Abstract
RNA ligases participate in repair, splicing, and editing pathways that either reseal broken RNAs or alter their primary structure. Bacteriophage T4 RNA ligase (gp63) is the best-studied member of this class of enzymes, which includes yeast tRNA ligase and trypanosome RNA-editing ligases. Here, we identified another RNA ligase from the bacterial domain—a second RNA ligase (Rnl2) encoded by phage T4. Purified Rnl2 (gp24.1) catalyzes intramolecular and intermolecular RNA strand joining through ligase-adenylate and RNA-adenylate intermediates. Mutational analysis identifies amino acids required for the ligase-adenylation or phosphodiester synthesis steps of the ligation reaction. The catalytic residues of Rnl2 are located within nucleotidyl transferase motifs I, IV, and V that are conserved in DNA ligases and RNA capping enzymes. Rnl2 has scant amino acid similarity to T4 gp63. Rather, Rnl2 exemplifies a distinct ligase family, defined by variant motifs, that includes the trypanosome-editing ligases and a group of putative RNA ligases encoded by eukaryotic viruses (baculoviruses and an entomopoxvirus) and many species of archaea. These findings have implications for the evolution of covalent nucleotidyl transferases and virus-host dynamics based on RNA restriction and repair.
T4 RNA ligase (1) is the founding member of a family of RNA end-joining enzymes involved in RNA repair, splicing, and editing pathways (2–8). T4 RNA ligase joins 3′ OH and 5′ PO4 RNA termini by means of three nucleotidyl transfer steps similar to those of DNA ligases (9–11). Step 1 is the reaction of RNA ligase with ATP to form a covalent ligase-(lysyl-N)–AMP intermediate. In step 2, the AMP is transferred to a 5′ PO4 RNA end to form an RNA-adenylate intermediate (AppRNA). In step 3, attack by an RNA 3′ OH on RNA-adenylate seals the ends and releases AMP. T4 RNA ligase can join two single-stranded RNA molecules without a complementary bridging polynucleotide. T4 RNA ligase can also catalyze intramolecular ligation to form a covalently closed RNA circle. RNA ligase has been a powerful tool in the synthesis of RNAs of defined sequence, RNA 3′ end modification, RNA 3′ end-labeling, RNA sequencing, and structural analysis (11).
During T4 infection in vivo, the RNA ligase, together with T4 polynucleotide kinase (Pnk), performs an RNA repair function that remodels and then seals broken tRNA ends. In Escherichia coli strains containing the prr locus, the host cell tRNALys is cleaved 5′ to the wobble position by a T4-induced anticodon nuclease. If Pnk and RNA ligase are not present, the synthesis of viral proteins is blocked by depletion of tRNALys, and the phage cannot replicate (2, 12–14). This pathway represents an RNA-based restriction system of host defense against a foreign invader. tRNA anticodon nuclease systems are present in other pathogenic bacteria (14). The bacterial toxins colicins D and E5 are also anticodon nucleases that attack specific tRNAs (15). Thus, RNA repair enzymology has broad significance as a means to combat, or recover from, damage to essential RNA molecules.
T4 RNA ligase is a 374-aa polypeptide encoded by gene 63 (16). The site of covalent adenylation has been mapped to Lys-99 (17, 18), which is located within a conserved motif (KxDG) that defines a superfamily of covalent nucleotidyl transferases embracing DNA ligases and mRNA capping enzymes (19, 20). DNA ligases and capping enzymes share a common tertiary structure composed of five conserved motifs (I, III, IIIa, IV, and V) responsible for nucleotide binding and catalysis (21–23). It has been suggested that DNA ligases and capping enzymes evolved from a common ancestral nucleotidyl transferase, possibly from an ancient RNA strand-joining enzyme. The structural basis for catalysis by RNA ligases is ill defined because few RNA ligase enzymes have been identified, cloned, and studied.
Sequence-based searches for RNA ligases similar to T4 gp63 identify yeast tRNA ligases (5) and a putative baculovirus ligase (24) as the only credible homologs. Thus, gp63-like RNA ligases have a narrow distribution in nature compared with the ubiquitous DNA ligases. This situation may reflect the lack of selection pressure to maintain the catalysis of RNA strand-transfer reactions after the establishment of DNA genomes. Alternatively, RNA ligases with a gp63-like mechanism are widely distributed, but we do not recognize them as such from sequence alone. For example, the recently identified Trypanosoma brucei RNA-editing ligases TbMP52 and TbMP48 (6–8) have little resemblance to the T4 gp63 RNA ligase, to the point that four iterations of a Psi-BLAST search with gp63 failed to identify the trypanosome RNA ligases and vice versa.
Here, we identify and characterize another RNA-joining enzyme from the bacterial domain—a second and previously unrecognized RNA ligase encoded by phage T4. The additional T4 RNA ligase is the product of gene 24.1 and will, henceforth, be referred to as Rnl2 to distinguish it from the original ligase gp63, which we rename Rnl1. Rnl2 exemplifies a family of RNA ligases, defined by variant nucleotidyl transferase motifs, that includes proteins encoded by archaea, eukarya, and eukaryotic viruses.
Materials and Methods
Recombinant T4 Rnl2.
The gp24.1 ORF was amplified by PCR from T4 DNA (a gift of Ken Kreuzer, Duke University, Durham, NC) with primers designed to introduce an NdeI restriction site at the start codon and a BamHI site 3′ of the stop codon. The PCR product was digested with NdeI and BamHI and inserted into pET16b (Novagen) to generate the plasmid pET-RNL2 encoding the T4 polypeptide fused to an N-terminal His-10 tag. Amino acid substitution mutations were introduced into the ORF by PCR with the two-stage overlap extension method (25). The inserts of the WT and mutant pET-RNL2 plasmids were sequenced completely to exclude the acquisition of unwanted changes during amplification and cloning.
pET-RNL2 plasmids were transformed into E. coli BL21(DE3). A 200-ml culture of E. coli BL21(DE3)/pET-RNL2 was grown at 37°C in LB medium containing 0.1 mg/ml ampicillin until the A600 reached 0.4. The culture was adjusted to 0.4 mM isopropyl β-d-thiogalactoside (IPTG), and incubation was continued at 37°C for 3 h. Cells were harvested by centrifugation, and the pellet was stored at −80°C. All subsequent procedures were performed at 4°C. Thawed bacteria were resuspended in 10 ml of buffer A [50 mM Tris⋅HCl, pH 7.5/0.25 mM NaCl/10% (wt/vol) sucrose]. Lysozyme and Triton X-100 were added to final concentrations of 50 μg/ml and a 0.1%, respectively. The lysate was sonicated to reduce viscosity and insoluble material was removed by centrifugation. The soluble extract was applied to a 1-ml column of Ni-NTA agarose (Qiagen, Chatsworth, CA) that had been equilibrated with buffer A containing 0.1% Triton X-100. The column was washed with 5 ml of the same buffer and then eluted stepwise with 2 ml of buffer B [50 mM Tris⋅HCl, pH 8.0/0.25 M NaCl/10% (vol/vol) glycerol] containing 0.05, 0.1, 0.2, and 0.5 M imidazole. The polypeptide compositions of the column fractions were monitored by SDS/PAGE. Rnl2 was recovered predominantly in the 0.1 M imidazole fraction, which contained 2–5 mg of protein. The WT and mutant Rnl2 preparations were stored at −80°C.
Adenylyltransferase Assay.
Standard reaction mixtures (20 μl) containing 50 mM Tris acetate (pH 6.5), 5 mM DTT, 1 mM MgCl2, 20 μM [α-32P]ATP, and Rnl2 as specified were incubated for 5 min at 37°C. The reactions were quenched with SDS, and the products were analyzed by SDS/PAGE. The ligase-[32P]AMP adduct was visualized by autoradiography of the dried gel and quantitated by scanning the gel with a PhosphorImager.
RNA Ligase Assay.
An 18-mer oligoribonucleotide (5′-AUUCCGAUAGUGACUACA) was 5′ 32P-labeled by using T4 polynucleotide kinase and [γ-32P]ATP and then purified by electrophoresis through a nondenaturing 20% polyacrylamide gel (26). RNA ligation reaction mixtures (10 μl) containing 50 mM Tris⋅acetate (pH 6.5), 5 mM DTT, 5 mM MgCl2, 1 pmol of 5′ 32P-labeled 18-mer RNA, and ATP and Rnl2 as specified were incubated for 15 min at 22°C. The reactions were quenched by adding 5 μl of 90% formamide/20 mM EDTA. The samples were electrophoresed through an 18% polyacrylamide gel containing 7 M urea in 45 mM Tris⋅borate/1 mM EDTA. The ligation products were visualized by autoradiography of the gel.
Results
Identification of a Second RNA Ligase Encoded by Bacteriophage T4.
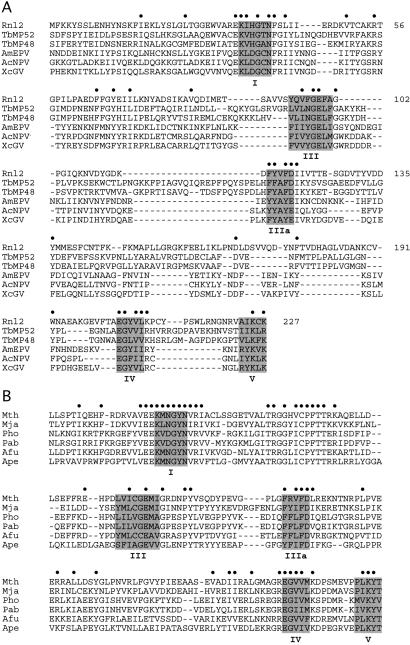
Although T4 gp63 and the T. brucei RNA-editing ligases display little sequence similarity and, thereby, seem to comprise distinct lineages, one can detect by visual inspection and manual alignment the presence of nucleotidyl transferase motifs I, III, IIIa, IV, and V in both types of RNA ligases (6, 7). The nucleotidyl transferase motifs of the T. brucei ligases are highlighted in Fig. 1. Motif I, which contains the active-site lysine, adheres to a consensus sequence KxHGxN in the trypanosome RNA ligases, which differs from the KxDGxR sequence characteristic of ATP-dependent DNA ligases and most mRNA capping enzymes (20, 27). A BLAST search with the T. brucei RNA ligases identified other proteins containing the variant KxHGxN motif I sequence, including the putative RNA-editing ligase ortholog of Leishmania (7, 8) and a protein of unknown function (gp24.1) encoded by phage T4. The 334-aa T4 gp24.1 polypeptide contains all five nucleotidyl transferase motifs in the usual order and with typical spacing between the motifs (Fig. 1A). Thus, we predicted that gp24.1 is a previously unappreciated second T4 RNA ligase, which we named Rnl2.
Figure 1.
Rnl2-like family of RNA ligases. (A) The amino acid sequence of T4 Rnl2 from residues 1–227 is aligned to the sequences of the RNA-editing ligases TbMP52 and TbMP48 and putative ligases from the poxvirus AmEPV and baculoviruses AcNPV and XcGV. (B) Alignment of Rnl2-like proteins from six species of archaea. Positions of side-chain identity/similarity in all of the polypeptides included in the respective alignments are indicated by dots (•). Nucleotidyl transferase motifs I, III, IIIa, IV, and V are highlighted in shaded boxes.
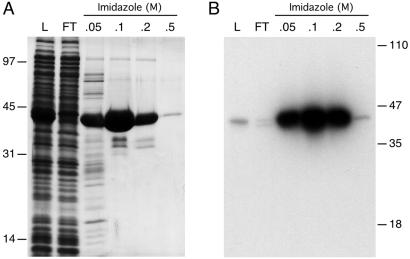
We expressed Rnl2 in E. coli as a His-10-tagged fusion and purified the 42-kDa recombinant protein from a soluble bacterial extract by adsorption to Ni-agarose and step-elution with 50, 100, and 200 mM imidazole (Fig. 2A). The adenylyltransferase activity of recombinant Rnl2 was evinced by label transfer from [α-32P]ATP to the Rnl2 polypeptide to form a covalent enzyme-adenylate adduct (Fig. 2B). Adenylyltransferase activity paralleled the elution profile of the Rnl2 protein during Ni-agarose chromatography. We conclude that recombinant Rnl2 is a covalent nucleotidyl transferase. The activities of Rnl2 were characterized in detail by using the 0.1 M imidazole eluate fraction.
Figure 2.
Purification and adenylyltransferase activity of T4 Rnl2. (A) Aliquots (10 μl) of the soluble lysate of isopropyl β-d-thiogalactoside-induced bacteria (L), the nickel-agarose flow-through (FT), and the indicated imidazole eluate fractions were analyzed by SDS/PAGE. The gel was stained with Coomassie blue dye. The positions and sizes (kDa) of marker polypeptides are shown on the left. (B) Reaction mixtures (20 μl) containing 50 mM Tris⋅HCl (pH 8.0), 5 mM DTT, 5 mM MgCl2, 0.17 μM [α-32P]ATP, and 1 μl of the indicated fractions were incubated for 5 min at 37°C. The reaction products were resolved by SDS/PAGE. An autoradiograph of the gel is shown.
Adenylyltransferase Reaction.
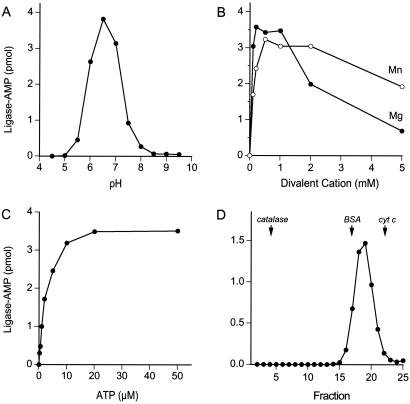
The adenylyltransferase activity displayed a bell-shaped pH profile with an optimum at pH 6.5 (Fig. 3A). Activity was virtually nil at pH ≤ 5.0 or ≥ 8.5. Rnl2 required a divalent cation cofactor to form the covalent adduct. MgCl2 and MnCl2 supported optimal activity at 0.2–1 mM and 0.5–2 mM concentrations, respectively (Fig. 3B). The yield of Rnl2-AMP complex was proportional to ATP concentration from 0.2 to 5 μM and reached saturation at 20 μM (Fig. 3C). Half-saturation was achieved at 2 μM ATP. We calculated that ≈70% of the Rnl2 protein was adenylated with 32P-AMP at saturating ATP. The remaining ≈30% of the Rnl2 preparation likely consists of preformed Rnl2-AMP intermediate (see below).
Figure 3.
Characterization of the adenylyltransferase reaction. (A) pH dependence. Reaction mixtures (20 μl) containing 50 mM buffer (either Tris⋅acetate, pH 4.5, 5.0, 5.5, 6.0, 6.5, 7.0, or Tris⋅HCl, pH 7.5, 8.0, 8.5, 9.0, 9.5), 5 mM DTT, 1 mM MgCl2, 20 μM [α-32P]ATP, and 200 ng of Rnl2 were incubated for 5 min at 37°C. The extent of Rnl2-AMP formation is plotted as a function of pH. (B) Divalent cation dependence. Reaction mixtures (20 μl) containing 50 mM Tris⋅acetate (pH 6.5), 5 mM DTT, 20 μM [α-32P]ATP, 200 ng of Rnl2, and MgCl2 or MnCl2 as specified were incubated for 5 min at 37°C. Rnl2-AMP formation is plotted as a function of divalent cation concentration. (C) ATP dependence. Reaction mixtures (20 μl) containing 50 mM Tris⋅acetate (pH 6.5), 5 mM DTT, 1 mM MgCl2, 200 ng of Rnl2, and [α-32P]ATP as specified were incubated for 5 min at 37°C. Rnl2-adenylate formation is plotted as a function of ATP concentration. (D) Glycerol gradient sedimentation. An aliquot (50 μg) of Rnl2 was mixed with 50 μg each of catalase, BSA, and cytochrome c and the mixture was applied to a 4.8-ml 15–30% glycerol gradient containing 50 mM Tris⋅HCl (pH 8.0), 2 mM DTT, 250 mM NaCl, 0.1% Triton X-100. The gradient was centrifuged at 50,000 rpm for 18 h at 4°C in a Beckman SW50 rotor. Fractions were collected from the bottom of the tube. Aliquots (1 μl) of the glycerol gradient fraction were assayed for adenylyltransferase activity. The activity profile is shown. The positions of the peak fractions of catalase, BSA, and cytochrome c are indicated by arrows.
The native size of Rnl2 was gauged by sedimentation through a 15–30% glycerol gradient. Marker proteins catalase (248 kDa), BSA (66 kDa), and cytochrome c (13 kDa) were included as internal standards. The adenylyltransferase activity sedimented as a single discrete peak between BSA and cytochrome c (Fig. 3D). The activity profile paralleled exactly the sedimentation profile of the Rnl2 polypeptide (not shown). We surmise that Rnl2 is a monomer in solution.
RNA Ligase Activity.
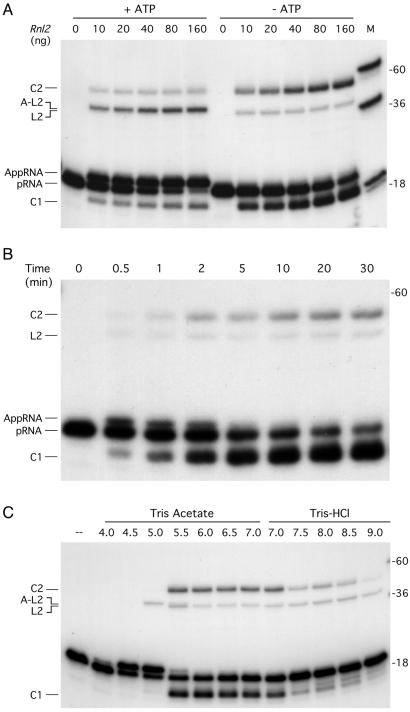
Rnl2 was placed in reaction with a 5′ 32P-labeled 18-mer RNA oligonucleotide and magnesium in the presence or absence of 1 mM ATP. In both cases, Rnl2 formed novel radiolabeled RNAs; however, the distribution of the products varied depending on whether ATP was present. When ATP was included, the predominant RNA product migrated ≈1 nt slower than the input 18-mer strand (Fig. 4A). This species, which was resistant to alkaline phosphatase (not shown), corresponded to the RNA-adenylate (AppRNA) generated by AMP transfer from Rnl2-AMP to the 5′ end of the input 18-mer RNA. In the absence of ATP, the major product (denoted C1 in Fig. 4A) migrated ≈2 nt faster than the input 18-mer strand. The C1 product, which was also resistant to alkaline phosphatase, corresponds to a covalently closed 18-mer circle formed by intramolecular ligation. Ligation in the absence of added ATP reflects the presence of preformed ligase-adenylate in the enzyme preparation.
Figure 4.
RNA ligase activity. (A) Protein titration. Reaction mixtures (10 μl) containing 50 mM Tris⋅acetate (pH 6.5), 5 mM DTT, 5 mM MgCl2, 1 pmol of 5′ 32P-labeled 18-mer RNA (pRNA), Rnl2 as specified, and either 1 mM ATP (+ATP) or no ATP (−ATP) were incubated for 15 min at 22°C. (B) Kinetics. A reaction mixture (100 μl) containing 50 mM Tris⋅acetate (pH 6.5), 5 mM DTT, 5 mM MgCl2, 10 pmol of pRNA substrate, and 800 ng of Rnl2 were incubated at 22°C. Aliquots (10 μl) were withdrawn at the times indicated and quenched immediately with formamide-EDTA. (C) pH-dependence. Reaction mixtures (10 μl) containing 50 mM buffer (either Tris⋅acetate, pH 4.5, 5.0, 5.5, 6.0, 6.5, or 7.0, or Tris⋅HCl, pH 7.0, 7.5, 8.0, 8.5, or 9.0), 5 mM DTT, 5 mM MgCl2, 1 pmol of pRNA substrate, and 80 ng of Rnl2 were incubated for 15 min at 22°C. Rnl2 was omitted from a control reaction (–). The reaction products were analyzed by PAGE along with a mixture of 5′ 32P-labeled DNA oligonucleotide size markers (18-mer, 36-mer, and 60-mer; lane M in A).
The Rnl2 reaction products also included higher molecular weight 32P-labeled species corresponding to a 36-mer linear dimer (L2), a 36-mer circular dimer (C2), and an adenylated 36-mer linear dimer (A-L2), respectively (Fig. 4A). As with the monomeric products, the formation of an adenylated linear dimer was favored in the presence of ATP. At saturating levels of enzyme in reactions containing ATP, 60% of the input substrate was converted to RNA-adenylate, 10% to adenylated linear dimer, 10% to monomer circle, and 5% to dimer circle. In the absence of ATP, the circular dimer was the preferred product over the linear dimer (Fig. 4A). (Note that the linear RNA molecules migrated 1–2 nt “longer” than DNA strands of identical length and sequence. The circular RNAs migrated aberrantly compared with linears.)
The inclusion of ATP promoted accumulation of AppRNAs and suppressed formation of ligated circles. The likely explanation for the ATP effect is that Rnl2 is prone to dissociate from the newly formed RNA-adenylate product of step 2, and that an immediate reaction with ATP to form ligase-adenylate precludes it from rebinding to the RNA-adenylate for subsequent catalysis of strand joining. Similar ATP trapping effects leading to the accumulation of high levels of the adenylated nucleic acid intermediate have been observed for DNA ligases when they react with a substrate containing a 1-nt gap (26, 28). Sugino et al. (10) showed that catalysis of step 3 by T4 Rnl1 was inhibited by ATP and suggested that the RNA intermediates dissociate from Rnl1 after RNA-adenylate is formed.
It was conceivable that the change in the Rnl2 product distribution in the presence of ATP actually reflected Rnl2-catalyzed joining of the 3′ OH of ATP to the 5′ PO4 of the 18-mer RNA to generate a slower-migrating product, pppApRNA. Two lines of experiments seem to exclude such a reaction. First, we found that other NTPs do not stimulate formation of the slower product at the expense of monomer circle, as was seen for ATP (not shown). If the 3′ OH of an NTP was being ligated to the 5′ PO4, then Rnl2 would not be expected to have specificity for ATP as the 3′ OH substrate. In contrast, the model suggested above (whereby Rnl2 dissociates after forming AppRNA and is trapped by covalent adenylylation) predicts that only ATP would stimulate formation of the slower species. Second, if the product was pppApRNA, then its electrophoretic mobility should be altered by treatment with alkaline phosphatase (which would remove three 5′ phosphates while preserving the 32P in the RNA chain). We observed no shift in mobility of the species we designated AppRNA after phosphatase treatment (not shown).
The strand-joining reaction of T4 Rnl2 in the absence of ATP required a divalent cation cofactor and was optimal at 0.5–5 mM MgCl2 (not shown). Although circles were the major product at each concentration tested (0.5, 1, 2, and 5 mM), the relative abundance of dimer circles was increased at 5 mM MgCl2. Similarly, the formation of AppRNA in the presence of 1 mM ATP required a divalent cation and was optimal at 0.5–5 mM MgCl2; here also, the adenylated linear dimer increased in abundance at 5 mM vs. 1 mM MgCl2 (not shown). T4 Rnl2 failed to either ligate or adenylate a 5′ 32P-labeled 18-mer DNA oligonucleotide substrate under the conditions that were permissive for RNA ligation and adenylation (not shown).
RNA-Adenylate Intermediate.
A kinetic analysis of the strand-joining reaction in the absence of ATP showed that RNA-adenylate was the first product formed (Fig. 4B). RNA-adenylate persisted from 0.5 to 2 min and then declined to undetectable levels by 5–10 min, concomitant with decay of the 18-mer substrate and formation of monomer and dimer circles (Fig. 4B). These results suggest that the RNA-adenylate is a genuine intermediate along the Rnl2 ligation reaction pathway.
Additional evidence implicating RNA-adenylate as a reaction intermediate emerged from an analysis of pH effects on the strand-joining reaction in the absence of ATP (Fig. 4C). The yields of the major circular products were optimal between pH 5.5 and 7.0. Reducing the pH to 5.0 or 4.5 completely suppressed the formation of circles, but dramatically stimulated the formation of RNA-adenylate (Fig. 4C). This result implies that the step of phosphodiester bond formation became rate-limiting at pH 4.5–5.0, such that the normally evanescent RNA-adenylate intermediate accumulated to high levels. Also, the transition from pH 5.5 to 5.0 resulted in the formation of adenylated linear dimers in lieu of nonactivated linear dimers (the ≈1-nt mobility difference between the A-L2 and L2 species is easily seen in Fig. 4C). Further reduction of the pH to ≤4.0 abrogated all activity of Rnl2. Raising the pH to ≥7.5 suppressed the intramolecular ligation reaction but did not diminish the low amounts of linear dimers formed by intermolecular joining (Fig. 4C).
Analysis of the 32P-labeled reaction products after enzymatic digestion and polyethyleneimine-cellulose thin layer chromatography confirmed that 5′ adenylated RNA was the major product formed by Rnl2 in the presence of ATP under standard conditions (pH 6.5). All of the 32P-label in the substrate RNA (which migrated as a smear near the chromatographic origin) was converted to 32Pi by alkaline phosphatase, whereas most of the product generated by Rnl2 with or without ATP resisted phosphatase digestion (Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org). Treatment with nuclease P1 (which cleaves the 3′ O–P bonds in the RNA) liberated 5′ AMP as the predominant labeled product from both the pRNA substrate and the ligation products formed in the absence of ATP (predominantly circles), but the P1 digestion product of the ligation products formed in the presence of ATP was different and migrated slower that 5′ AMP. Combined digestion with alkaline phosphatase and nuclease P1 converted all of the label in the minus-ATP ligation products to 32Pi, yet the majority of the plus-ATP ligation product resisted this dual treatment and comigrated the material generated by nuclease P1 alone (Fig. 7). This material corresponds to the phosphatase resistant 5′ AppA dinucleotide liberated by digestion of AppRNAs with nuclease P1. Quantitation of the chromatogram indicated that 75% of the label in the plus-ATP product was converted to AppA, a value that agrees with the 70% fraction of adenylated RNA products determined by PAGE (i.e., 60% adenylated monomer and 10% adenylated linear dimer).
Mutational Analysis of Rnl2 Identifies Residues Essential for Catalysis.
The presence of the nucleotidyl transferase motifs in Rnl2 raised the question of whether and how they contribute to RNA ligase function. To address this question, we introduced Ala substitutions at Lys-35 and His-37 in motif I, Glu-204 in motif IV, and Lys-225 and Lys-227 in motif V. In addition, we replaced His-37 with Asp, which is the side chain present at the equivalent position in motif I of T4 Rnl1 and virtually all DNA ligases and RNA capping enzymes.
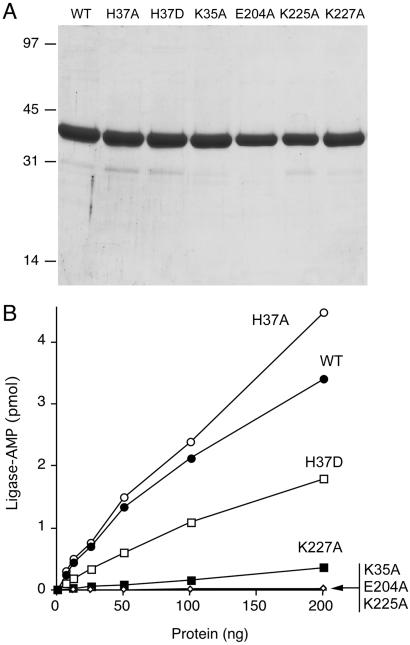
The K35A, H37A, H37D, E204A, K225A, and K227A mutants were expressed in bacteria and purified from soluble bacterial lysates by Ni-agarose chromatography (Fig. 5A). The adenylyltransferase activities were assayed by protein titration (Fig. 5B). The extent of ligase-adenylate formation by WT Rnl2 was proportional to input protein up to 200 ng of protein. The K35A mutant was inert over the same range. This result is consistent with the Rnl2 motif I lysine being the site of covalent NMP attachment, as it is in other members of the polynucleotide ligase/capping enzyme superfamily. The H37A change in motif I had no deleterious effect on formation of ligase-adenylate, whereas the introduction of Asp had a modest effect (2-fold decrement). The adenylation reaction was abrogated by the E204A change in motif IV and the K225A mutation in motif V. The K227A change in motif V reduced adenylyltransferase activity to 5% of the wild-type level (Fig. 5B).
Figure 5.
Mutational effects on adenylyltransferase activity. (A) Aliquots (4 μg) of the Ni-agarose preparations (0.1 M imidazole eluates) of wild-type Rnl2 and the indicated mutants were analyzed by SDS/PAGE. The Coomassie blue-stained gel is shown. (B) Reaction mixtures contained 50 mM Tris·acetate (pH 6.5), 5 mM DTT, 1 mM MgCl2, 20 μM [α-32P]ATP, and wild-type or mutant Rnl2, as specified. Ligase-adenylate formation is plotted as a function of input Rnl2 protein.
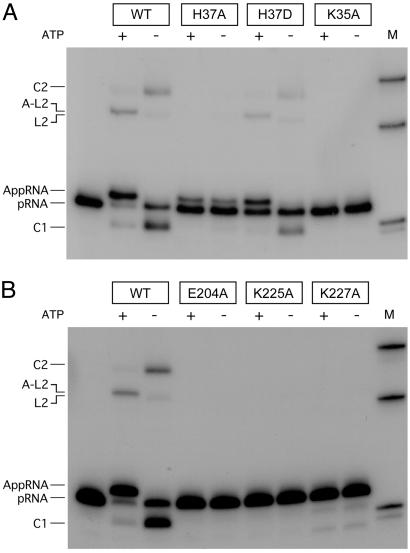
Mutational effects on RNA ligation in the presence and absence of ATP are shown in Fig. 6. Mutants K35A, E204A, and K225A failed to form either ligated RNAs or RNA-adenylate. This finding is in keeping with their inability to perform the initial ligase adenylation step in the reaction pathway upon which subsequent RNA transformations depend. K227A, which had feeble Rnl2 adenylation activity, formed only trace amounts of ligated RNA that required prolonged exposure of the gel to see (not shown).
Figure 6.
Mutational effects on RNA strand joining. Reaction mixtures contained 50 mM Tris⋅acetate (pH 6.5), 5 mM DTT, 5 mM MgCl2, 1 pmol of pRNA substrate, 80 ng of WT or mutant Rnl2, and either 1 mM ATP (+) or no ATP (−). The products were resolved by PAGE and visualized by autoradiography. 32P-labeled DNA markers (18-mer, 36-mer, and 60-mer) were run in lane M.
H37A, which was fully active in step 1 ligase adenylation, was capable of transferring the adenylate to the 5′ PO4 of RNA to form RNA-adenylate (in the presence or absence of ATP) but was selectively impaired at the step of phosphodiester bond formation (Fig. 6). The introduction of Asp at position 37 rectified the strand-joining defect of the H37A mutant. The spectrum of H37D reaction products resembled that of WT Rnl2 with regard to the predominance of the RNA-adenylate in the presence of ATP and of the circular monomer in the absence of ATP (Fig. 6). These findings show that functional groups within the nucleotidyl transferase motifs of Rnl2 play essential roles at different steps of the RNA ligase reaction pathway.
Discussion
T4 Encodes a Second RNA Ligase.
We have shown here that Rnl2, the product of T4 gene 24.1, is an RNA-specific polynucleotide ligase that catalyzes intramolecular and intermolecular RNA strand joining through ligase-adenylate and RNA-adenylate intermediates. Cyclization of an 18-mer RNA was the favored outcome of the Rnl2 strand-joining reaction, rather than formation of linear multimers. This preference likely reflects proximity of the intramolecular 3′ OH terminus to the active site. The same preference for cyclization is displayed by T4 Rnl1 with substrates of similar size (10). RNA-adenylate was the predominant product formed by Rnl2 in the presence of 1 mM ATP, an effect likely caused by release of the enzyme from the step 2 product and its immediate reaction with ATP to generate ligase-adenylate, as discussed above. Rnl2 also formed high levels of the RNA-adenylate intermediate in the absence of ATP when the reaction conditions were shifted to more acidic pH (4.5–5.0). Similar effects are observed for E. coli and T4 DNA ligases, whereby the normally fleeting DNA-adenylate intermediate can be trapped at acidic pH (29). Such findings suggest that the chemical mechanism of phosphodiester bond formation is generally similar in RNA and DNA ligases, even though their substrate specificities (RNA vs. DNA, single-strand vs. double-strand) are different.
To understand the structural requirements for RNA ligation, we initiated an Ala-scanning mutational analysis of selected residues in the nucleotidyl transferase motifs of Rnl2. Our results implicate motif I residues Lys-35 and His-37 as essential catalysts of step 1 (ligase adenylation) and step 3 (phosphodiester synthesis), respectively. The requirement for Lys-35 of Rnl2 for overall ligation, and step 1 in particular, is consistent with mutational data for the equivalent lysine of T4 Rnl1 (18). Thus, we infer that Lys-35 is the site of AMP attachment to Rnl2. The requirement for Rnl2 motif I residue His-37 is intriguing, given that most other polynucleotide ligases have an Asp at this position. The H37A mutation caused a specific block to step 3 (without any apparent impact on step 1) that resulted in accumulation of RNA adenylate, from which we surmise that His-37 is not essential for catalysis of step 2. Strand-joining activity is restored by the H37D substitution, attesting to the functional flexibility of this position of motif I in Rnl2. It is noteworthy that whereas conservative mutations of the motif I aspartate of T4 Rnl1 (to Asn, Ser, or Glu) also had no effect on step 1, such changes abolished step 2 and step 3 of the ligation pathway (18). The motif I Asp is essential for step 2 catalysis by Chlorella virus DNA ligase and is also implicated in step 3 (30, 31). It would seem that Rnl2 is less reliant on this motif I side chain for catalysis of step 2 than is Rnl1 and DNA ligase.
Here, we provide evidence that conserved residues in motifs IV and V are essential for the activity of an RNA ligase. The corresponding side chains were shown to be essential for the activities of DNA ligases (31, 32) and mRNA capping enzymes (33, 34). The present results hint that the structural basis for nucleotidyl transfer is at least partially conserved among RNA ligases, DNA ligases, and mRNA capping enzymes. Such conservation is consistent with the speculation that RNA-joining enzymes that evolved during a primordial RNA/protein world are the ancestors of present-day DNA ligases and mRNA capping enzymes (20).
Rnl2 Exemplifies an RNA Ligase Family Found in All Phylogenetic Domains.
Although functional groups in motifs I, IV, and V are essential for catalysis by Rnl2, there is little overall conservation of primary structure between Rnl2 and the family of RNA ligases that includes T4 Rnl1 and fungal tRNA ligases. Instead, Rnl2 belongs to a distinct class of RNA ligases that includes the RNA editing ligases of kinetoplastid protozoa (Fig. 1A). A BLAST search for additional proteins related to T4 Rnl2 revealed a previously uncharacterized family of putative Rnl2-like ligases encoded by archaea and eukaryotic DNA viruses (baculoviruses and a poxvirus; Fig. 1). The additional members of the expanded Rnl2-like ligase family all contain nucleotidyl transferase motifs I, III, IIIa, IV, and V. The distinctive and defining feature of this family is the strict conservation of an asparagine located 5 amino acids downstream of the motif I lysine. Motif I in this family adheres to a consensus sequence Kx[H,D,N]GxN.
The eukaryotic viral branch of the Rnl2-like family is typified by the HE65 protein of the Autographa californica nuclear polyhedrosis virus (NPV). AcNPV is the prototype of the baculoviruses, which are nuclear DNA viruses that infect diverse arthropod hosts (35). Other baculoviruses that encode an Rnl2-like protein include Heliocoverpa armigera NPV, Bombyx mori NPV, and Xestia c-nigrum granulovirus (Fig. 1A). Little is known about the 553-aa baculovirus HE65 protein, except that its gene is transcribed at early times during virus infection. A 524-aa Rnl2-like protein is also encoded by the Amsacta moorei entomopoxvirus (AmEPV), a cytoplasmic poxvirus that infects caterpillars (36). It is notable that Rnl2-like putative ligases in the eukaryotic domain are apparently restricted to viruses (baculo and entomopox) and protozoa (Trypanosoma and Leishmania) that spend all or part of their life cycles in an arthropod host. We speculate that lateral gene transfer within the athropod vector may have played a role in the spread of the Rnl2-like ligases among these seemingly unrelated pathogens.
Rnl2-like proteins are distributed widely among archaeal proteomes, including those of Methanobacterium thermoautotrophicum, Methanococcus jannaschii, Pyrococcus horikoshii, Pyrococcus abyssi, Archaeoglobus fulgidus, and Aeropyrum pernix (Fig. 1B), as well as Methanosarcina acetivorans, Methanosarcina mazei, and Methanopyrus kandleri (not shown). The archaeal Rnl2-like protein are of fairly uniform size (369–395 aa) and are significantly smaller than the archaeal ATP-dependent DNA ligases (37), which vary from 556 to 619 aa in length.
Possible Biological Roles of Rnl2 and Rnl2-Like Proteins.
We can deduce indirectly from the deletion analysis of gene 24 by Engman and Kreuzer (38) that the neighboring gp24.1 gene encoding Rnl2 is nonessential for phage replication in a standard laboratory E. coli B strain. The RNA-joining activity of Rnl1 (gp63) is also not essential for phage replication in E. coli B, but is required, together with Pnk, to reverse the tRNALys restriction defense mounted by bacteria carrying the prr locus. Different bacterial strains in the wild may carry different tRNA restriction systems against which Rnl2 is specifically suited to act. Alternatively, Rnl2 may participate in RNA-editing reactions akin to those performed by the Rnl2-like enzymes of trypanosomes.
The coexistence of two RNA ligases in the same organism, with complementary functions in related biological pathways, has clear precedent in T. brucei, where the TbMP52 RNA ligase activity is specifically required for the editing pathway in which uridylates are deleted from pre-mRNAs (39). The second ligase TbMP48 is implicated in the pathway of uridylate insertion (39). The AcNPV baculovirus is another case where two (putative) RNA ligases inhabit the same biological niche. One of the AcNPV RNA ligases (the Orf86 product) resembles T4 Rnl1, whereas the second baculovirus RNA ligase resembles T4 Rnl2. Given that the AcNPV Orf86 protein seems to be a fusion of two enzymatic domains corresponding to T4 Pnk and T4 Rnl1, we speculate that the RNA repair enzyme system of bacteriophage T4 is recapitulated in its entirety in AcNPV. To extend the analogy further, we wonder whether eukaryotic organisms can respond to virus infection by triggering scission of essential RNAs, to which the virus must respond by catalyzing their repair.
Archaea have a clear need for an RNA ligase to participate in the removal of introns from their tRNAs. Archaea do not encode a recognizable homolog of yeast tRNA ligase, even though they do possess a tRNA splicing endonuclease that resembles the fungal endonuclease (3). The Rnl2-like proteins of archaea are plausible candidates for a ligase component of the archaeal tRNA splicing machinery.
Supplementary Material
Abbreviation
- NPV
nuclear polyhedrosis virus
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Silber R, Malathi V G, Hurwitz J. Proc Natl Acad Sci USA. 1972;69:3009–3013. doi: 10.1073/pnas.69.10.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amitsur M, Levitz R, Kaufman G. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abelson J, Trotta C R, Li H. J Biol Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- 4.Gonzales T N, Sidruaski C, Dörfler S, Walter P. EMBO J. 1999;18:3119–3132. doi: 10.1093/emboj/18.11.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Q, Teplow D, Lee T D, Abelson J. Biochemistry. 1990;29:6132–6138. doi: 10.1021/bi00478a004. [DOI] [PubMed] [Google Scholar]
- 6.Schnaufer A, Panigrahi A K, Panicucci B, Igo R P, Salavati R, Stuart K. Science. 2001;291:2159–2162. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- 7.McManus M T, Shimamura M, Grams J, Hajduk S L. RNA. 2001;7:167–175. doi: 10.1017/s1355838201002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rusche L N, Huang C E, Piller K J, Hemann M, Wirtz E, Sollner-Webb B. Mol Cell Biol. 2001;21:979–989. doi: 10.1128/MCB.21.4.979-989.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cranston J W, Silber R, Malathi V G, Hurwitz J. J Biol Chem. 1974;249:7447–7456. [PubMed] [Google Scholar]
- 10.Sugino A, Snopek T J, Cozarelli N R. J Biol Chem. 1978;252:1732–1738. [PubMed] [Google Scholar]
- 11.Uhlenbeck O C, Gumport R I. Enzymes. 1982;15:31–58. [Google Scholar]
- 12.Amitsur M, Morad I, Kaufmann G. EMBO J. 1989;8:2411–2415. doi: 10.1002/j.1460-2075.1989.tb08371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penner M, Morad I, Snyder L, Kaufmann G. J Mol Biol. 1995;249:857–868. doi: 10.1006/jmbi.1995.0343. [DOI] [PubMed] [Google Scholar]
- 14.Kaufmann G. Trends Biochem Sci. 2000;25:70–74. doi: 10.1016/s0968-0004(99)01525-x. [DOI] [PubMed] [Google Scholar]
- 15.Tomita K, Ogawa T, Uozumi T, Watanabe K, Masaki H. Proc Natl Acad Sci USA. 2000;97:8278–8283. doi: 10.1073/pnas.140213797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snopek T J, Wood W B, Conley M P, Chen P, Cozarelli N R. Proc Natl Acad Sci USA. 1977;74:3355–3359. doi: 10.1073/pnas.74.8.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thogerson H C, Morris H R, Rand K N, Gait M J. Eur J Biochem. 1985;147:325–329. doi: 10.1111/j.1432-1033.1985.tb08753.x. [DOI] [PubMed] [Google Scholar]
- 18.Heaphy S, Singh M, Gait M J. Biochemistry. 1987;26:1688–1696. doi: 10.1021/bi00380a030. [DOI] [PubMed] [Google Scholar]
- 19.Shuman S, Schwer B. Mol Microbiol. 1995;17:405–410. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. [DOI] [PubMed] [Google Scholar]
- 20.Shuman S. Prog Nucleic Acid Res Mol Biol. 2000;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- 21.Subramanya H S, Doherty A J, Ashford S R, Wigley D B. Cell. 1996;85:607–615. doi: 10.1016/s0092-8674(00)81260-x. [DOI] [PubMed] [Google Scholar]
- 22.Håkansson K, Doherty A J, Shuman S, Wigley D B. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 23.Odell M, Sriskanda V, Shuman S, Nikolov D. Mol Cell. 2000;6:1183–1193. doi: 10.1016/s1097-2765(00)00115-5. [DOI] [PubMed] [Google Scholar]
- 24.Durantel D, Croizier L, Ayres M D, Croizier G, Possee R D, Lopez-Ferber M. J Gen Virol. 1998;79:629–637. doi: 10.1099/0022-1317-79-3-629. [DOI] [PubMed] [Google Scholar]
- 25.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 26.Shuman S. Biochemistry. 1995;34:16138–16147. doi: 10.1021/bi00049a029. [DOI] [PubMed] [Google Scholar]
- 27.Ho C K, Shuman S. Proc Natl Acad Sci USA. 2001;98:3050–3055. doi: 10.1073/pnas.061636198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sriskanda V, Shuman S. Nucleic Acids Res. 1988;26:4618–4625. doi: 10.1093/nar/26.20.4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harvey C L, Gabriel T F, Wilt E M, Richardson C C. J Biol Chem. 1971;246:4253–4530. [PubMed] [Google Scholar]
- 30.Sriskanda V, Shuman S. Nucleic Acids Res. 1998;26:525–531. doi: 10.1093/nar/26.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sriskanda V, Shuman S. Nucleic Acids Res. 2002;30:903–911. doi: 10.1093/nar/30.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sriskanda V, Shuman S. J Biol Chem. 2002;277:9661–9667. doi: 10.1074/jbc.M110613200. [DOI] [PubMed] [Google Scholar]
- 33.Shuman S, Liu Y, Schwer B. Proc Natl Acad Sci USA. 1994;91:12046–12050. doi: 10.1073/pnas.91.25.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S P, Deng L, Ho C K, Shuman S. Proc Natl Acad Sci USA. 1997;94:9573–9578. doi: 10.1073/pnas.94.18.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ayres M D, Howard S C, Kuzio J, Lopez-Ferber M, Possee R D. Virology. 1994;202:586–605. doi: 10.1006/viro.1994.1380. [DOI] [PubMed] [Google Scholar]
- 36.Bawden A L, Glassberg K J, Diggans J, Shaw R, Farmerie W, Moyer R W. Virology. 2000;274:120–139. doi: 10.1006/viro.2000.0449. [DOI] [PubMed] [Google Scholar]
- 37.Sriskanda V, Kelman Z, Hurwitz J, Shuman S. Nucleic Acids Res. 2000;28:2221–2228. doi: 10.1093/nar/28.11.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engman H W, Kreuzer K N. Gene. 1993;123:69–74. doi: 10.1016/0378-1119(93)90541-a. [DOI] [PubMed] [Google Scholar]
- 39.Huang C E, Cruz-Reyes J, Zhelonkina A G, O'Hearn S, Wirtz E, Sollner-Webb B. EMBO J. 2001;20:4694–4704. doi: 10.1093/emboj/20.17.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.