Abstract
Oxysterols regulate cholesterol homeostasis through liver X receptor (LXR; cholesterol-lowering)- and sterol regulatory element-binding protein (SREBP; cholesterol-raising)-mediated signaling pathways. Previously we reported that the hypocholesterolemic agent LY295427 (4α-allylcholestan-3α-ol) reverses oxysterol-mediated suppression of SREBP processing. We now report that LY295427 increases expression of insulin-induced gene-1 (INSIG-1) and restores SREBP processing in cells treated with oxysterols. In cells overexpressing the INSIG-1 gene, by contrast, SREBP processing is suppressed and oxysterol regulation is disrupted. SREBP processing is not restored by addition of LY295427, but is restored by increasing the levels of SREBP cleavage-activating protein (SCAP). These findings suggest that the INSIG-1 protein alters sterol balance by modulating SREBP processing jointly with SCAP. To test whether the action of oxysterols on SREBP processing is mediated through endogenous INSIG-1 protein, we used RNAi to lower the expression of the INSIG-1 gene, and found that reduced INSIG-1 protein levels caused the loss of SREBP regulation by oxysterols. We conclude that: (i) INSIG-1 gene expression is suppressed by oxysterols; (ii) LY295427 treatment counters the suppressive effects of oxysterols on SREBP processing, resulting in the expression of the INSIG-1 gene; and (iii) INSIG-1 gene expression affects SREBP processing. Taken together, these data suggest that INSIG-1 plays a critical role in regulating cholesterol concentrations in the cell.
LY295427 (4α-allylcholestan-3α-ol) was identified for its ability to activate the low-density lipoprotein receptor (LDLR) promoter in the presence of oxysterols (1). Normally, sterol treatment reduces cellular cholesterol levels by decreasing LDLR gene expression. Administration of LY295427 to hypercholesterolemic hamsters reduces plasma LDL levels by 70% (2). Results from experiments that measured the binding of 25-hydroxycholesterol (25HC) in lysates generated from livers of rats fed LY295427 suggest that LY295427 treatment enhances levels of uncharacterized oxysterol binding proteins (3).
We previously reported that LY295427 treatment reverses suppression of sterol regulatory element-binding protein (SREBP) processing mediated by oxysterols, but not by LDL or fatty acids (4). The hypocholesterolemic (lowering plasma cholesterol) effects resulting from LY295427 treatment are due to activation of SREBP-regulated genes, such as LDLR, and a concomitant increase in LDL metabolism (4). These data, together with work from other laboratories (1, 2), suggest that LY295427 represents a previously uncharacterized class of cholesterol-lowering drugs that exerts effects through SREBP to enhance clearance of plasma LDL by LDLR when cholesterol concentrations are elevated.
The pioneering work of Brown and Goldstein (5) has shown that the proteolytic processing required to generate an active nuclear form of SREBP from an inactive, membrane-bound precursor is exquisitely sensitive to sterol concentrations in the cell. Sterol loss results in the translocation of SREBP from the endoplasmic reticulum (ER) to the Golgi, where it is cleaved by site 1 and site 2 proteases (S1P, S2P; refs. 6 and 7). The cleaved fragment induces transcription of LDLR and cholesterol biosynthetic enzymes, resulting in enhanced cellular cholesterol levels. When sterol concentrations are elevated, SREBP is complexed with an SREBP cleavage activating protein (SCAP) and maintained as an inactive precursor in the ER membrane (8, 9). Determining how LY295427 reverses the effects of oxysterols in cells may lead to a better understanding of the molecular mechanisms involved in sterol-mediated suppression of SREBP processing.
We report here that LY295427 selectively diminishes the efficacy of oxysterols in mediating the SREBP and liver X receptor (LXR) signaling pathways that regulate cellular cholesterol levels. We also find that insulin-induced gene-1 (INSIG-1), initially described as an insulin-regulated gene that is highly expressed in developing or regenerating liver (10–12), is not expressed in cells treated with 25HC. Addition of LY295427 to cells treated with oxysterols results in expression of the INSIG-1 gene and restoration of SREBP processing. Our data suggest that INSIG-1 plays an important role in maintaining cholesterol homeostasis.
Materials and Methods
Materials.
LY295427 was a gift from Julio Medina (Tularik, South San Francisco, CA) and Robert Gadski (Eli Lilly); 24(S), 25-Epoxycholesterol was a gift from E. J. Corey (Harvard University, Cambridge, MA); and 9-cis-retinoic acid and arachadonic acid were purchased from Sigma. Arachadonic acid was bound to BSA (13). Oxysterols were purchased from Steraloids, Wilton, NH. Gal-LXRα and luciferase reporter plasmids were provided by David Mangelsdorf (University of Texas Southwestern Medical Center). Sodium mevalonate, sodium compactin, lipoprotein-deficient serum (LPPS), and delipidated FCS were prepared as described (14, 15).
cDNA Microarray, Northern Analyses, and Cloning.
SV589 cells were grown in DMEM containing 10% (vol/vol) FCS and 100 units/ml penicillin, 100 μg/ml streptomycin sulfate at 37°C, and 5% CO2. Cells were seeded on day 0 at a density of 2 × 106 cells per 15-cm plate on day 1, the cells were switched to medium A (DMEM/10% LPPS/10 μg/ml cholesterol/50 μM sodium mevalonate/50 μM sodium compactin) supplemented with 1 μg/ml 25HC ± 20 μM LY295427. After 24 h, cells were harvested and total RNA was isolated (Trizol Reagent, Life Technologies, Grand Island, NY). Poly(A)+ mRNA (Oligotex spin columns, Qiagen, Valencia, CA) was subjected to hybridization analyses using a microarray representing ≅10,000 human cDNAs (Incyte Genomics, St. Louis). A second analysis of the same poly(A)+ mRNA was performed on a custom human cDNA chip arrayed at Tularik. Northern analysis was performed as described (4). cDNA was synthesized using poly(A)+ mRNA from SV589 cells treated with LY295427 and 25HC (GIBCO/BRL). Human INSIG-1 was obtained using a GC melt kit (CLONTECH) before PCR, then introduced into an expression plasmid driven by a CMV promoter. Probes encoded human INSIG-1 (GenBank accession no. U96876), rat cyclophilin (GenBank accession no. M19533), and LDLR (GenBank accession no. NM000527).
Transient Transfections.
HEK293 cells were grown in DMEM supplemented with 10% (vol/vol) FCS and 100 units/ml penicillin, 100 μg/ml streptomycin sulfate at 37°C, and 8% CO2. A day before transfection, cells were seeded at 25,000 per well per 96-well plate in DMEM/10% (vol/vol) FCS. To measure LXR activation, expression plasmids encoding Gal4-LXRα (CMX-Gal4-hLXRα) was cotransfected with a Gal4-DNA response element linked to a luciferase reporter (TK-MH100 × 3-LUC; ref. 16). To measure SREBP processing, cells were transfected with plasmids encoding chimeric alkaline-phosphatase–SREBP (CMV-PLAP–SREBP-2) and SCAP (CMV–SCAP), and assayed as described (7). Data were normalized to a β-galactosidase expression. Data are presented as mean relative light units from triplicate assays ± SEM.
Preparation of Nuclear Extracts and Immunoblotting.
HepG2 cells were grown in DMEM supplemented with 10% (vol/vol) FCS and 100 units/ml penicillin, 100 μg/ml streptomycin at 37°C, and 5% CO2. On day 1, the medium was switched to medium A and supplemented with 1 μg/ml 25HC ± 20 μM LY295427. After 24 h, cells were harvested and separated into membrane and nuclear fractions (4). Protein concentrations were determined using a micro BCA kit (Pierce). Proteins were immunoblotted with a mouse monoclonal antibody against the NH2 terminus of human SREBP-2 (IgG-1D2; ref. 4).
RNA Interference.
Twenty-one-nucleotide RNA oligonucleotides directed against human INSIG-1 or luciferase (17) were obtained from Oligos Etc., Guilford, CT. The siRNA sequences targeting human INSIG-1 (GenBank accession no. U96876) correspond to the coding regions (nucleotides 417–437, 702–722, or 853–873) relative to the first nucleotide of the start codon. HEK293 cells were transfected with siRNA on day 1 and day 2 by using Oligofectamine (Invitrogen). On day 3, the medium was switched to medium A supplemented with 1 μg/ml 25HC ± 20 μM LY295427. After 8 h, cells were harvested and either RNA or protein was harvested (4).
Results
LY295427 Treatment Reverses Oxysterol-Mediated Effects in Cells.
In previous work we demonstrated that LY295427 treatment reverses suppression of SREBP processing mediated by oxysterols, but not by LDL-delivered cholesterol (4). To determine whether the reversal of oxysterol-mediated regulation by LY295427 is specific to SREBP, we tested the effects of LY295427 treatment on activation of LXR (Fig. 1). We examined LXR because this receptor is activated by oxysterols (16, 18, 19), and regulates genes involved in maintaining cholesterol levels in the cell, including SREBP-1c (20).
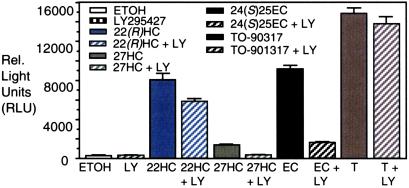
Figure 1.
LY295427 selectively reverses only oxysterol-mediated signaling pathways. HEK293 cells were transfected with plasmids encoding Gal4-LXRα and a luciferase reporter (16). After transfection, cells were treated with ethanol, 20 μM LY295427, or 10 μM of the following oxysterols: 22(R)-hydroxycholesterol [22(R)HC], 27-hydroxycholesterol (27HC), 24(S),25-epoxycholesterol [24(S),25EC], or 1 μM T0–901317 in the absence or presence of 20 μM LY295427. After 20 h cells were harvested and assayed (16).
Cells were transfected with a plasmid encoding a fusion protein of Gal4 and the LXR ligand-binding domain along with a response element linked to a luciferase reporter. Cells were treated with oxysterols, or a nonsterol activator of LXR in the presence or absence of LY295427. Activation of LXR was assessed by measuring luciferase activity.
Addition of vehicle or LY295427 to cells did not activate the receptor. Addition of LY295427 in combination with oxysterols resulted in decreased activation of LXR relative to addition of oxysterols alone. Activation of LXR by 22(R)HC was reduced less than 20% with LY295427 treatment, whereas activation by 27HC or 24(S),25EC was reduced 4- to 5-fold. In contrast, LY295427 treatment did not affect LXR activation by T0–901317, a non-sterol activator (21). Reduction of LXR activation by oxysterols, but not by T0–901317, in the presence of LY295427 is significant because it suggests that the mechanism through which LY295427 acts to selectively diminish the efficacy of oxysterols is not mediated through LXR.
INSIG-1 Gene Expression Is Regulated by Oxysterols and LY295427.
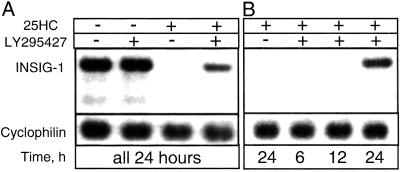
Reversal of oxysterol-mediated effects in cells following LY295427 treatment requires a minimum of 6 h (4), suggesting that the effect may require gene transcription. We used microarray analysis to determine whether treating cells with LY295427 and 25HC would result in up-regulation of genes involved in modulating oxysterol action (Table 1). In addition to the predicted up-regulation of known SREBP-regulated and sterol-responsive genes, INSIG-1 was the most highly up-regulated gene in two independent screens using different microarrays containing approximately 10,000 cDNAs. Sequence analysis of the INSIG-1 protein suggests that it contains transmembrane domains (10). Northern analysis showed that the INSIG-1 gene was highly expressed in cells grown in serum containing low sterol concentrations and treated with vehicle or LY295427 alone. INSIG-1 gene expression was completely repressed by addition of 25HC, and this repression was reversed with LY295427 treatment between 12 and 24 h (Fig. 2 A and B).
Table 1.
Gene chip analysis: The effect of adding LY295427 to cells treated with oxysterols
| Fold Δ Chip 1/2 | Gene name | Function |
|---|---|---|
| 6/8 | Insulin-induced gene-1 (INSIG-1) | Insulin regulation? |
| 7/ND | Methylsterol oxidase (ERG25) | Cholesterol synthesis |
| 7/6 | Stearoyl-CoA desaturase (SCD-1) | Fatty acid synthesis |
| 7/ND | Isopentenyl diphosphate isomerase | Isoprenoid synthesis |
| 5/3 | Famesyl-diphosphate famesyltransferase | Isoprenoid synthesis |
| 5/1 | Mevalonate kinase | Isoprenoid synthesis |
| 4/3 | HMGCoA reductase | Cholesterol synthesis |
| 4/3 | Squalene epoxidase | Cholesterol synthesis |
| ND/3 | LDLR | Cholesterol homeostasis |
| ND/3 | 7-Dehydrocholesterol Δ 7-reductase | Cholesterol synthesis |
| 3/2 | Mevalonate diphosphate decarboxylase | Isoprenoid synthesis |
| 2/2 | Famesyl diphosphate synthase | Isoprenoid synthesis |
| 2/1 | NAD(P)-dependent steroid dehydrogenase-like | Unknown |
| 2/1 | ATP citrate lyase | Fatty acid/CH synthesis |
| ND/2 | SREBP2 | Cholesterol homeostasis |
| ND/2 | Isopentyl-diphosphate Δ isomerase | Isoprenoid synthesis |
| ND/2 | Sterol-C4-methyl oxidase-like | Unknown |
Figure 2.
Northern analysis showing that LY295427 induces INSIG-1 gene expression in the presence of 25-hydroxycholesterol (25HC) within 12–24 h. (A) SV589 cells were treated with ethanol (lane 1), 20 μM LY295427 (lane 2), 1 μg/ml 25HC (lane 3), or 1 μg/ml 25HC and 20 μM LY295427 (lane 4), and harvested after 24 h. (B) Cells were treated with 1 μg/ml 25HC (lane 1), or 1 μg/ml 25HC and 20 μM LY295427 (lanes 2–4), and harvested 6 h (lane 2), 12 h (lane 3), or 24 h (lanes 1 and 4) later.
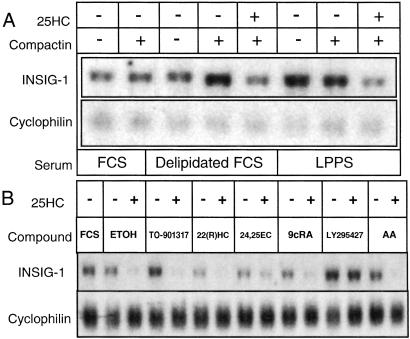
To characterize sterol-mediated regulation of INSIG-1 gene expression, we treated cells with compactin, an inhibitor of de novo cholesterol biosynthesis, in the absence or presence of 25HC (Fig. 3A). We also grew cells in sera with varied lipid content to establish conditions under which INSIG-1 gene expression is the most stringently controlled. Northern analysis revealed that the INSIG-1 gene was moderately expressed in cells grown in FCS, regardless of compactin treatment. FCS was used because it has high lipid content. By contrast, compactin treatment of cells grown in delipidated FCS resulted in up-regulation of the INSIG-1 gene, which was reversed with 25HC. Cells grown in LPPS, in which sterol and triglyceride concentrations are also low (15), resulted in INSIG-1 gene up-regulation that was even more pronounced with compactin treatment. Addition of 25HC to these cells reduced INSIG-1 gene expression, demonstrating that exogenous sterols negatively regulate the INSIG-1 gene. The finding that INSIG-1 mRNA levels are increased in the presence of compactin when exogenous lipid levels are low indicates that INSIG-1 gene expression is sensitive to endogenous sterol concentrations.
Figure 3.
Regulation of INSIG-1 gene expression. (A) INSIG-1 gene expression is sensitive to cellular sterol concentrations. SV589 cells were grown in medium supplemented with 10% of either FCS (lanes 1 and 2), delipidated FCS (lanes 3–5), or LPPS (lanes 6–8). Cells received 25HC, or compactin as indicated. (B) LY295427 action is not linked to signaling pathways mediated by LXR, RXR, or fatty acids. SV589 cells were grown in medium supplemented with 10% FCS alone (lane 1) or with ethanol (ETOH, lanes 2 and 3), 1 μM TO-901317 (lanes 4 and 5), 10 μM 22(R)HC (lanes 6 and 7), 10 μM 24,25EC (lanes 7 and 8), 1 μM 9-cis-retinoic acid (9cRA, lanes 10 and 11), 20 μM LY295427 (lanes 12 and 13), or 100 μM arachadonic acid (AA, lanes 14 and 15) in the absence or presence of 1 μg/ml 25HC as indicated.
To determine whether INSIG-1 gene expression and LY295427 action are linked by a signaling pathway other than cholesterol, we assayed for changes in INSIG-1 gene expression in cells treated with activators of the LXR, retinoid X receptor (RXR), or fatty acid-signaling pathways (Fig. 3B). Cells were treated with either of these compounds alone, or in the presence of 25HC. We reasoned that reversal of the 25HC-mediated suppression of the INSIG-1 gene by any of these compounds would aid in determining the mechanism of LY295427 and the biology involved. Cells were grown in LPPS because it showed the most pronounced regulation of INSIG-1 by sterols (Fig. 3A).
We observed that T0–901317, 9-cis-retinoic acid (9cRA) or arachadonic acid (AA) treatment did not significantly alter INSIG-1 mRNA levels in the absence of sterols, or reverse the loss of INSIG-1 gene expression like LY295427 when 25HC was added (Fig. 3B). These data suggest that INSIG-1 gene expression is not directly regulated by the nuclear receptors LXR or RXR, or by fatty acids, and that LY295427 action is also not mediated through these signaling pathways. By contrast, the suppression by oxysterols that activate LXR, 22(R)HC, and 24(S),25EC was further abolished by combined treatment with 25HC. The finding that LY295427 was the only compound tested that could reverse the suppression of INSIG-1 gene expression by 25HC supports a selective mechanism of LY295427 action independent of other pathways that regulate lipid metabolism.
SREBP Processing Is Repressed by Overexpressing INSIG-1 Protein and Is Not Restored by LY295427.
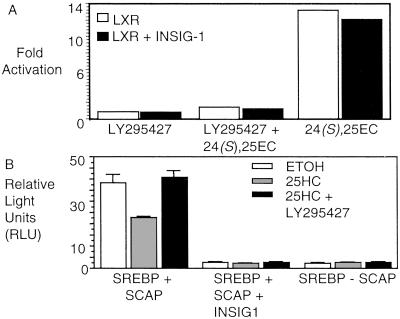
The initial experiments reported above indicate that addition of LY295427 restores INSIG-1 gene expression in the presence of 25HC, and also reverses oxysterol-mediated activation of LXR. To determine whether these findings are linked—i.e., whether LY295427 reverses the effects of oxysterols by restoring INSIG-1 gene expression—we tested the effect of overexpressing the INSIG-1 gene on oxysterol-mediated activation of LXR or suppression of SREBP processing (Fig. 4 A and B). The ability to overexpress INSIG-1 protein in cells in the absence of LY295427 allowed us to separate their effects.
Figure 4.
Overexpression of INSIG-1 protein represses SREBP processing, but not LXR activation. (A) INSIG-1 protein does not inhibit oxysterol-mediated activation of LXRα. HEK293 cells were transfected with plasmids encoding Gal4-LXRα and a luciferase reporter with or without an expression plasmid for INSIG-1. Cells were treated with ethanol, 20 μM LY295427, 20 μM LY295427 and 10 μM 24(S),25EC, or 10 μM 24(S),25EC 6–8 h later. After 20 h, cells were harvested and assayed. Activation is expressed as fold activation determined by the division of relative light units in the sterol treated samples by those treated with ethanol. (B) INSIG-1 protein represses SREBP processing. HEK293 cells were transfected with plasmids encoding a chimeric alkaline-phosphatase SREBP-2 (PLAP–SREBP-2) and SCAP with or without a plasmid expressing INSIG-1. Cells were treated with ethanol or 1 μg/ml 25HC, or 1 μg/ml 25HC and 20 μM LY295427.
We cloned the INSIG-1 gene and expressed it behind a constitutive CMV promoter. Cells were transfected with plasmids necessary for evaluating LXR activation with or without the expression plasmid for INSIG-1. Cells were either treated with 24(S),25EC in the absence or presence of LY295427. We found that INSIG-1 protein overexpression did not alter either the activation of LXR by 24(S),25EC, or the suppression of oxysterol-induced activation by LY295427 (Fig. 4A), suggesting that the mechanism of LY295427 action is distinct from the mechanism of the INSIG-1 protein.
We also examined the effects of overexpressing INSIG-1 protein on SREBP processing by using a transfection assay designed to measure SREBP processing in cells that has been described (7). Briefly, SREBP processing is assessed by transfection of plasmids encoding SCAP and a chimeric SREBP protein containing placental alkaline phosphatase (PLAP) fused to the amino terminus of SREBP-2. Proteolytic processing of the PLAP–SREBP chimera results in the cleavage of the amino-terminal portion of the protein containing the enzyme that is excreted into the medium. Alkaline phosphatase activity is assayed by luminescence and is indicative of SREBP processing in the cell.
We observed that when endogenous INSIG-1 is expressed in normal, oxysterol-responsive cells, SREBP is processed (Figs. 2 and 6A). As in normal cells, addition of 25HC to cells overexpressing PLAP–SREBP and SCAP reduces SREBP processing, and LY295427 treatment restores processing (Fig. 4B). Similar results were obtained in SV589 cells (data not shown). By contrast, in cells overexpressing INSIG-1 in combination with PLAP–SREBP and SCAP, SREBP processing is completely abolished, regardless of whether 25HC or LY295427 were added (Fig. 4B).
Figure 6.
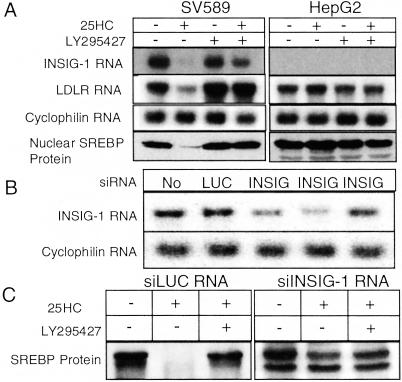
INSIG-1 protein mediates SREBP regulation by sterols. (A) Northern and Western analysis of SV589 and HepG2 cells treated with ethanol (lane 1), 1 μg/ml 25HC (lane 2), 20 μM LY295427 (lane 3), or 1 μg/ml 25HC and 20 μM LY295427 (lane 4). (B) Northern analysis of HEK293 cells not transfected (lane 1), transfected with siRNAs targeting luciferase (lane 2), or INSIG-1 [lane 3, nucleotides (nts) 417–427; lane 4, nts 702–722; lane 5, nts 853–873] on days 1 and 2 by using Oligofectamine. On day 3, the medium was switched to medium A (see Materials and Methods). Total RNA was isolated and 10-μg samples were analyzed by Northern analysis. Probes encoded INSIG-1 or cyclophilin. (C) Western analysis of HEK293 cells transfected with the most potent siRNA against INSIG-1 used in B (lane 4, nts 702–722). On day 3, cells were fed medium A supplemented with ethanol (lanes 1 and 4), 1 μg/ml 25HC (lanes 2 and 5), or 1 μg/ml 25HC + 20 μM LY295427 (lanes 3 and 6).
These conflicting effects observed with endogenous and overexpressed INSIG-1 on SREBP processing may be due to high INSIG-1 protein levels interfering with SCAP function. This explanation is supported by the observation that repression of SREBP processing occurs in the absence of oxysterols when either INSIG-1 is overexpressed, or when SCAP is not overexpressed (Fig. 4B). The lack of a functional SREBP–SCAP complex resulting from INSIG-1 overexpression would also explain why addition of LY295427 did not rescue SREBP processing in the presence of oxysterols. Alternatively, the potent suppression of SREBP processing following INSIG-1 overexpression may indicate that INSIG-1 plays an important role in regulating the basal activity of the SCAP–SREBP complex.
Repression of SREBP Processing Through the INSIG-1 Protein Is Reversed by Increasing SCAP Levels.
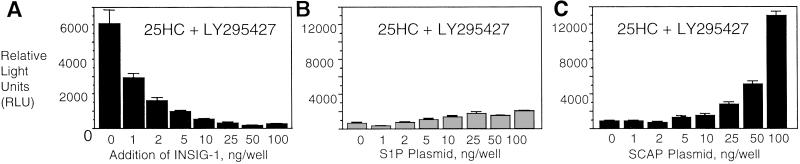
Site 1 protease (S1P) and SCAP are two proteins that are responsible for regulating SREBP processing. We reasoned that overexpression of these proteins might reverse the suppressive effects of INSIG-1 protein on SREBP processing. We first performed a dose–response assay to identify the minimum amount of INSIG-1 plasmid needed to adequately suppress SREBP processing (Fig. 5A). This concentration was used in subsequent assays when we tested the effect of overexpressing S1P or SCAP on SREBP processing in the presence of INSIG-1 protein. We found that SREBP processing was restored by overexpression of SCAP (Fig. 5C), but not S1P (Fig. 5B), in cells overexpressing INSIG-1, suggesting that the INSIG-1 protein and SCAP jointly regulate SREBP processing, either by directly interacting with SREBP or through a common factor. Results from these overexpression studies may indicate that the ratio of SCAP to INSIG-1 levels in the cell are critical for maintaining sterol responsiveness and subsequent cholesterol balance. Because SCAP overexpression results in the loss of sterol regulation in some cell types (unpublished observations), it should be noted that at the SCAP concentrations used, SREBP processing is still sterol-regulated (data not shown). The inability of LY295427 to reverse the repressive effects of INSIG-1 protein overexpression on SREBP processing in the presence of 25HC suggests that the INSIG-1 protein and LY295427 exert their opposing effects (i.e., INSIG-1 protein decreases SREBP processing, whereas LY295427 increases it in the presence of oxysterols) through competing pathways.
Figure 5.
SREBP processing can be reversed by overexpression of SCAP protein, but not by treatment with LY295427. (A) HEK293 cells were transfected (7) with plasmids encoding PLAP–SREBP-2 and SCAP, and increasing concentrations of an INSIG-1 expression plasmid. Cells were treated with 1 μg/ml 25HC and 20 μM LY295427 6–8 h later. (B) Cells were transfected with plasmids encoding PLAP–SREBP-2 and SCAP, and 5 ng per well INSIG-1 along with increasing concentrations of a plasmid encoding either S1P or SCAP. Cells were treated with 1 μg/ml 25HC and 20 μM LY295427.
Oxysterol Regulation of SREBP Is Mediated by Endogenous INSIG-1 Protein.
In SV589 (immortalized human fibroblasts) cells SREBP processing is tightly regulated by oxysterol concentrations while in HepG2 cells, regulation is less sensitive and less consistent (unpublished observations). To determine whether the difference in oxysterol regulation of SREBP processing between these two cell types can be attributed to the level of INSIG-1 protein, we treated cells with oxysterols in the absence or presence of LY295427, and then assayed for INSIG-1 or LDLR gene expression and SREBP processing (Fig. 6A).
We observed that SV589 cells treated with 25HC showed decreased expression of INSIG-1 and LDLR mRNA by Northern analysis, and decreased SREBP processing by Western analysis (Fig. 6A). Treatment with LY295427 restored gene expression and SREBP protein processing in SV589 cells. In contrast, when HepG2 cells were not oxysterol-responsive, INSIG-1 gene expression was undetectable by Northern analysis, and addition of LY295427 had no effect on restoring INSIG gene expression. LDLR gene expression and SREBP processing were not regulated by oxysterols in these cells. The lack of INSIG-1 gene expression in these HepG2 cells may explain their inability to properly regulate SREBP processing, and is consistent with the suggestion that normal levels of INSIG-1 protein are required for maintaining oxysterol-induced regulation of SREBP.
RNAi-Mediated Reduction of INSIG-1 Gene Expression Abolishes Oxysterol Regulation of SREBP Processing.
To further support the role of the INSIG-1 protein as a oxysterol-induced regulator of SREBP, we silenced the expression of the INSIG-1 gene in HEK293 cells by using RNA interference (RNAi) (17). Northern analysis shows that cells transfected with a siRNA duplex targeting firefly luciferase did not affect the expression of INSIG-1 mRNA. Cells transfected with three different siRNA oligos designed to silence the INSIG-1 gene varied in their ability to suppress INSIG-1 gene expression (Fig. 6B), with two siRNAs showing substantial reductions in the level of INSIG-1 mRNA.
Cells were treated with the most potent siRNA, and then the effect of adding 25HC and LY295427 on SREBP processing was measured. Previously, we showed that addition of 25HC reduced SREBP processing, whereas addition of 25HC in combination with LY295427 reverses repression and restores processing (4). Addition of siRNA targeted to luciferase did not alter the effect of 25HC or LY295427 treatment on SREBP processing in cells (Fig. 6C). In striking contrast, addition of siRNA targeted to the INSIG-1 gene prevented loss of SREBP processing on addition of 25HC. This result supports the conclusion that oxysterol-mediated regulation of SREBP processing depends on expression of the INSIG-1 protein.
Discussion
Effects of LY295427 and the INSIG-1 Protein on SREBP Processing.
We previously demonstrated that addition of LY295427 to cells treated with 25HC restores SREBP processing, resulting in the expression of genes that enhance cholesterol levels when sterol concentrations are already elevated (Table 1; ref. 4). We now show that addition of LY295427 also causes an increase in the expression of the INSIG-1 gene in cells treated with oxysterols (Table 1, Fig. 2). When INSIG-1 protein is overexpressed, processing of SREBP is suppressed and insensitive to oxysterol concentrations in the cell. Under these conditions, LY295427 treatment does not restore SREBP processing (Fig. 4B). When INSIG-1 gene expression is inhibited through RNAi, SREBP processing also becomes unresponsive to oxysterols (Fig. 6C). Thus, it appears that the INSIG-1 protein is a critical component regulating SREBP processing and sterol concentrations in the cell, and that regulation of cellular INSIG-1 protein levels is essential for proper oxysterol-mediated regulation. Our data suggest that when oxysterol concentrations in the cell are low, the INSIG-1 gene is expressed and SREBP is processed (Figs. 2 and 6A). As oxysterol levels increase, SREBP processing is suppressed and INSIG-1 levels decrease, suggesting a feedback loop for SREBP processing in which INSIG-1 plays a critical role.
LXR is an oxysterol receptor that also plays a role in maintaining cholesterol homeostasis, but our experiments suggest that it does not mediate the effects of LY295427, or regulate INSIG-1 gene expression or protein function.
A Model for LY295427 and INSIG-1 Protein Action.
SREBP plays a critical role in regulating cholesterol homeostasis. Although it is well known that oxysterols inhibit the processing of SREBP, the molecular mechanisms through which this occurs have not been elucidated because the binding of sterols to SCAP or SREBP has not been observed. The failure to demonstrate a direct interaction between sterols and these proteins suggests that cholesterol-dependent membrane fluidity may play a role, or that sterol sensing may require other proteins. Our data are best interpreted in terms of sterol sensing involving other proteins.
It is known that SREBP directly interacts with SCAP. SCAP has a domain that is believed to be involved in sterol sensing because overexpression of this domain (22), or introduction of point mutations (23, 24) in this region abolish the ability of sterols to suppress SREBP processing. One explanation for this result is that SCAP normally binds to another protein, and that overexpression of the SCAP sterol sensing domain prevents this protein from taking part in the regulation of SREBP processing. It has been reported that a class of synthetic steroidal and nonsteroidal hypolipidemic compounds interact with the sterol sensing domain of SCAP (25); however, these observations do not exclude the possibility that sterols bind to an additional sterol sensing protein that interacts with SCAP. INSIG-1 appears to be a membrane-bound protein (10), and represses SREBP processing in the presence of sterols, making it a candidate interacting protein for SCAP or SREBP.
One model for how the INSIG-1 protein may function is that on expression, the INSIG-1 protein binds to the SREBP–SCAP complex in the ER membrane and allows SREBP to be processed. Our finding that repression of SREBP processing in cells that overexpress INSIG-1 protein could be reversed by increasing SCAP levels is consistent with INSIG-1 interacting with SCAP (Fig. 5C). INSIG-1 gene expression and function are sensitive to sterol content (Figs. 2, 3, and 6C). The INSIG-1 gene is expressed when oxysterol levels are low and SREBP is processed (Figs. 2 and 6A). Conversely, as oxysterol levels increase, INSIG-1 gene expression is silenced and SREBP processing is suppressed. Our data support that the INSIG-1 protein mediates the suppressive effects of oxysterols on SREBP processing because reduction of INSIG-1 protein levels by RNAi results in the loss of SREBP regulation by oxysterols (Fig. 6C). INSIG-1 protein may either bind sterols directly or bind to an additional protein that directly senses sterol content.
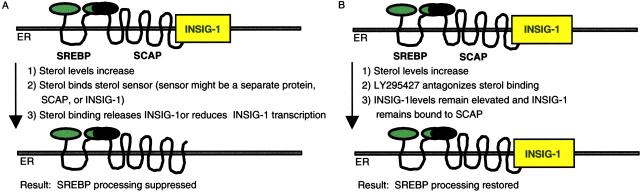
Details of our model are outlined in Fig. 7. We propose that the INSIG-1 protein mediates suppression of SREBP by oxysterols through interaction with an unidentified sterol sensor that could be SCAP, INSIG-1, or another protein. When sterol concentrations in the cell are low, the INSIG-1 protein is expressed and interacts with SREBP–SCAP, allowing SREBP to move from the ER to the Golgi for processing by S1P and S2P. The processed form of SREBP enters the nucleus, resulting in the up-regulation of genes involved in cholesterol synthesis and uptake. As oxysterol levels increase, they bind to the sterol sensor and cause INSIG-1 protein to be released or INSIG-1 gene expression to be reduced. Reduction in INSIG-1 protein function may cause SREBP–SCAP to be retained in the ER membrane, resulting in the suppression SREBP processing, and the down-regulation of the INSIG-1 gene and other genes that are regulated by SREBP to lower sterol concentrations in the cell. LY295427 treatment would reverse these effects by antagonizing sterol binding, and maintaining INSIG-1 gene expression (Fig. 7B).
Figure 7.
Proposed models for INSIG-1 protein and LY295427 action. Effect of oxysterols and INSIG-1 on SREBP processing in the absence (A) and presence (B) of LY295427.
Summary.
INSIG-1 gene expression is regulated by naturally occurring oxysterols and the synthetic hypocholesterolemic agent, LY295427. The INSIG-1 protein modulates SREBP processing, and its function is sensitive to oxysterol concentrations. These results indicate that the INSIG-1 protein is a critical component of the sterol-sensing system that regulates cholesterol levels in the cell.
Acknowledgments
I thank Daphne Head and Christine Alvarez for skilled assistance, Dr. David Russell for initiating this project and for his guidance and insights throughout the work, Dr. Joe Goldstein and Dr. Mike Brown for generously providing reagents and for stimulating scientific discussions, and Robert Gadski and Julio Medina for LY295427. I thank the reviewers for their thoughtful comments. This work was supported by National Institutes of Health Grant HL-20948.
Abbreviations
- INSIG-1
insulin-induced gene-1
- SREBP
sterol regulatory element-binding protein
- SCAP
SREBP cleavage activating protein
- LDLR
low-density lipoprotein receptor
- LXR
liver X receptor
- 25HC
25-hydroxycholesterol
- ER
endoplasmic reticulum
- LPPS
lipoprotein-deficient serum
- PLAP
placental alkaline phosphatase
Note Added in Proof.
Yang et al. (26) have reported that overexpressed INSIG-1 directly interacts with the sterol-sensing domain of SCAP, and that binding is enhanced in the presence of sterols. Based on this result, the loss of INSIG-1 as proposed in our model is consistent with decreased transcription of the INSIG-1 gene in the presence of sterols.
References
- 1.Lin H, Rampersaud A A, Archer R A, Pawlak J M, Beavers L S, Schmidt R J, Kauffman R F, Bensch W R, Bumol T F, Apelgren L D, et al. J Med Chem. 1995;38:277–288. doi: 10.1021/jm00002a010. [DOI] [PubMed] [Google Scholar]
- 2.Bensch W R, Gadski R A, Bean J S, Beavers L S, Schmidt R J, Perry D N, Murphy A T, McClure D B, Eacho P I, Breau A P, et al. J Pharmacol Exp Ther. 1999;289:85–289. [PubMed] [Google Scholar]
- 3.Bowling N, Metter W F, Gadski R A, McClure D B, Schreyer T, Dawson P A, Vlahos C J. J Lipid Res. 1996;37:2586–2598. [PubMed] [Google Scholar]
- 4.Janowski B A, Shan B, Russell D W. J Biol Chem. 2001;276:45408–45416. doi: 10.1074/jbc.M108348200. [DOI] [PubMed] [Google Scholar]
- 5.Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rawson R B, Zelenski N G, Nijhawan D, Ye J, Sakai J, Hasan M Z, Chang T Y, Brown M S, Goldstein J L. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 7.Sakai S J, Rawson R B, Espenshade P J, Cheng D, Seegmiller A C, Goldstein J L, Brown M S. Mol Cell. 1998;2:505–514. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- 8.Nohturfft A, DeBose-Boyd R A, Scheek S, Goldstein J L, Brown M S. Proc Natl Acad Sci USA. 1999;96:11235–11240. doi: 10.1073/pnas.96.20.11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nohturfft A, Yabe D, Goldstein J L, Brown M S, Espenshade P J. Cell. 2000;102:315–323. doi: 10.1016/s0092-8674(00)00037-4. [DOI] [PubMed] [Google Scholar]
- 10.Diamond R H, Du K, Lee V M, Mohn K L, Haber B A, Tewari D S, Taub R. J Biol Chem. 1993;268:15185–15192. [PubMed] [Google Scholar]
- 11.Peng Y, Schwarz E J, Lazar M A, Genin A, Spinner N B, Taub R. Genomics. 1997;43:278–284. doi: 10.1006/geno.1997.4821. [DOI] [PubMed] [Google Scholar]
- 12.Bortoff K D, Zhu C C, Hrywna Y, Messina J L. Endocrine. 1997;2:199–207. doi: 10.1007/BF02778142. [DOI] [PubMed] [Google Scholar]
- 13.Hannah V C, Ou J, Luong A, Goldstein J L, Brown M S. J Biol Chem. 2001;276:4365–4372. doi: 10.1074/jbc.M007273200. [DOI] [PubMed] [Google Scholar]
- 14.Brown M S, Faust J R, Goldstein J L. J Biol Chem. 1978;253:1121–1128. [PubMed] [Google Scholar]
- 15.Goldstein J L, Basu S K, Brown M S. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 16.Janowski B A, Grogan M J, Jones S A, Wisely G B, Kliewer S A, Corey E J, Mangelsdorf D J. Proc Natl Acad Sci USA. 1999;96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elbashir S M, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature (London) 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 18.Janowski B A, Willy P J, Devi T R, Falck J R, Mangelsdorf D J. Nature (London) 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 19.Lehmann J M, Kliewer S A, Moore L B, Smith-Oliver T A, Oliver B B, Su J L, Sundeth S S, Winegar D A, Blanchard D E, Spencer T A, Willson T M. J Biol Chem. 1997;272:3137–3140. doi: 10.1074/jbc.272.6.3137. [DOI] [PubMed] [Google Scholar]
- 20.Repa J J, Liang G, Ou J, Bashmakov Y, Lobaccaro J M, Shimomura I, Shan B, Brown M S, Goldstein J L, Mangelsdorf D J. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Repa J J, Turley S D, Lobaccaro J A, Medina J, Li L, Lustig K, Shan B, Heyman R A, Dietschy J M, Mangelsdorf D J. Science. 2000;289:1524–1529. doi: 10.1126/science.289.5484.1524. [DOI] [PubMed] [Google Scholar]
- 22.Yang T, Goldstein J L, Brown M S. J Biol Chem. 2000;275:29881–29886. doi: 10.1074/jbc.M005439200. [DOI] [PubMed] [Google Scholar]
- 26.Yang T, Espenshade P J, Wright M E, Yabe D, Gong Y, Aebersold R, Goldstein J L, Brown M S. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 23.Nohturfft A, Brown M S, Goldstein J L. Proc Natl Acad Sci USA. 1998;95:12848–12853. doi: 10.1073/pnas.95.22.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hua X, Norhturfft, Goldstein J L, Brown M S. Cell. 1996;87:425–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- 25.Grand-Perret T, Bouillot A, Perrot A, Commans S, Walker M, Issandou M. Nat Med. 2001;7:1332–1338. doi: 10.1038/nm1201-1332. [DOI] [PubMed] [Google Scholar]
- 27.Yang T, Espenshade P J, Wright M E, Yabe D, Gong Y, Aebersold R, Goldstein J L, Brown M S. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]