. If CheY is phosphorylated at one pole and CheY∼P decays with the rate constant measured for the attractant response, k ≈ 2 s−1, then its concentration will not be uniform over the length of the cell. At steady state, the distance from the pole over which the concentration of CheY∼P falls by 1/e ≈ 1/3, is (D/k)1/2 ≈ 2 μm, as observed (29). Thus, given the ultrasensitivity of the motor response shown in Fig. 2B, the rotational bias might vary significantly with the distance from the pole. This probably sets an upper limit on the allowable rate for CheY∼P hydrolysis in the cytoplasm. We know that the disparity in rotational bias is not very great, because fully adapted wild-type cells spend most of their time swimming at top speed (30), which occurs only when all of the filaments spin counterclockwise in one coherent bundle, while some tumbles involve clockwise rotation of all of the flagella (31).
Supplementary Material
Acknowledgments
We thank David Blair and Sandy Parkinson for providing us with bacterial strains and N. Le Novére for helpful comments on the model of allosteric transitions. This work was supported by a grant from the National Institutes of Health and by the Rowland Institute for Science.
Abbreviations
- CFP
cyan fluorescent protein
- FRET
fluorescence resonance energy transfer
- IPTG
isopropyl β-d-thiogalactoside
- YFP
yellow fluorescent protein
References
- 1.Maddock J R, Shapiro L. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 2.Sourjik V, Berg H C. Mol Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- 3.Falke J J, Bass R B, Butler S L, Chervitz S A, Danielson M A. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourret R B, Stock A M. J Biol Chem. 2002;277:9625–9628. doi: 10.1074/jbc.R100066200. [DOI] [PubMed] [Google Scholar]
- 5.Segall J, Manson M, Berg H C. Nature (London) 1982;296:855–857. doi: 10.1038/296855a0. [DOI] [PubMed] [Google Scholar]
- 6.Khan S, Castellano F, Spudich J L, McCray J A, Goody R S, Reid G P, Trentham D R. Biophys J. 1993;65:2368–2382. doi: 10.1016/S0006-3495(93)81317-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jasuja R, Keyoung J, Reid G P, Trentham D R, Khan S. Biophys J. 1999;76:1706–1719. doi: 10.1016/S0006-3495(99)77329-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cluzel P, Surette M, Leibler S. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 9.Sourjik V, Berg H C. Proc Natl Acad Sci USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyawaki A, Tsien R Y. Methods Enzymol. 2000;327:472–500. doi: 10.1016/s0076-6879(00)27297-2. [DOI] [PubMed] [Google Scholar]
- 11.Wouters F S, Verveer P J, Bastiaens P I H. Trends Cell Biol. 2001;11:203–211. doi: 10.1016/s0962-8924(01)01982-1. [DOI] [PubMed] [Google Scholar]
- 12.Guzman L-M, Belin D, Carson M J, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scharf B E, Fahrner K A, Berg H C. J Bacteriol. 1998;180:5123–5128. doi: 10.1128/jb.180.19.5123-5128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang H, Blair D F. J Bacteriol. 1995;177:3485–3495. doi: 10.1128/jb.177.12.3485-3495.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao R, Amsler C D, Matsumura P, Khan S. J Bacteriol. 1996;178:258–265. doi: 10.1128/jb.178.1.258-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bornhorst J A, Falke J J. J Gen Physiol. 2001;118:693–710. doi: 10.1085/jgp.118.6.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alon U, Camarena L, Surette M G, Aguera y Arcas B, Liu Y, Leibler S, Stock J B. EMBO J. 1998;17:4238–4248. doi: 10.1093/emboj/17.15.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Förster T. Ann Phys. 1948;2:55–75. [Google Scholar]
- 19.Tsien R Y. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 20.Zacharias D A, Violin J D, Newton A C, Tsien R Y. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Swanson R V, Simon M I, Weis R M. Biochemistry. 1995;34:14626–14636. doi: 10.1021/bi00045a003. [DOI] [PubMed] [Google Scholar]
- 22.Gegner J A, Dahlquist F W. Proc Natl Acad Sci USA. 1991;88:750–754. doi: 10.1073/pnas.88.3.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim C, Jackson M, Lux R, Khan S. J Mol Biol. 2001;307:119–135. doi: 10.1006/jmbi.2000.4389. [DOI] [PubMed] [Google Scholar]
- 24.Monod J, Wyman J, Changeux J-P. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 25.Duke T A, Le Novére N, Bray D. J Mol Biol. 2001;308:541–553. doi: 10.1006/jmbi.2001.4610. [DOI] [PubMed] [Google Scholar]
- 26.Stewart R C. Biochemistry. 1997;36:2030–2040. doi: 10.1021/bi962261k. [DOI] [PubMed] [Google Scholar]
- 27.Levit M N, Liu Y, Stock J B. Biochemistry. 1999;38:6651–6658. doi: 10.1021/bi982839l. [DOI] [PubMed] [Google Scholar]
- 28.Blat Y, Gillespie B, Bren A, Dahlquist F W, Eisenbach M. J Mol Biol. 1998;284:1191–1199. doi: 10.1006/jmbi.1998.2224. [DOI] [PubMed] [Google Scholar]
- 29.Segall J E, Ishihara A, Berg H C. J Bacteriol. 1985;161:51–59. doi: 10.1128/jb.161.1.51-59.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berg H C, Brown D A. Nature (London) 1972;239:500–504. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- 31.Turner L, Ryu W S, Berg H C. J Bacteriol. 2000;182:2793–2801. doi: 10.1128/jb.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.