Abstract
The inherent chemical properties of RNA molecules are expanded by posttranscriptional modification of specific nucleotides. Pseudouridine (ψ), the most abundant of the modified bases, features an additional imino group, NH1, as compared with uridine. When ψ forms a Watson–Crick base pair with adenine in an RNA helix, NH1 is positioned within the major groove. The presence of ψ often increases thermal stability of the helix or loop in which it is found [Hall, K. B. & McLaughlin, L. (1992) Nucleic Acids Res. 20, 1883–1889]. X-ray crystal structures of transfer RNAs [e.g., Arnez, J. & Steitz, T. (1994) Biochemistry 33, 7560–7567] have depicted water molecules bridging ψNH1 groups and nearby phosphate oxygen atoms, but direct evidence for this interaction in solution has not been acquired. Toward this end, we have used a rotating-frame Overhauser effect spectroscopy-type NMR pulse sequence with a CLEAN chemical-exchange spectroscopy spin-lock pulse train [Hwang, T.-L., Mori, S., Shaka, A. J. & van Zijl, P. C. M. (1997) J. Am. Chem. Soc. 119, 6203–6204] to test for ψNH1–water cross-relaxation effects within two RNA helices: (i) a complementary duplex, in which ψ is not associated with structural change, and (ii) an RNA duplex representing the eukaryotic pre-mRNA branch-site helix from Saccharomyces cerevisiae, in which a conserved ψ extrudes the branch-site adenosine from the helix. Our data implicate a water–ψNH1 hydrogen bond both in stabilizing the complementary helix and in favoring formation of the unique structure of the branch-site helix.
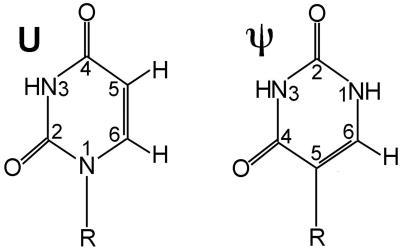
By 1950, it was known that RNA extracted from cells contained a small percentage of a fifth base, thought to be 5-methylcytosine (1). Later, this fifth nucleoside, which accounted for ≈4% of nucleotides in yeast transfer (t)RNA, was identified as 5-ribosyluracil, an isomer of uridine (U) (2–5). Pseudouridine (abbreviated ψ) since has been found to be the most abundant modified base in tRNA, ribosomal RNA, and small nuclear (sn)RNA. Modification of U to ψ, catalyzed by a site-specific pseudouridylase, involves scission of the nucleoside's glycosidic bond, followed by rotation of the base about its 3–6 axis, and reattachment through the carbon at the 5-position of the ring to form the only natural nucleoside with a C—C base–sugar bond. The resulting modified base, which can form a Watson–Crick base pair with adenine, features an additional ring nitrogen atom that is protonated at physiological pH (NH1; Fig. 1; reviewed in ref. 6).
Figure 1.
Schematic structures of U and ψ bases. R, ribose. The ψ base is a uracil rotated about the 3–6 ring axis, so that is has a C—C base–sugar linkage and an additional protonated ring nitrogen.
The presence of this modified base has been associated with an increase in thermal stability of secondary structure (7) and has been shown to decrease the motion of neighboring bases in molecular dynamics simulations (8), which may explain its prevalence in structural RNAs. For example, ψ are clustered within the peptidyl transferase center of the ribosome (9), are conserved within regions of snRNAs that are involved in RNA–RNA interactions (10), and have been implicated in spliceosome assembly (11).
Although the presence of ψ generally is not associated with major structural change (reviewed in ref. 6), we recently have found a major exception in the eukaryotic splicing apparatus: a highly conserved ψ in the region of U2 snRNA that pairs with the intron to form the precursor (pre)-mRNA branch-site helix induces a dramatic change in conformation and modest increase in thermal stability, as compared with its unmodified analogue (12, 36). The presence of ψ results in extrusion of an adenosine, the 2′OH of which is the nucleophile in the first step of splicing, from the helix into a position that may facilitate its role in splicing catalysis. Such studies emphasize the biological importance of modified bases and may describe a means by which RNA-mediated chemical activity is enabled through site-specific, posttranscriptional modification.
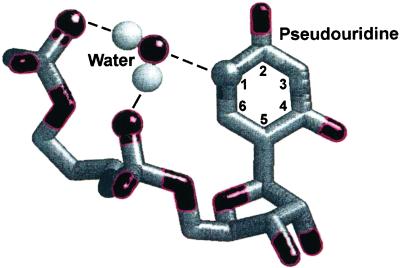
The mechanism by which ψ stabilizes (and, in the case of the pre-mRNA branch site, modifies) local RNA architecture remains unclear. Stabilization of RNA secondary structure by ψ has been hypothesized to result from additional hydrogen bonds involving the ψNH1 (13) or from more favorable stacking interactions between ψ and neighboring bases (14). Corroborating the first hypothesis is the observation of water-mediated hydrogen bonds between the NH1 proton and phosphate oxygen atoms in crystal structures of tRNA molecules (ref. 15; Fig. 2) and molecular dynamics simulations (8). Water-mediated hydrogen bonds involving ψNH1 have been postulated to stabilize ψ-containing helices in solution (13, 16), but no direct evidence for their existence has been obtained. Experiments supporting the stacking hypothesis showed that 5-methyl U, which cannot form hydrogen bonds in the major groove, at the base of a stem loop was as stable as the native ψ modification (14). In contrast with all these studies, incorporation of ψ into single-stranded regions and loops, in some cases, can be destabilizing (17).
Figure 2.
Schematic view of a water-mediated hydrogen-bonding interaction between ψ and water. Oxygen atoms are colored red. Here, the water molecule is bridging an interaction between the ψNH1 group and phosphate oxygen atoms belonging to its own and the 5′ nucleotide's phosphate group (6).
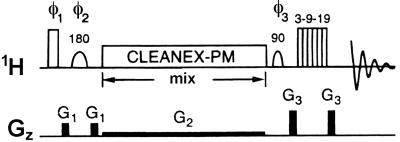
We have sought to understand the molecular basis for stabilization by ψ in a complementary RNA duplex and a duplex representing the eukaryotic pre-mRNA branch-site helix. We have accomplished this by use of an NMR experiment designed to identify RNA protons involved in chemical exchange with water molecules whose transient association with the RNA is sufficiently long to effect cross-relaxation. This experiment, called CLEANEX-PM (pulse sequence shown in Fig. 3; ref. 18), previously has been used to examine interactions between proteins and surface hydration water molecules. Despite a large difference in the structural, and perhaps functional, role of ψ in each of these contexts, we demonstrate the presence of a cross-relaxation-induced nuclear Overhauser effect (NOE) between the ψNH1 proton and water protons in each duplex. This finding supports the model of a water-mediated hydrogen bond involving ψ as the source of added stabilization or structural change.
Figure 3.
CLEANEX-PM pulse sequence (18). Shaped pulses are selective for water. The CLEANEX-PM windowless pulse train consists of: 135° (x) 120° (−x), 110° (x), 110° (−x), 120° (x), 135° (−x). Shaped pulses are 180° pulses selective for water. The phases of the pulses are cycled as follows: φ1 = x; φ2 = x, y, −x, −y; φ3 and rec. = x, −x, x, −x. A selective 90° water flip-back pulse (19) enables a higher repetition rate for the pulse sequence, and any residual water is suppressed by the WATERGATE (20) water-suppression scheme. The long, low-power (≈0.1 Gauss/cm) gradient applied during the spin lock prevents radiation damping (21) during this period.
Materials and Methods
Design and Synthesis of Molecular Constructs.
Two RNA duplexes containing ψ bases were designed for study, a 9-bp complementary duplex (ψ9 + 9) consisting of the sequences (5′-GGU GψA GUA-3′) and (5′-UAC UAC ACC-3′) and a model duplex representing the branch-site U2 snRNA–intron pairing from the yeast spliceosome (ψ9 + 10) consisting of the sequences (5′-GGU GψA GUA-3′) and (5′-UAC UAA CAC C-3′) (Fig. 4). The only difference between the two duplexes, shown as the extra A (underlined) in ψ9 + 10, denotes the branch-site adenosine, which is the nucleophile in the first step of spliceosome-mediated splicing of pre-mRNA. The oligomers were obtained from Dharmacon Research (Boulder, CO; refs. 22 and 23) and deprotected according to company protocols. Samples were ethanol-precipitated, partially desalted by three washes of the pelleted RNA with 80% ethanol, and lyophilized to dryness. Oligomers then were resuspended in diethyl pyrocarbonate-treated water, and strand concentration was measured from absorbance at 260 nm. Equimolar amounts of strands were combined to obtain duplexes of the 9-bp complementary molecule (ψ9 + 9) or the model branch site (ψ9 + 10), respectively. Duplexes were lyophilized to dryness and resuspended in 250 μl of NMR buffer consisting of 10 mM sodium phosphate, pH 6.4, 50 mM sodium chloride, and 0.1 mM EDTA in 90% H2O/10% 2H2O (99.96%; Cambridge Isotope Laboratories, Cambridge, MA). Final concentration of RNA duplexes in NMR samples was ≈1 mM. Microvolume NMR tubes (Shigemi, Allison Park, PA) were used for all NMR data collection.
Figure 4.
RNA duplexes used for investigation of water–ψ interactions. The complementary duplex (ψ9 + 9) is shown in A, and the branch-site construct (ψ9 + 10) is shown in B.
NMR Experiments.
A 500-MHz Varian Inova spectrometer (Department of Chemistry and Biochemistry, Florida State University, Tallahassee) was used for data collection. Spectra acquired at 5°C were referenced to the water resonance at 5.08 ppm. NOESY spectra of exchangeable protons were collected for each sample by using excitation sculpting water-suppression techniques (ES-NOESY; ref. 24). ES-rotating-frame Overhauser effect spectroscopy (ES-ROESY) experiments were performed with 150-ms mixing times and ≈3-kHz spin-lock fields (B1). Experiments featuring a CLEANEX-PM spin-lock (18) composite pulse scheme (Fig. 3) then were applied to the same samples.
Acquisition parameters for the CLEANEX-PM pulse sequence were optimized for application to RNA as compared with proteins (18). CLEANEX-PM experiments were performed with mixing times of 5, 50, 100, 150, 200, 250, 300, 350, 500, 750, and 1,000 ms to assess the buildup rate of each ROE. Buildups for each duplex were performed at temperatures of 5, 10, 15, 20, 25, 30, and 35°C to assess the effect of increased exchange rates on ROEs. Additional mixing time arrays were collected for ψ9 + 9 at temperatures of 40°C and 45°C.
Results
Assignment of RNA Protons Interacting with Water.
Assignment of proton resonances in ψ9 + 9 was achieved by standard methods from homonuclear spectra (25). Chemical shifts for ψ9 + 10, also assigned from homonuclear spectra, were reported previously (12). The NH1 resonance of ψ was identified in NOESY spectra by its up-field position (10.55 ppm) and by the cross-peak it displayed to its own H6 proton resonance. In NOESY spectra of ψ9 + 9, but not in spectra of ψ9 + 10, the ψ NH1 proton resonance also displayed very weak cross-peaks to A23 H2, G5 H1′, and A7 amino proton resonances, suggesting a more stacked environment for the ψ base in ψ9 + 9 as compared with ψ9 + 10.
Spectra showed strong NOEs between the water resonance and imino protons ψNH1 and U4 NH3 of the ψ9 + 9 duplex. Weaker cross-peaks were observed between water and the other imino resonances in the molecule. In an effort to identify the type of magnetization transfer indicated by the NOE, ROESY spectra then were collected to determine the phase of these cross-peaks relative to the diagonal. Excitation sculpting techniques (24) were used for water suppression, so as not to obscure cross-peaks between water and imino protons. Strong, positive cross-peaks, having the same phase as the diagonal, were seen between U4 NH3 and the water resonance and between ψNH1 and the water resonance in the NOESY and ROESY spectra of the ψ9 + 9 duplex. Weaker, positive cross-peaks also were observed in spectra of the ψ9 + 9 duplex between water and the other imino proton resonances.
The same NOESY and ROESY experiments described above were performed with the ψ9 + 10 duplex. NOEs observed in spectra of ψ9 + 10 were the same as those observed between the water resonance and the ψNH1 and U4 imino protons in spectra of ψ9 + 9. As seen in the ψ9 + 9 ROESY spectra, comparatively weaker, positive cross-peaks also were observed to the other imino proton resonances of ψ9 + 10.
Cross-Relaxation Between RNA and Water Protons in a Complementary Duplex.
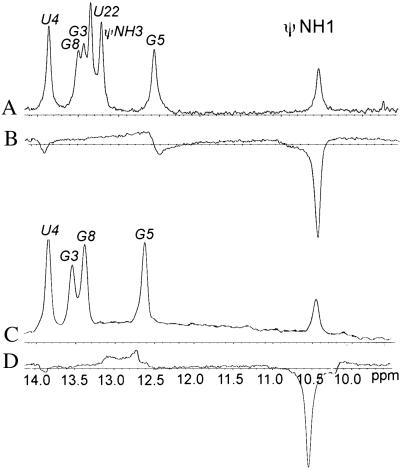
Positive ROESY cross-peaks between imino protons and water in the spectrum of the complementary duplex indicate exchange with water but do not exclude contribution from an ROE interaction induced by cross-relaxation that is obscured by the exchange-induced ROE. To distinguish ROEs arising from pure chemical exchange from those having a cross-relaxation component, a CLEANEX-PM experiment (18) was used. In spectra collected for the ψ9 + 9 duplex molecule at 5°C and a 150-ms mixing time, a sharp negative peak was seen at the resonance corresponding to ψNH1 at 10.55 ppm (Fig. 5), indicating cross-relaxation induced between this proton and nearby water protons. A small, negative peak also was observed at approximately 13.9 ppm, indicating a lesser interaction between U4 NH3 and water.
Figure 5.
Imino proton regions of one-dimensional spectra of ψ9 + 10 and the 9-bp complementary duplex. Resonance assignments are labeled above the peaks. Traces A and B correspond to a one-dimensional spectrum with excitation sculpting water suppression (24) and a CLEANEX-PM spectrum (18), respectively, of the complementary duplex, and traces C and D are the equivalent spectra of the branch-site duplex.
To rule out the possibility that the negative peak corresponding to the ψNH1 proton of ψ9 + 9 is not an exchange-relayed NOE, we note that the CLEANEX pulse sequence (Fig. 3) suppresses exchange-relayed effects. In addition, any small contributions from such effects are unlikely in this case because there are no exchangeable protons within 4 Å of the ψ NH1 proton in an A form helix.
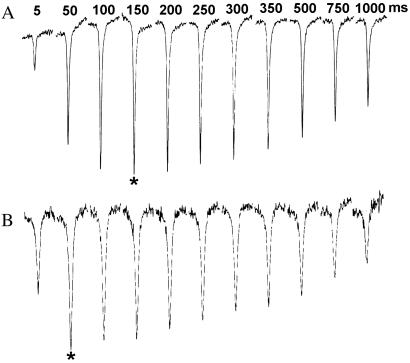
To assess the rate of buildup of the ψNH1–water cross-relaxation-induced NOEs in the ψ9 + 9 duplex, CLEANEX-PM spectra were collected at various mixing times and temperatures. These experiments showed that the ψNH1-negative ROE builds up faster with increasing temperature (Fig. 6). The resonance starts to broaden substantially at temperatures exceeding 30°C and is absent in spectra collected at temperatures higher than 40°C. This result likely is due to the increased exchange rates of the ψNH1 proton with water at higher temperatures. It was a general observation that most resonances in the CLEANEX spectrum became narrower with increased temperature, up to about 40°C, consistent with a faster tumbling rate for ψ9 + 9. This was observed for the small, negative U4 NH3 resonance and the Watson–Crick-paired ψNH3 proton resonance that is observed only at temperatures above 10°C. The peak intensity of the ψNH3 resonance increases with the temperature. Such an observation is consistent with the increase of exchange rates as temperature is increased. Taken together, these data from traditional ROESY and CLEANEX-PM spectroscopy have established that the ψNH1 proton in a complementary RNA duplex is exchanging with the bulk solvent on a time scale faster than the correlation time of the molecule and is cross-relaxing with water, entirely consistent with a water-mediated hydrogen bond in the major groove.
Figure 6.
ψNH1 resonance in CLEANEX spectra of the complementary ψ9 + 9 duplex at mixing times of 5, 50, 100, 150, 200, 250, 300, 350, 500, 750, and 1,000 ms. The array shown in A was collected at 5°C, and the array shown in B was collected at 25°C. Asterisks mark the peaks with the greatest intensity in each array, indicating the approximate optimum buildup time for the ψNH1–water NOE at each temperature.
Cross-Relaxation Between RNA and Water Protons in the Branch-Site Duplex.
CLEANEX spectra were recorded for the model branch-site duplex (ψ9 + 10) under identical conditions used for the complementary duplex (ψ9 + 9). Spectral features observed in CLEANEX spectra of the ψ9 + 10 at 5°C, using a mixing time of 150 ms, were nearly identical to those observed in spectra of the ψ9 + 9 duplex. Specifically, we observed a negative peak at 10.55 ppm, indicating cross-relaxation between ψNH1 and water protons. For comparison, this experiment also was performed under identical conditions with the unmodified branch-site duplex, which contains a U in place of ψ. No strong negative resonances were observed in the imino region for the unmodified duplex.
We then measured intensities of the negative peak at different temperatures. As for the complementary duplex, buildup of the negative NOE for ψNH1 in the ψ9 + 10 is faster at higher temperatures. At temperatures greater than 30°C, the ψNH1 resonance becomes broad and disappears, as do the other imino resonances, on denaturation of the duplex (Tm for the duplex is 41.8°C; ref. 12). This behavior contrasts with broadening observed for the ψNH1 resonance of ψ9 + 9, which disappears from the spectrum well below the melting temperature of a 9-bp complementary duplex (26).
As found for the complementary duplex, suppression of exchange-relayed NOEs by the pulse sequence (18) and topology of the duplex (36) make it highly unlikely that other exchangeable protons contribute to the negative NOE we observe. Spatial relationships seen within the major groove of the branch-site duplex (36), specifically, the orientation of the ψNH1 with respect to its own phosphate oxygen atoms, suggest that both the dimensions and the presentation of donor and acceptor groups would accommodate a water molecule. We therefore conclude that, as seen for the complementary duplex, a water-mediated hydrogen bond involving ψNH1 is likely to be present.
Discussion
NOESY experiments detect through-space transfer of magnetization between nearby protons. In principle, NOESY experiments can be used to detect cross-relaxation between protons of biomolecules and bound water molecules. In practice, however, observation of such interactions is difficult because: (i) cross-peaks between protons of water and RNA often are obscured as a result of water-suppression techniques (27), and (ii) NOEs involving water arise from two different physical phenomena, cross-relaxation and chemical exchange (28). Whereas a nonexchangeable proton resonance exhibiting a cross-peak to water in a NOESY spectrum usually is due to cross-relaxation effects between that proton and a bound water molecule, an exchangeable proton often will have a cross-peak to the bulk water frequency from pure chemical exchange. It is important to distinguish between the two phenomena because only those water molecules participating in hydrogen bonds with RNA will be sufficiently long-lived to exhibit cross-relaxation.
We observe the prominent ψNH1 proton resonance as relatively narrow lines in exchangeable proton spectra of both the complementary duplex and the branch-site duplex. The up-field chemical shift, as compared with imino protons in Watson–Crick base pairs (7, 14, 29), implies that the base is in the anti position relative to its ribose (the NH3 imino participates in a canonical base pair, not the NH1), and NH1 is positioned within the major groove of an A form helix. Imino resonances in rapid exchange with the solvent, e.g., imino protons of base pairs on duplex termini, are not visible in spectra (30). The appearance of the NH1 proton resonances suggests that these protons are slowed from exchange, perhaps by hydrogen bonding. Hall and McLaughlin found that longitudinal relaxation times for NH1 protons of ψ residues within complementary duplexes were comparable to those measured for the ψNH3 protons, which are involved in Watson–Crick base pairs (29). These data support the hypothesis of ψNH1 involvement in a hydrogen bond as a source of increased thermal stability. However, the geometry of an A form helix makes the possibility of a direct hydrogen bond between ψNH1 and an RNA acceptor unlikely (29). A water-mediated hydrogen bond involving this proton, as proposed by Davis and Poulter (13), would provide an explanation for the slow exchange rate of ψNH1 in RNA helices.
Cross-peaks in NOESY spectra represent contributions from several types of interactions with water, but the source (e.g., whether a particular NOE is derived from chemical exchange, cross-relaxation, or relayed exchange) cannot be determined from this experiment alone. Data from NOESY experiments also can be ambiguous because molecules with certain correlation times (ωoτc ≈ 1, where ωo is the Larmor frequency and τc is the correlation time) may have NOE enhancements close to zero (reviewed in ref. 31). ROESY spectra, however, because of the nature of the rotating-frame experiment, always have a positive NOE enhancement between 0.38 and 0.68, so it never approximates zero. Cross-peaks arising from cross-relaxation will be 180° out of phase from the diagonal and, thus, can be distinguished from NOEs arising from chemical exchange, which have the same phase as the diagonal (32). ROESY experiments have been used effectively to identify tightly bound water molecules in the core of proteins (33) and in stabilization of DNA duplexes (27) and triplexes (34). A limitation of traditional ROESY experiments is that positive and negative contributions to an NOE from cross-relaxation and chemical exchange, respectively, may cancel each other.
The CLEANEX-PM was developed as a tool to investigate the role of surface-hydration water molecules in protein structure (18), and, to our knowledge, has not been used previously to examine nucleic acid–water interactions. The hard 90°–G1-selective 180°–G1 pulse combination excites water protons but dephases all other magnetization, thereby allowing specific detection of solute–water interactions. CLEANEX filters out cross-relaxation effects from tightly bound water molecules, which exchange much more slowly than the overall correlation time of the molecule, leaving only interactions between protons in the biomolecule and more loosely associated (more rapidly exchanging) waters. Thus, ROEs arising even partially from cross-relaxation effects appear as a negative peak in a one-dimensional spectrum with an intensity proportional to specific characteristics of the interaction, including exchange rate, distance, and local motion, whereas an NOE arising from pure chemical exchange will appear as a positive peak. Our experiments combined NOESY experiments to verify interaction between ψNH1 and water, followed by a ROESY experiment to determine that the ψNH1–water interaction has a substantial element of chemical exchange and a CLEANEX ROESY to evaluate the role of cross-relaxation in the interaction.
Stabilization of a Complementary RNA Duplex by a Water-Mediated Hydrogen Bond.
Our data unequivocally demonstrate cross-relaxation between the ψNH1 proton and nearby water protons in the complementary duplex, consistent with a water-mediated hydrogen bond. Broadening and disappearance of the negative ψNH1 resonance well below the predicted melting temperature for a similar sequence that contains a U instead of ψ (26) suggests that the NOE between ψNH1 and water is dominated by chemical exchange at higher temperatures and that the water probably does not aid in stabilization of the duplex at these temperatures.
This strong evidence for a water-mediated hydrogen bond involving ψNH1 provides a mechanism by which the presence of ψ stabilizes RNA helices. The free-energy contribution from a water-mediated hydrogen bond was estimated from changes in thermodynamic parameters after substitution of modified bases in an RNA–hairpin loop to be in the range of −0.5 kcal/mol (35). However, considering that a number of other interaction changes may accompany such substitutions, such values are only approximate. An alternative mechanism for stabilization by ψ was presented by Yarian et al. (14), who noted that 1-methyl-ψ was just as stable as ψ at the base of an RNA loop and more stable than U. Because 1-methyl-ψ cannot form a hydrogen bond in the major groove, Yarian et al. concluded that stabilization resulted from improved base stacking. We do not necessarily consider the two models mutually exclusive: it is possible that the spatial context of the ψ is different in the middle of a helix and when closing a loop, which could result in different interactions, or that stacking interactions of the added methyl group coincidentally provide energetic contributions similar to the hydrogen bond they replace.
Pseudouridine is not always found to be a source of stabilization. For example, ψNH1 resonances belonging to ψ residues in single-stranded or loop regions of molecules are sometimes broad, or not visible at all (14, 17). It is perhaps significant that ψ incorporation in such regions of RNA secondary structure is destabilizing rather than stabilizing, emphasizing that ψ residues may have various structural roles depending on the particular context.
Water–ψ Interactions in the Pre-mRNA Branch-Site Helix.
As an important step in assembly of the eukaryotic spliceosome, a consensus sequence of the pre-mRNA's intron pairs with the U2 snRNA to form a short helix with an unpaired adenosine residue. The mechanism by which the 2′OH of the branch-site adenosine residue is positioned for nucleophilic activity is of major interest in understanding the structural biology of spliceosomal splicing. An intriguing feature of this pairing is the apparently absolute phylogenetic conservation of a ψ residue in U2 snRNA nearly opposing the branch site. We recently have solved the structure of the ψ-modified (ψ9 + 10) and unmodified branch-site helices by solution NMR techniques (Protein Data Bank ID codes 1LPW and 1LMV, respectively) and have found that ψ at the conserved site in the U2 snRNA strand induces a dramatic structural change in the branch-site helix as compared with its unmodified counterpart, marked by extrusion of the unpaired adenosine into the minor groove of the helix, where it is stabilized by formation of a base triple (36). Kinking of the backbone results in exposure of the 2′OH into a position in which it may be more accessible for nucleophilic activity. The ψ does not form a Watson–Crick base pair with the opposing A but is partially stacked on it, i.e., there is actually less stacking of this ψ on its neighbors than in the unmodified analogue. We also observed a change in free energy of duplex formation of −0.7 ± 0.1 kcal/mol on incorporation of ψ in the branch-site helix as compared with its unmodified counterpart. As in the case of the RNA–hairpin loop studied by SantaLucia et al. (35), it is not possible to correlate this change in ΔG with a single interaction because of numerous structural differences between the modified and unmodified analogues. By comparison, the unmodified duplex has the extra adenosine stacked within the A form helix, with the U pairing with both the branch-site adenosine and its adenosine 5′ neighbor (36). The question arose of how ψ, which differs from U primarily in the presence of an additional NH group, favors formation of such a markedly different structure. We noted that the orientation and dimensions in the vicinity of ψNH1 in the branch-site structural model provide space to accommodate a water molecule.
From the rate of buildup and decay of the NOE between ψNH1 and water (Fig. 6), we can extract qualitative information about the lifetime of the water molecule in the major groove. The degree of NOE enhancement depends on both the correlation time and molecular motions, so it is not possible to assign a precise value from these measurements alone. From traditional ROESY experiments, we observe that the water molecule in the major groove has a residence time that is less than the correlation time of either duplex, which is likely to be less than a few nanoseconds. The buildup and decay profiles of the ψNH1–water NOE indicate that the water molecule has a slightly longer residence time in the branch-site duplex than in the complementary duplex. This conclusion also is supported by the observation that the negative ψNH1 resonance in spectra of the branch-site duplex does not broaden substantially until the temperature approaches the melting transition of the duplex, whereas the corresponding resonance in the complementary duplex broadens out well before the melting temperature is reached.
Data presented here strongly suggest the presence of a water-mediated hydrogen bond involving ψNH1 in the branch-site duplex. We therefore propose that the water-mediated hydrogen bond involving the unpaired ψ is likely to contribute to stabilization of the observed architectural features in the branch-site helix (36). These findings are significant because they establish a molecular basis for the structural change induced by ψ at the spliceosomal branch site, one that may have important biological implications.
These results provide insight into the biological roles of ψ, many of which are conserved in specific positions throughout structural RNAs. In the absence of ψ, both the complementary and branch-site helices adopt A form helical geometry. Our evidence suggests that ψ conserved in complementary helical regions of biological RNAs contribute to local stability (but no change in global structure) by formation of an additional hydrogen bond. A conserved ψ in the branch-site duplex, associated with formation of a markedly different architecture, also appears to mediate its effect by hydrogen bond formation. We speculate that this is not an isolated case and that numerous, other ψ-mediated enhancements of structure and activity soon will be found in biological RNAs. Although the ψ–water interactions appear to be similar for both molecules examined in this study, fascinatingly, one ψ acts to stabilize an existing local architecture (in the complementary duplex), and the other ψ actively stabilizes an alternative conformation for the branch-site adenosine.
Acknowledgments
N.L.G. dedicates this paper to Prof. David Mauzerall on the occasion of his 70th birthday. We thank Drs. Tim Logan and Greg Wylie for helpful discussions. This research was funded partially by Public Health Service Grant RO1-GM54008 (to N.L.G.). M.I.N. is a recipient of a Molecular Biophysics fellowship funded by a National Science Foundation predoctoral training grant.
Abbreviations
- snRNA
small nuclear RNA
- ROESY
rotating-frame Overhauser effect spectroscopy
- NOE
nuclear Overhauser effect
- ψ
pseudouridine
- CLEANEX-PM
CLEAN chemical-exchange spectroscopy
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1LPW and 1LMV).
References
- 1.Wyatt G R. Nature (London) 1950;166:237–238. doi: 10.1038/166237b0. [DOI] [PubMed] [Google Scholar]
- 2.Yu C-T, Allen F W. Biochim Biophys Acta. 1959;32:393–406. doi: 10.1016/0006-3002(59)90612-2. [DOI] [PubMed] [Google Scholar]
- 3.Scannel J P, Crestfield A M, Allen F W. Biochim Biophys Acta. 1959;32:406–412. doi: 10.1016/0006-3002(59)90613-4. [DOI] [PubMed] [Google Scholar]
- 4.Cohn W E. Biochim Biophys Acta. 1959;32:569–571. doi: 10.1016/0006-3002(59)90644-4. [DOI] [PubMed] [Google Scholar]
- 5.Cohn W E. J Biol Chem. 1960;235:1488–1498. [PubMed] [Google Scholar]
- 6.Charette M, Gray M W. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 7.Hall K B, McLaughlin L. Biochemistry. 1991;30:1795–1801. doi: 10.1021/bi00221a010. [DOI] [PubMed] [Google Scholar]
- 8.Auffinger P, Westhof E. In: Modification and Editing of RNA. Grosjean H, Benne R, editors. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 103–112. [Google Scholar]
- 9.Bakin A, Lane B G, Ofengand J. Biochemistry. 1994;33:13475–13483. doi: 10.1021/bi00249a036. [DOI] [PubMed] [Google Scholar]
- 10.Gu J, Patton J R, Shimba S, Reddy R. RNA. 1996;2:909–918. [PMC free article] [PubMed] [Google Scholar]
- 11.Yu Y-T, Shu M-D, Steitz J A. EMBO J. 1998;17:5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newby M I, Greenbaum N L. RNA. 2001;7:833–845. doi: 10.1017/s1355838201002308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis D, Poulter C. Biochemistry. 1991;30:4223–4231. doi: 10.1021/bi00231a017. [DOI] [PubMed] [Google Scholar]
- 14.Yarian C S, Basti M M, Cain R J, Ansari G, Guenther R H, Sochacka E, Czerwinska G, Malkiewicz A, Agris P F. Nucleic Acids Res. 1999;27:3543–3549. doi: 10.1093/nar/27.17.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnez J, Steitz T. Biochemistry. 1994;33:7560–7567. doi: 10.1021/bi00190a008. [DOI] [PubMed] [Google Scholar]
- 16.Davis D. Nucleic Acids Res. 1995;23:5020–5026. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meroueh M, Grohar P J, Qiu J, SantaLucia J, Scaringe S A, Chow C S. Nucleic Acids Res. 2000;28:2075–2083. doi: 10.1093/nar/28.10.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang T-L, Mori S, Shaka A J, van Zijl P C M. J Am Chem Soc. 1997;119:6203–6204. [Google Scholar]
- 19.Grzesiek S, Bax A. J Am Chem Soc. 1993;115:12593–12594. [Google Scholar]
- 20.Sklenar V, Piotto M, Leppik R, Saudek V. J Magn Reson A. 1993;102:241–245. [Google Scholar]
- 21.Bloembergen N, Pound R V. Phys Rev. 1954;95:8–12. [Google Scholar]
- 22.Scaringe S A, Wincott F E, Caruthers M H. J Am Chem Soc. 1998;120:11820–11821. [Google Scholar]
- 23.Scaringe S A. Methods Enzymol. 2000;317:3–18. doi: 10.1016/s0076-6879(00)17003-x. [DOI] [PubMed] [Google Scholar]
- 24.Callihan D, West J, Kumar S, Schweitzer B, Logan T. J Magn Reson B. 1996;112:82–85. doi: 10.1006/jmrb.1996.0114. [DOI] [PubMed] [Google Scholar]
- 25.Wijmenga S S, Mooren M M W, Hilbers C W. In: NMR of Macromolecules: A Practical Approach. Roberts G C K, editor. Oxford: Oxford Univ. Press; 1993. pp. 217–258. [Google Scholar]
- 26.Xia T, SantaLucia J, Jr, Burkard M E, Kierzek R, Schroeder S J, Jiao X, Cox C, Turner D. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 27.Kubinec M G, Wemmer D E. J Am Chem Soc. 1992;114:8739–8740. [Google Scholar]
- 28.van de Ven F J M, Janssen H G J M, Graslund A, Hilbers C W. J Magn Reson. 1988;79:221–235. [Google Scholar]
- 29.Hall K B, McLaughlin L. Nucleic Acids Res. 1992;20:1883–1889. doi: 10.1093/nar/20.8.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel D J, Hilbers C W. Biochemistry. 1975;14:2651–2656. doi: 10.1021/bi00683a014. [DOI] [PubMed] [Google Scholar]
- 31.Cavanagh J, Fairbrother W J, Palmer A G, Skelton N J. Protein NMR Spectroscopy. New York: Academic; 1996. pp. 288–290. [Google Scholar]
- 32.Bothner-By A A, Stevens R L, Lee J M, Warren C D, Jeanloz R W. J Am Chem Soc. 1984;106:811–813. [Google Scholar]
- 33.Otting G, Wuthrich K. J Am Chem Soc. 1989;111:1871–1875. [Google Scholar]
- 34.Radhakrishnan I, Patel D J. Structure (London) 1994;2:395–405. doi: 10.1016/s0969-2126(00)00041-1. [DOI] [PubMed] [Google Scholar]
- 35.SantaLucia J, Kierzek R, Turner D H. Science. 1992;256:217–219. doi: 10.1126/science.1373521. [DOI] [PubMed] [Google Scholar]
- 36. Newby, M. I. & Greenbaum, N. L. (2002) Nat. Struct. Biol., in press. [DOI] [PubMed]