Abstract
Conventional kinesin has a double-headed structure consisting of two motor domains and moves processively along a microtubule using the two heads cooperatively. The movement of single and multiple truncated heads of Drosophila kinesin was measured using a laser trap and nanometer detecting apparatus. Single molecules of single-headed kinesin bound to the microtubules with a 3.5 nm biased displacement toward the plus end of the microtubule. The position of these single-headed kinesin molecules bound to a microtubule did not change until they had dissociated, indicating that single kinesin heads utilize nonprocessive movement processes. Two molecules of single-headed kinesin moved continuously along a microtubule with a lower velocity and force than that of single molecules of double-headed kinesin. The biased binding of the heads determines the directionality of movement, whereas two molecules of single-headed kinesin move continuously without dissociation from a microtubule.
INTRODUCTION
Conventional kinesin is an intracellular vesicle transporter. Kinesin forms homodimers with two motor heads (Bloom et al., 1988; Kuznetsov et al., 1988). The double-headed kinesin moves processively along a microtubule via the cooperative interaction of both heads by utilizing the energy of adenosine 5′-triphosphate (ATP) hydrolysis (Howard et al., 1989). The processivity of single molecules of kinesin ensures high-energy efficiency of vesicle transport and economical protein usage. The unidirectional processive movement has been explained by a hand-over-hand mechanism in which the two heads step alternatively (Asbury et al., 2003; Higuchi et al., 2004; Howard, 2001; Kaseda et al., 2003; Yildiz et al., 2004). It is difficult to distinguish the role of each head during the movement of double-headed kinesin. The movement of each individual head has not been elucidated.
Single-headed conventional kinesin (SHC-kinesin) was genetically constructed using a combination of two previously reported methods (Berliner et al., 1995; Hancock and Howard, 1998). In one method the SHC-kinesin molecules were prepared as monomer heads by truncating the tail section so it did not form the coiled-coil (Berliner et al., 1995). The other method utilized heterodimers by coexpressing full length kinesin with the kinesin tails minus the head portion (Hancock and Howard, 1998). The velocity of the SHC-kinesins was lower than that of double-headed kinesin (Hancock and Howard, 1998; Inoue et al., 2001; Young et al., 1998). The distance the microtubule moved on the SHC-kinesin molecules became shorter as the number of SHC-kinesin molecules interacting with the microtubules in the in vitro motility assay was reduced (Berliner et al., 1995; Hancock and Howard, 1998; Young et al., 1998). This result indicates that the processivity of SHC-kinesin is very low or nonexistent. Hancock and Howard (1998) suggested that 4–6 molecules of SHC-kinesin are needed for continuous movement along a microtubule for distances >300 nm without detachment in an in vitro motility assay. In contrast, Inoue et al. (2001) showed that single molecules of SHC-kinesin move processively along microtubules using a fluorescent imaging technique with video resolution. To clearly show whether single molecules of SHC-kinesin moved processively along a microtubule, recordings at a high spatial resolution were required.
Monomeric motors of the kinesin superfamily, KIF1A or Unc104, provide useful information about the behavior of motor heads of conventional kinesin when interacting with a microtubule, and vice versa. Multiple molecules of KIF1A and Unc104 exhibited continuous rapid movement (Okada et al., 2003; Tomishige et al., 2002). The term ‘processive’ has been adopted for the movement of individual motor proteins and ‘continuous’ for the movement of ensembles of motors. Okada and Hirokawa (1999) also showed that single molecules of KIF1A moved processively along a microtubule. Processive movement of KIF1A is enhanced by the electrostatic interaction between the K-loop and the microtubule because the K-loop deleted KIF1A did not move processively beyond the spatial resolution of the image analysis system of ∼50 nm (Okada and Hirokawa, 1999, 2000). KIF1A bound to the microtubule moved toward the plus end of the microtubule with a displacement of 2.8 nm (Okada et al., 2003). The direction of the processive movement of KIF1A is determined by the biased binding. Measuring the displacement of the biased binding of single SHC-kinesin molecules is an important issue to address.
The unidirectional processive movement of single molecules of double-headed kinesin and multiple molecules of monomeric kinesin is an important process used to translocate large vesicles along a microtubule in a cell. To understand the movement of single-headed and double-headed kinesin, we evaluated the movement of single and multiple molecules of SHC-kinesin with nanometer accuracy using a laser trap. Single molecules of SHC-kinesin bound to a microtubule with a definite bias for the plus end of the microtubule and dissociated from a microtubule without exhibiting processive movement. There was no significant displacement of SHC-kinesin molecules bound to the microtubules. Two molecules of SHC-kinesin moved continuously with lower force and velocity than single molecules of double-headed kinesin.
MATERIALS AND METHODS
Protein
SHC-kinesin and double-headed kinesin consist of 351 and 411 amino acid residues from the N-termini of Drosophila kinesin with biotin carboxyl career protein (BCCP) fused at the C-termini. The expression and purification systems have been reported previously by Iwatani and co-workers (Berliner et al., 1995; Huang and Hackney, 1994; Iwatani et al., 1999). Tubulin was purified from porcine brain and the microtubules were labeled with tetramethylrhodamine succinimidyl ester (Molecular Probes, Eugene, OR). The minus end of the microtubules was marked with a higher concentration of rhodamine-labeled tubulin (Howard and Hyman, 1993).
Laser trap nanometry
SHC-kinesin molecules were bound to the polystyrene beads (0.2 μm diameter, Molecular Probes, Eugene, OR) via streptavidin (Sigma, St. Louis, MO; Inoue et al., 1997) and were trapped by an infrared laser (λ = 1064 nm) positioned near a fluorescence-labeled microtubule. Details of the laser trap system and apparatus have been reported previously (Nishiyama et al., 2001; Svovoda et al., 1993). All assays were performed using a solution containing 25 mM K-Acetate, 1 mM EGTA, 4 mM MgCl2, 10 μM Taxol, 0.12 mg/ml casein, and 20 mM K-HEPES (pH 7.2) with an added oxygen scavenger system (0.14 M 2-mercaptoethanol, 20 mM glucose, 20 μg/ml catalase, 100 μg/ml glucose oxydase) at 25°C ± 1°C.
Data analysis
The bead displacements were recorded at a sampling rate of 20 kHz with a bandwidth of 10 kHz. The force produced by SHC-kinesin was calculated from the bead displacement multiplied by the trap stiffness (35 fN/nm) that was determined from the variance of the thermal fluctuations of a trapped bead by the equipartion of thermal theory (Kojima et al., 1997). The velocity, taking into account the attenuation factor, was derived from the low pass filter at 5 Hz. The attenuation factor for correcting the displacement of the SHC-kinesin fragments was evaluated from (Kt + Kp)/Kp, where Kt is the stiffness of the optical trap and Kp is that of the bead-to-glass linkage which is the series of linkages of a bead, the SHC-kinesin molecules, a microtubule, and the surface of a glass slide (Kojima et al., 1997; Svoboda et al., 1993). The total stiffness, Kp + Kt of the linkage and the trap, was also calculated from the variance of the bead fluctuations when kinesin interacted with the microtubule.
The displacement at each binding event of single SHC-kinesin molecules to the microtubule was calculated from the displacement multiplied by the attenuation factor. The attenuation factor, 1.72 ± 0.02 (mean ± SE, n = 162), at the displacement of −5 to 5 nm was not a significant difference from 1.69 ± 0.02 (n = 122) at the displacement of −10 to −5 and 5 to 10 nm before attenuation. This factor decreased slightly to 1.62 ± 0.02 (n = 116) at <−10 and >10 nm before attenuation, or <−17 and >17 nm after attenuation.
The velocity of each displacement trace was derived from the slope of the displacement which had been smoothed by a low pass filter of 5 Hz and multiplied by the attenuation factor. The average values of the attenuation factors during continuous movement were 1.7 at low loads of ∼1 pN and 1.2 at high loads of >5 pN.
RESULTS
Minimum number of SHC-kinesin molecules required for continuous movement
To make the SHC-kinesin-beads, various concentrations of SHC-kinesin were mixed with a constant concentration of the polystyrene beads, 0.2 μm in diameter, in a solution (∼0.01%, v/v or ∼60 pM). Beads coated with SHC-kinesin were trapped using optical tweezers and then placed on a microtubule. A reduction in Brownian motion of an SHC-kinesin-bead was considered to be an interaction with a microtubule (Veigel et al., 1999). The ratio of the number of SHC-kinesin-beads interacting with the microtubules to the total number of beads observed at each concentration of SHC-kinesin was determined. Interaction times shorter than 50 ms have not been included to avoid any artifacts resulting from the binding of molecules other than the SHC-kinesin molecules to a microtubule.
In the presence of 1 mM adenosine 5′-(β-γ-imido)triphosphate (AMP-PNP), SHC-kinesin-beads bound to a microtubule for a long time, i.e., for more than several seconds (Fig. 1 a). At low concentrations of SHC-kinesin and in the presence of ATP (6 μM), SHC-kinesin-beads bound to and dissociated from a microtubule repeatedly without exhibiting continuous movement (Fig. 1 b). Fig. 1, c and d, shows the continuous movement of SHC-kinesin-beads at high and low ATP concentrations.
FIGURE 1.
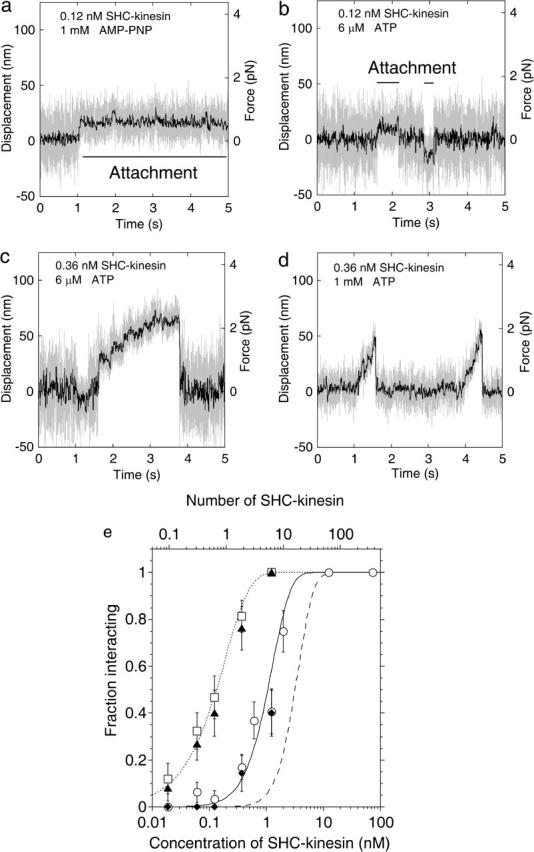
Behavior of the SHC-kinesin. (a–d) Force generation of SHC-kinesin. Data without low pass filter (shaded traces) and having passed through a 50 Hz low pass filter (black traces). Attachment in the presence of 1 mM AMP-PNP (a) and sequential attachment and detachment at 6 μM ATP (b) at SHC-kinesin concentration of 0.12 nM. Continuous movement at 6 μM (c) and 1 mM ATP (d) at SHC-kinesin concentration of 0.36 nM. (e) The ratio of the SHC-kinesin-beads interacting with a microtubule to all beads measured is shown as a fraction of those interacting (vertical axis). In this study, an interacting event is when the variance decreased to half its initial value for longer than 50 ms. Rectangles indicate the ratio of the beads binding to microtubules at 1 mM AMP-PNP. Triangles indicate the ratio of the beads interacting with microtubules at 6 μM ATP. The average number of SHC-kinesin molecules on a bead was calculated by fitting the Poisson distribution (dotted curve) to the data at 1 mM AMP-PNP and is shown on an upper abscissa. The ratios of the beads moving continuously at 1mM ATP and 6 μM ATP are shown in the circles and diamonds, respectively. Error bars are expressed as ±(p(1 − p)/n)1/2, where p is the fraction interacting and n is the number of the measuring beads (n = 19 ∼ 42 at each point; Svoboda and Block, 1994). The moving ratios were simulated when two or more (solid curve) or three or more (dashed curve) SHC-kinesin molecules on a bead interacted simultaneously with a microtubule (see Appendix).
In the presence of AMP-PNP, single molecules of SHC-kinesin on the beads should bind to a microtubule for long periods of time because of the strong binding (Endow and Higuchi, 2000; Huang and Hackney, 1994). Thus, the ratio of the interacting beads to the total number of beads indicates the ratio of the beads to which one or more molecules of SHC-kinesin had bound to a microtubule. Assuming SHC-kinesins are distributed randomly on the beads, the number of molecules on the bead can be described by a Poisson distribution (Svoboda and Block, 1994). According to the Poisson distribution, the probability that there are one or more SHC-kinesin molecules on a bead can be described by the curve, 1 − exp(−C/C0), where C is the concentration of SHC-kinesin (nM) and C0 is the concentration when the average number of SHC-kinesin molecules on a bead is one (SHC-kinesin concentration of 0.2 nM). The ratio of the beads interacting with a microtubule increased with an increase in C (Fig. 1 e, rectangles on the dotted line). Therefore, the average number of SHC-kinesins bound to a bead assuming the Poisson distribution of the molecules on beads can be calculated (Fig. 1 e, upper horizontal axis).
At ATP concentrations of 6 μM, the ratio of the beads interacting with a microtubule was almost the same as that of the beads in the presence of AMP-PNP (Fig. 1 e, triangles and rectangles). At low ATP and low SHC-kinesin concentrations (<∼1 nM), the ratio of the beads moving continuously was considerably lower than that of the beads interacting with a microtubule (Fig. 1 e, triangles and diamonds). This indicates that single molecules of SHC-kinesin bind to and dissociate from a microtubule without processive movement, although multiple molecules can move continuously.
In the presence of 1 mM ATP, the ratio of the beads interacting with a microtubule was lower than that of the beads in the presence of 6 μM ATP (Fig. 1 e, triangles and circles). The time from binding to dissociation (the cycle time) was too short to detect and is discussed later. The ratio of the beads interacting with a microtubule was almost the same as that of the beads moving continuously in the presence of 6 μM ATP (Fig. 1 e, circles and diamonds).
The minimum number of SHC-kinesin molecules required for the continuous movement on a bead was determined by comparing the ratio of the beads moving continuously with the curves obtained from a simulation model. In this model, the distances among the SHC-kinesin molecules were calculated by considering the geometry of the spherical bead and SHC-kinesin construct and testing whether molecules on the bead interacted simultaneously with the microtubule (see Appendix). The ratio of the continuous movement in the presence of 1 mM ATP (circles in Fig. 1 e) and the number of interacting molecules was in good agreement with the solid curve (Fig. 1 e), which shows that two or more molecules of SHC-kinesin simultaneously interact with a microtubule. The data, however, could not be fitted to the curve that describes three or more molecules of SHC-kinesin interacting with the microtubule (dashed curve in Fig. 1 e). This result suggests that two molecules of SHC-kinesin are able to support continuous movement.
Binding and dissociation of SHC-kinesin
Single molecules of SHC-kinesin bound to and dissociated from a microtubule without processive movement (Figs. 1 b and 2 a). The dwell time and the directionality when SHC-kinesin bound to a microtubule were analyzed. The event where the noise (the variance of displacement) was reduced to less than half for >50 ms was considered to be a binding event (Fig. 2 a). Time (tB) from the binding to the dissociation was measured to determine the binding time (Fig. 2 a). The histograms of tB showed an exponential decay with a time constant of 99 ms at 6 μM ATP and 380 ms at 1.2 μM ATP (Fig. 2 b).
FIGURE 2.
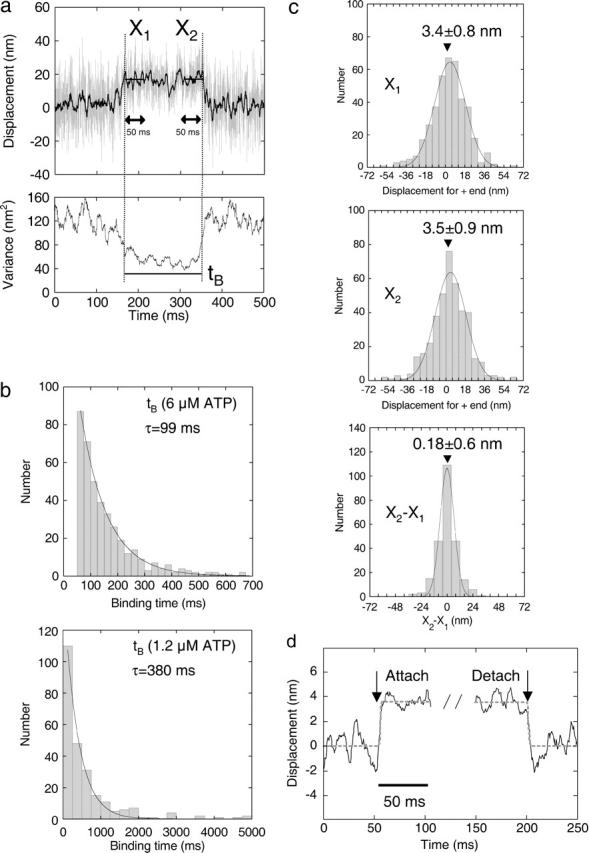
Analysis of the biased binding at 6 μM ATP. (a) The time course of the displacement and its variance of SHC-kinesin-beads are shown in the top and bottom panels, respectively. X1 and X2 indicate the average displacements from the trap center for 50 ms after binding and before dissociation. tB is binding time from the initial binding to dissociation. (b) The histograms of the binding time in the presence of 6 μM ATP (top, n = 400) and 1.2 μM ATP (bottom, n = 245), respectively. (c) The histograms of X1 (n = 400), X2 (n = 400), and X1 − X2 (n = 242 for tB > 100 ms) could be fitted to Gaussian curves with peaks at X1 = 3.4 (mean ± SE = 0.8) nm, X2 = 3.5 (mean ± SE = 0.9) nm, and X2 − X1 = 0.18 (mean ± SE = 0.6) nm. (d) Ensemble-averaged traces at binding and detachment by single molecules of SHC-kinesin. Each trace (through 200 Hz low pass filter) for 50 ms before and after the binding and detachment was fitted by rectangle curve represented as d = a ×[1 + exp{−4(t − b)}]−1 + c, where d is displacement, t is time, a is the step size, and b and c are fitting parameters. Black solid curves are the ensemble traces synchronized at the binding and the detachment, and shaded dashed curves are rectangle curves with step displacement of 3.5 ∼ 3.6 nm.
The histograms of the displacement are shown in Fig. 2 c. X1 and X2 show the mean displacement from the trap center for a 50 ms period after binding and before dissociation. This allowed the directionality of binding to be measured. The microtubule polarity was determined by marking the minus end with a strong fluorescence dye (Howard and Hyman, 1993). Microtubules with opposite directionalities were selected to minimize any artifacts associated with the shift. The minus ends of 50% of the microtubules were oriented to the upper region on the TV display and the other 50% to the lower region. The histograms of X1 and X2 show Gaussian distributions which have peaks biased to the plus end of the microtubule. The values of X1 and X2 were 3.4 ± 0.8 nm (mean ± SE, n = 400) and 3.5 ± 0.9 nm (n = 400), respectively. These values did not depend on the direction that the microtubules were oriented on the TV display; the displacements at the opposite directionalities of microtubules were 3.1 ± 1.1 and 3.8 ± 1.1 nm for X1 and 3.2 ± 1.2 and 3.8 ± 1.1 nm for X2. The average step size of each bead was 2–6 nm with a standard error of 2 nm (n = 100), indicating that no continuous movement had been included in the steps upon binding. As a result, single molecules of SHC-kinesin bound to a microtubule toward the plus end of the microtubule with a 3.5 nm bias. The displacement from X1 to X2, 0.18 ± 0.6 nm (n = 242, tB ≥ 100 ms), was not significantly different from zero (Fig. 2 c). Furthermore, the ensemble displacement was analyzed by synchronizing the traces at both binding and detachment (n = 71, tB > 100 ms; deCastro et al., 2000; Okada et al., 2003). Displacements were considered significant if they were >1.5 (±2 SD) nm within ∼3 ms after binding. No such displacements were observed. These results indicate that single molecules of SHC-kinesin bind to microtubules in the direction of the plus end and do not move during binding.
Movement of multiple molecules of SHC-kinesin
The ratio of the beads moving continuously for distances >20 nm after binding to a microtubule (>2 steps of 8 nm) increased with an increase in the concentration of SHC-kinesin. The continuous movement of the SHC-kinesin and the processive movement of double-headed kinesin at the various concentrations are shown in Fig. 3 a. The beads coated with SHC-kinesin moved continuously for 70–200 nm and then detached from the microtubule. The frequency of force generation increased as the SHC-kinesin concentration increased from 0.36 nM to 2.0 nM. At an SHC-kinesin concentration of 12 nM, the period when the beads dissociated from the microtubules decreased dramatically. Beads that detached from the microtubule quickly reattached to the microtubule before it had returned to the center of laser trap at zero force and then once again showed continuous movement. Single molecules of double-headed kinesin at a concentration of 0.1 nM moved processively for ∼200 nm and generated stall forces of 7–8 pN as reported previously (Inoue et al., 1997; Iwatani et al., 1999).
FIGURE 3.
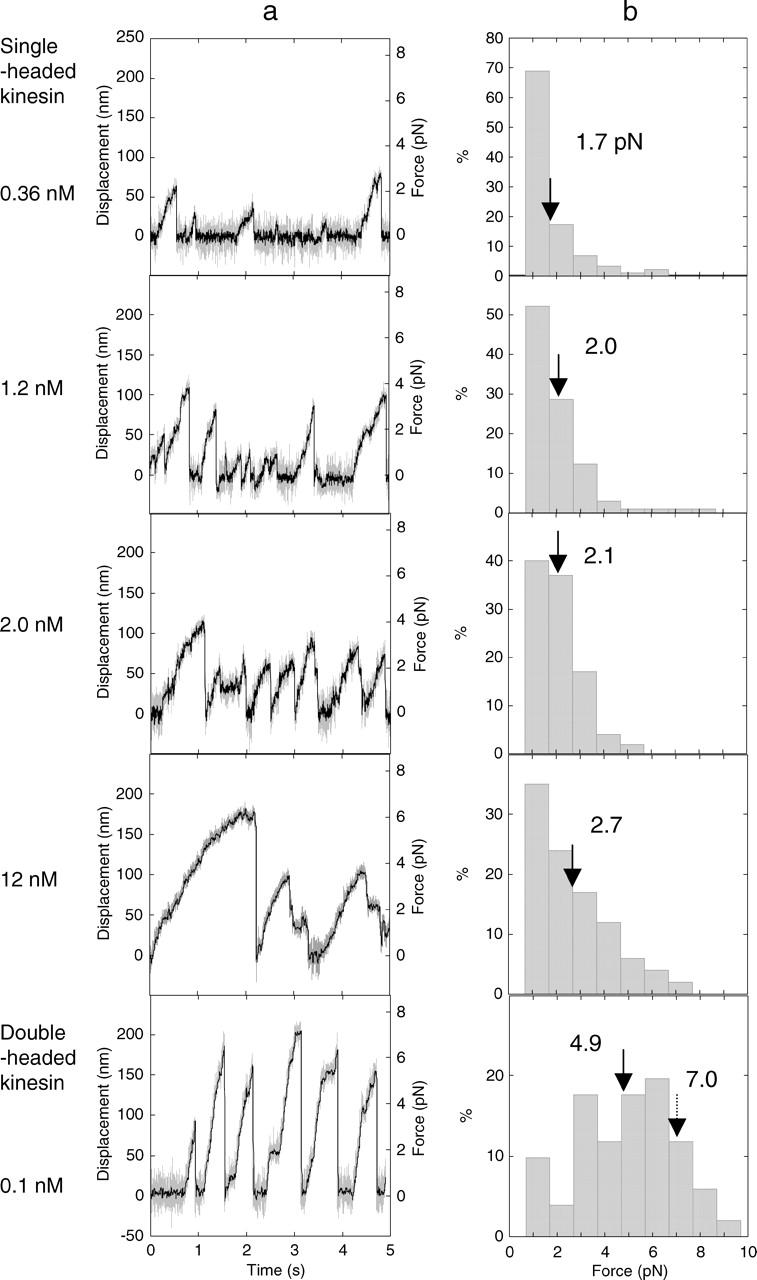
Force generation of the SHC-kinesin. (a) Force generation of SHC-kinesin at concentrations of 0.36, 1.2, 2.0, and 12 nM from top to bottom panels and that of single molecules of double-headed kinesin at concentration of 0.12 nM (lowest panel). (b) The histograms of the generated force of multiple molecules of SHC-kinesin and single molecules of double-headed kinesin. Solid arrows indicate the average forces, and broken arrow indicates the stall force.
The histograms of the force generated by SHC-kinesin and double-headed kinesin are shown in Fig. 3 b. The force was measured just before kinesin detached from the microtubules. The force population of SHC-kinesin in the histograms decreased almost exponentially with the increase in force. The mean force at an SHC-kinesin concentration of 0.36 nM was 1.7 pN, and this increased to 2.7 pN at a concentration of 12 nM. The force population of double-headed kinesin was increased and then decreased with an increase in the force (bottom in Fig. 3 b). The stall force, at which kinesin did not move >10 nm over a time frame of 100 ms, was 7 ∼ 8 pN. The mean force of 4.9 pN, however, was lower because the forces before movement ceased have been included.
The force-velocity relationships for two molecules of SHC-kinesin at <1.2 nM and for single molecules of double-headed kinesin are shown in Fig. 4 a. The highest 10% of forces (>7 pN) for double-headed kinesin which corresponded to the stall force recorded have been analyzed (Iwatani et al., 1999; Kojima et al., 1997). Therefore, the highest 10% of forces were also analyzed for SHC-kinesin. At low forces of ∼1pN, the velocity of SHC-kinesin was ∼400 nm/s. This value was approximately half of the velocity obtained for double-headed kinesin, ∼750 nm/s. The sliding velocity of double-headed kinesin decreased slightly with forces up to 3 pN and dramatically at forces >3 pN. The sliding velocity of SHC-kinesin decreased in an almost linear fashion.
FIGURE 4.
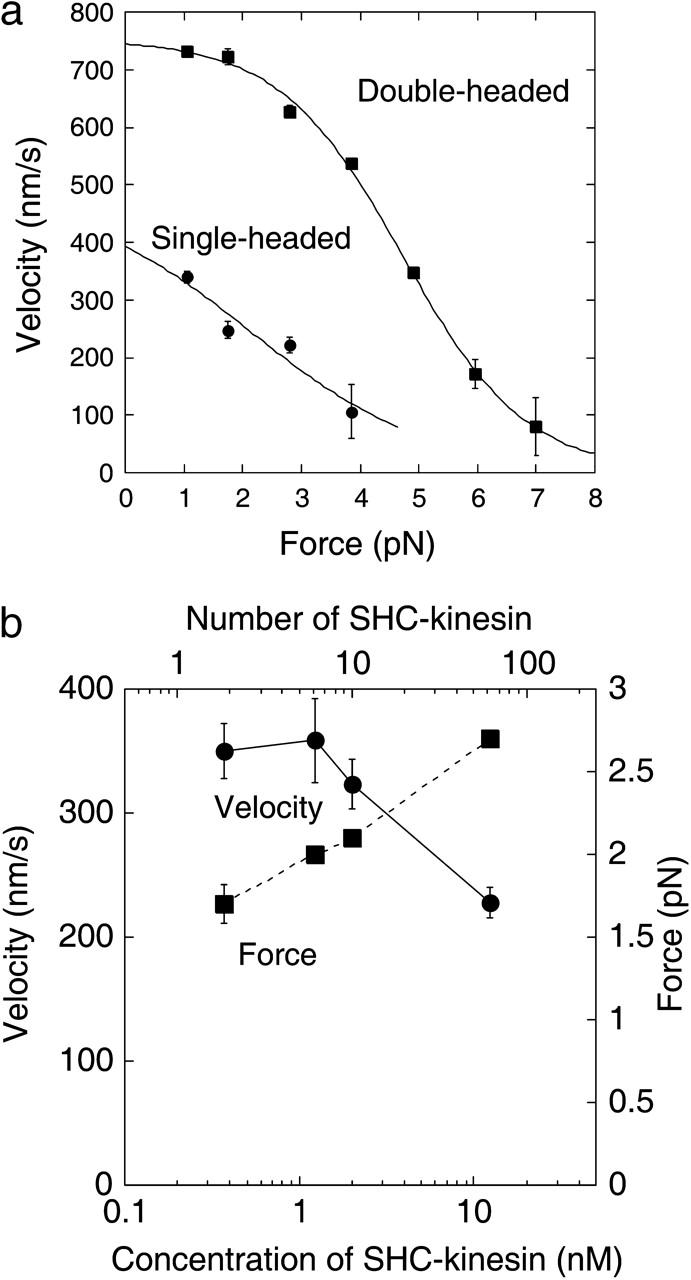
Movement of the SHC-kinesin. (a) Relationship between velocity and force of two SHC-kinesin molecules and single double-headed kinesin molecules are shown in circles (n = 21) and rectangles (n = 15), respectively. The curves could be well fitted to the equation, V = 8/{a + b × exp(kF)}, (V, velocity (nm/s); F, force (pN); a, b, and k are fitting parameters; Nishiyama et al., 2002). (b) Relationship between the number of SHC-kinesin molecules bound to a bead and velocity (circles) and force (rectangles), (n = 87 ∼ 100 at each point).
The force and velocity changed with the concentration of SHC-kinesin (Fig. 4 b). The mean force increased gradually from 1.7 to 2.7 pN with an increase in the concentration of SHC-kinesin from 0.36 nM to 12 nM. The velocity of SHC-kinesin was recorded at small forces of ∼1 pN. The velocity remained constant at ∼350 nm/s at SHC-kinesin concentrations between 0.36 and 2.0 nM and then decreased to ∼230 nm/s at a concentration of 12 nM.
DISCUSSION
Nonprocessive movement of single molecules of SHC-kinesin
In this study, the movement of single molecules of SHC-kinesin expressed as monomers (Berliner et al., 1995) was measured by the laser trap and nanometry system. Single and multiple molecules of SHC-kinesin bound to small beads, 0.2 μm in diameter, interacted with a microtubule. The position of the beads was detected with high spatiotemporal resolution to accurately determine the movement of SHC-kinesin.
The number of SHC-kinesin molecules bound to a bead was estimated using the Poisson distribution of the molecules on the bead. Single molecules of SHC-kinesin on a bead bound to and dissociated from a microtubule without undergoing processive movement. If single molecules of SHC-kinesin moved processively, the ratio of the beads moving continuously in the presence of ATP should be the same as that binding in the presence of AMP-PNP because single molecules of SHC-kinesin bind strongly to the microtubules in the presence of AMP-PNP. In this instance, the curve did not match the ratio of the beads moving continuously (Fig. 1 e, dotted curve, and diamonds and circles), indicating the lack of processive movement of single molecules of SHC-kinesin.
In the in vitro motility assay, the run lengths of the microtubules moving on SHC-kinesins bound to the glass slides became shorter and even too short to detect when low concentrations of SHC-kinesin were used (Hancock and Howard, 1998; Young et al., 1998). This result indicates that the processivity of single molecules of SHC-kinesin is very low or nonexistent. However, the video analysis system had a limited spatial resolution of ∼300 nm, thus the number of steps <38 (≈300 nm/8 nm) could not be counted (Hancock and Howard, 1998). In this study, only one step could be detected by using the optical tweezers apparatus with high spatial resolution. Steps longer than 8 nm in length, however, were not observed during the binding of SHC-kinesin to the microtubule. Using traces that had been averaged, there was no significant displacement from the binding to the dissociation of SHC-kinesin (0.18 ± 0.60 nm; Fig. 2 c). This result clearly indicates that single molecules of SHC-kinesin did not take any additional steps once they had bound to the microtubules. Thus, it appears that single molecules of SHC-kinesin bound to and dissociated from a microtubule upon the hydrolysis of ATP without undergoing any stepwise movement.
Biased binding and dissociation of SHC-kinesin
At low SHC-kinesin concentrations (<0.1 nM), beads were observed to bind to and then dissociate from a microtubule (Fig. 1 b) at low concentrations of ATP (Fig. 1 e, triangles and diamonds). At ATP concentrations of 6 and 1.2 μM, the time constants for binding were 99 and 380 ms, respectively. These values are in agreement with the cycle time of ATP hydrolysis by single molecules of SHC-kinesin which had been calculated using a kinetic parameter, Km=43 μM ATP, to be 101 and 460 ms, respectively (Huang and Hackney, 1994). The histograms of the binding time showed an exponential decay (Fig. 2 b), indicating that the hydrolysis of single ATP molecules could be approximately described by a first order reaction. This kinetic analysis supported the observation that the binding and dissociation would be performed within a single turnover of ATP by single molecules of SHC-kinesin. The binding of SHC-kinesin to a microtubule in the presence of 1 mM ATP was not observed because the binding time was too brief to detect (12 ms, estimated from the ATPase rate at 1mM ATP).
SHC-kinesin binds to a microtubule when in the ADP binding state because ADP release is a rate limiting step in the absence of microtubules (Hackney, 1995). After ADP release, SHC-kinesin binds strongly to a microtubule in a nucleotide free state. The SHC-kinesin and microtubule complex then hydrolyze ATP into ADP·Pi. After Pi is released, the SHC-kinesin in the ADP binding state dissociates from the microtubule because, in this state, it has a low affinity to the microtubules (Hackney, 1995). Thus, the binding, hydrolysis, and detachment of SHC-kinesin to a microtubule can be described as follows:
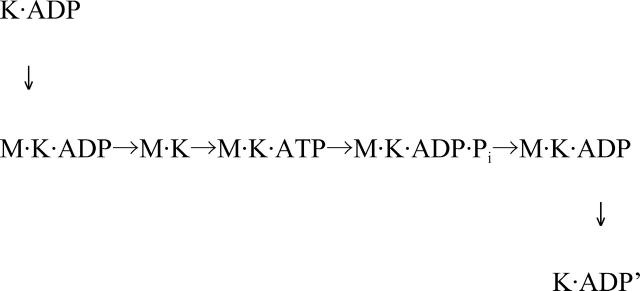 |
where M and K indicate a microtubule and an SHC-kinesin, respectively.
SHC-kinesin bound to a microtubule in the direction biased toward the plus end of 3.5 nm (Fig. 2 c). The 3.5-nm biased displacement was generated at the time of binding or within 30 ms of binding to a microtubule (Fig. 2 d). Since the ATP free state was calculated to be ∼100 ms at an ATP concentration of 6 μM from biochemical result (Huang and Hackney, 1994), the bias displacement occurred before the binding of ATP, that is, when it was bound to the microtubule or when ADP was released. The average 3.5 nm biased displacement was due to the translational movement of the SHC-kinesin head along a microtubule and an angular change of SHC-kinesin neck. Okada et al. (2003) evaluated the influence of the angular change of the neck linker of KIF1A by measuring the displacement of the beads bound to the N- and C-termini of KIF1A, respectively. The same relation between the steps and force of both the N- and C-termini indicates that the angular change of the neck linker did not affect the biased binding of KIF1A. The contribution of angular change will also be small on SHC-kinesin.
Continuous movement by multiple molecules of SHC-kinesin
The minimum number of SHC-kinesin molecules needed for continuous movement is two (Fig. 1 e). To move processively, it is essential that kinesin molecules have the directional displacement and do not dissociate from the microtubules. Directional movement by biased binding could be produced when two or more SHC-kinesin molecules attached to a bead. In this case, one molecule could produce the directional movement, whereas the other prevents the kinesin from dissociating from the microtubule (Tomishige et al., 2002). Inoue et al. (2001) observed the movement of single molecules of the fluorescently labeled SHC-kinesin with or without BCCP. They suggested that BCCP had some affinity for the microtubule preventing it from dissociating. In our experiment, it is possible that BCCP did not bind to the microtubule because the BCCP of SHC-kinesin at the C-termini was bound to a bead via streptavidin. As a result, the nonprocessive movement of single molecules of SHC-kinesin could be observed.
Both the velocity and force of two molecules of SHC-kinesin were lower than those of double-headed kinesin (Fig. 4 a). The reason for this reduction could be that the position and direction of SHC-kinesin binding to microtubules is not optimal because of the random binding to the beads. The probability that backward steps occurred (8–20 nm) was <6% for double-headed kinesin, and that increased to ∼20% for SHC-kinesin with a 1–2 pN load (data not shown). This increment in the frequency of backward movement would also contribute to the slower velocity.
Inoue et al. (1997) reported that the velocity and force of multiple molecules of SHC-kinesin in the absence of BCCP was comparable to that of double-headed kinesin. It has been suggested that SHC-kinesin attached to a small molecule of biotin, ∼0.5 nm in diameter, can bind to two sites on the single molecules of streptavidin, forming a pseudo-double-headed kinesin (Inoue et al., 1997). A BCCP of SHC-kinesin, ∼3 nm in diameter, should sterically block another SHC-kinesin binding to neighboring binding sites on an avidin because the distance between binding sites was ∼1 nm (Weber et al., 1989). Okada and colleagues suggested that the single-headed kinesin with BCCP on the avidin did not form a dimer because the Fab fragment of the antibody with a single binding site to kinesin had similar binding and movement ratios to those of streptavidin (Okada et al., 2003). In this study, two molecules of SHC-kinesin bound randomly to a bead. This resulted in the production of low forces and beads that moved at a slow velocity because the heads were not able to interact with the microtubule in the optimum orientation.
The force and the sliding velocity remained almost constant when the average number of SHC-kinesin molecules on a bead was between 1 and 10 (Fig. 4 b). The number of molecules interacting simultaneously with the microtubule was calculated to be ∼2 (occasionally 3) from the geometry of kinesin and the microtubules (Fig. 1 e, solid and dashed curves). Therefore, two molecules of SHC-kinesin produced a force of ∼2 pN in average.
When ∼60 molecules of SHC-kinesin were bound to a bead, the force increased and the velocity of sliding decreased (Fig. 4 b). The reason for the large forces is the number of molecules interacting simultaneously with a microtubule increases to >3. The decrease in velocity may be explained by an increase in the number of SHC-kinesin interacting with a microtubule in an unsuitable orientation and/or position. The slow velocity of the microtubules interacting with SHC-kinesin has also been observed in the motility assays by our lab (∼120 nm/s, data not shown) and others (Hancock and Howard, 1998; Inoue et al., 2001; Young et al., 1998). This result suggests that SHC-kinesin molecules interact with the microtubules in a less than optimum orientation which may inhibit the motility.
Model of the unidirectional movement by SHC-kinesin
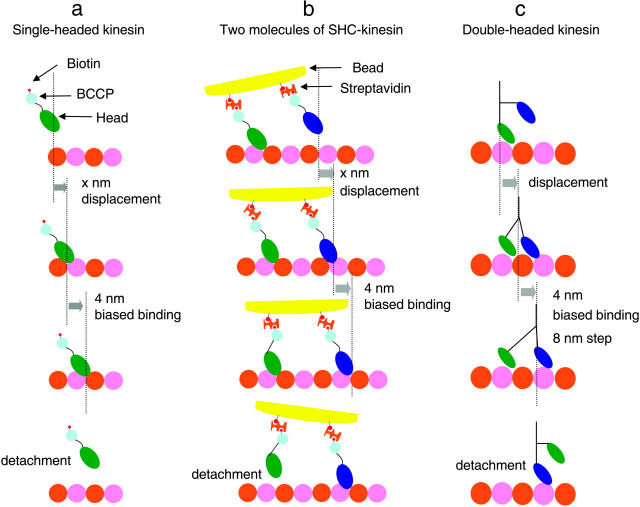
Single molecules of SHC-kinesin in the ADP binding state bind to tubulin with a displacement of x nm as a result of Brownian motion (Fig. 5 a). Then SHC-kinesin moves an average of ∼4 nm toward the plus end of the microtubule as biased binding. The SHC-kinesin and microtubule complex binds an ATP molecule to be hydrolyzed, and then SHC-kinesin in the ADP state dissociates from the microtubule.
FIGURE 5.
Movement models of SHC-kinesin and double-headed kinesin. (a) Single molecules of SHC-kinesin. (b) Two molecules of SHC-kinesin. (c) Single molecules of double-headed kinesin. Details are described in the text.
The movement of two molecules of SHC-kinesin begins with the state where the rear head binds to a microtubule. In contrast, the front head is in the ADP binding state and is positioned over the microtubule (Fig. 5 b). The front head attaches to the microtubule with displacement of x nm to the plus end. The displacements, x, of the heads are considered to be positive because of the neck linker docking to the head or structural changes that occur during ATP hydrolysis (Kikkawa et al., 2001; Rice et al., 1999). The front head then releases ADP and binds strongly to the microtubule. At the same time, the ∼4 nm biased displacement occurs to the plus end of the microtubule. Whereas the front head binds to a microtubule and produces force, the rear head is dissociated from the microtubule. The rear head then attaches to the microtubule with a displacement to the plus end as a result of the biased binding (data not shown). Finally, the front head dissociates from the microtubule once it is in the ADP binding state. Alternatively, the role of the front head could be exchanged with that of the rear head. Repetitions of these cycles generate the continuous movement. The regular 8 nm steps typically observed with double-headed kinesin were not observed for SHC-kinesin (data not shown). The distance between the heads and the SHC-kinesin interactions with the protofilaments most likely take steps of varying size but those other than 8 nm.
Double-headed kinesin motility begins with the state in which the front head with ADP attached is positioned over a microtubule and the rear head without nucleotides is bound to the microtubule (Hancock and Howard, 1999; Fig. 5 c). The front head attaches to a microtubule with a displacement toward the plus end of the microtubule. Either before or after the displacement, the single-head moves by ∼4 nm toward the plus end as biased binding and completes the 8 nm step, suggesting the existence of substeps (Nishiyama et al., 2001). The rear head is pulled forward by the front head and then dissociates from the microtubule (Uemura and Ishiwata, 2003). Repetitions of these cycles generate the unidirectional processive movement of 8 nm by the hand-over-hand mechanism (Asbury et al., 2003; Crevel et al., 1999; Higuchi et al., 2004; Kaseda et al., 2003; Yildiz et al., 2004).
According to our model, two molecules of SHC-kinesin and single molecules of double-headed kinesin move through the biased binding of single molecules of SHC-kinesin. Therefore, ‘bias binding’ is the key mechanism for the movement of motor proteins.
Acknowledgments
We thank Dr. N. Sasaki for helpful discussion and Dr. J. West for critically reading this manuscript. This work was supported by Grants-in-Aid for Scientific Research in Priority Areas from the Japan MEXT (H.H.).
APPENDIX
Model simulation for determining the minimum number of SHC-kinesin molecules required on a bead for continuous movement
The minimum number of SHC-kinesin molecules required for the continuous movement was determined using a model calculation, when n equals the number of molecules bound randomly on a bead, 200 nm in diameter. The probability, q(n, m), of n molecules bound on a bead out of an average number of molecules (m) was represented by mne−m/n! according to a Poisson distribution. A bead is a sphere, and the length between the BCCP-biotin and the microtubule binding site is relatively short (<10 nm, considering the configurations of SHC-kinesin and BCCP; Rice et al., 1999; Vale and Milligan, 2000). Thus, there is a limited distance between SHC-kinesin molecules, so this will limit the number of molecules that can simultaneously interact with a microtubule.
If the minimum number of molecules is two, the distances between every two molecules among n molecules can be calculated and the minimum distance between molecules, dmin, determined. The number of beads where dmin was shorter than the threshold distance d0 (nm) where two SHC-kinesin molecules were able to interact simultaneously with a microtubule were counted. The probabilities, p2(n, dmin < d0), are given by the number of the beads (dmin < d0) divided by the calculated number of beads (1000 trials at each n in this work). The total probability, P2(m) = Σq(n, m)·× p2(n, dmin < d0), n = 2–∞. The best value of d0 was obtained by fitting P2(m) to the experimental data to give a value of 48–62 nm (R = 0.98, 0.95 for d0 = 48, 62 nm). From this d0 value, the possible area where two SHC-kinesin molecules interact simultaneously with a microtubule was calculated to be 6%–10% of the total area of a bead. Thus, the minimum distance required for an SHC-kinesin to interact with a microtubule from the bead surface was calculated to be 3–5 nm. This is consistent with the result obtained for KIF1A (Okada et al., 2003).
The model simulation for the minimum number of three molecules is as follows. All combinations (nC3 ways) of three molecules out of a total of n molecules were selected. The number of beads on which the selected three molecules interacted simultaneously with a microtubule was counted. The probability, p3(n, d0, l0), where d0 (nm) is the threshold distance between SHC-kinesins and l0 (nm) is the width of microtubule, was given by the number of beads counted divided by the total number of the calculated beads (1000 trials at each n, in this work). The total probability, P3(m), that three molecules interacted simultaneously with a microtubule is represented by P3(m) = Σq(n) × p3(n, d0, l0), n = 3–∞. When l0 = 25 nm (diameter of microtubules) and d0 < 48–62 nm (the minimum distance for an SHC-kinesin to interact with a microtubule from the bead surface is 3–5 nm), P3(m) did not fit the experimental data. Even when d0 = 105 nm where the distance between the microtubule and the bead surface is 15 nm, i.e., longer than the length of SHC-kinesin, the simulated curve did not fit to the experimental data. These results indicate that the minimum number of molecules required for the continuous movement is two but not three.
Seiji Kakuta's present address is Material Technology Division of Aomori Industrial Research Center, Aomori 030-0113, Japan.
References
- Asbury, C. L., A. N. Fehr, and S. M. Block. 2003. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 302:2130–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berliner, E., E. C. Young, K. Anderson, H. K. Mahtani, and J. Gelles. 1995. Failure of a single-headed kinesin to track parallel to microtubule protofilaments. Nature. 373:718–721. [DOI] [PubMed] [Google Scholar]
- Bloom, G. S., M. C. Wagner, K. K. Pfister, and S. T. Brady. 1988. Native structure and physical properties of bovine brain kinesin and identification of the ATP-binding subunit polypeptide. Biochemistry. 27:3409–3416. [DOI] [PubMed] [Google Scholar]
- Crevel, I., N. Carter, M. Schliwa, and R. Cross. 1999. Coupled chemical and mechanical reaction steps in a processive Neurospora kinesin. EMBO J. 18:5863–5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deCastro, M. J., R. M. Fondecave, L. A. Clark, C. F. Schmidt, and R. J. Stewart. 2000. Working strokes by single molecules of the kinesin-related microtubule motor ncd. Nat. Cell Biol. 2:724–729. [DOI] [PubMed] [Google Scholar]
- Endow, S. A., and H. Higuchi. 2000. A mutant of the motor protein kinesin that moves in both directions on microtubules. Nature. 406:913–916. [DOI] [PubMed] [Google Scholar]
- Hackney, D. D. 1995. Highly processive microtubule-stimulated ATP hydrolysis by dimeric kinesin head domains. Nature. 377:448–450. [DOI] [PubMed] [Google Scholar]
- Hancock, W. O., and J. Howard. 1998. Processivity of the motor protein kinesin requires two heads. J. Cell Biol. 140:1395–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, W. O., and J. Howard. 1999. Kinesin's processivity results from mechanical and chemical coordination between the ATP hydrolysis cycles of the two motor domains. Proc. Natl. Acad. Sci. USA. 96:13147–13512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, H., C. E. Bronner, H. W. Park, and S. A. Endow. 2004. Rapid double 8-nm steps by a kinesin mutant. EMBO J. 23:2993–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, J. 2001. Mechanics of Motor Proteins and the Cytoskeleton. Sinauer Associates, Sunderland, MA.
- Howard, J., A. J. Hudspeth, and R. D. Vale. 1989. Movement of microtubules by single kinesin molecules. Nature. 342:154–158. [DOI] [PubMed] [Google Scholar]
- Howard, J., and A. A. Hyman. 1993. Preparation of marked microtubules for the assay of the polarity of microtubule-based motors by fluorescence microscopy. Methods Cell Biol. 39:105–113. [DOI] [PubMed] [Google Scholar]
- Huang, T.-G., and D. D. Hackney. 1994. Drosophila kinesin minimal motor domain expressed in Escherichia coli. J. Biol. Chem. 269:16493–16501. [PubMed] [Google Scholar]
- Inoue, Y., A. H. Iwane, T. Miyai, E. Muto, and T. Yanagida. 2001. Motility of one-headed kinesin molecules along microtubules. Biophys. J. 81:2838–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, Y., Y. Y. Toyoshima, A. H. Iwane, S. Morimoto, H. Higuchi, and T. Yanagida. 1997. Movements of truncated kinesin fragments with a short or an artificial flexible neck. Proc. Natl. Acad. Sci. USA. 94:7275–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani, S., A. H. Iwane, H. Higuchi, Y. Ishii, and T. Yanagida. 1999. Mechanical and chemical properties of cysteine-modified kinesin molecules. Biochemistry. 38:10318–10323. [DOI] [PubMed] [Google Scholar]
- Kaseda, K., H. Higuchi, and K. Hirose. 2003. Alternate fast and slow stepping of a heterodimeric kinesin molecule. Nat. Cell Biol. 5:1079–1082. [DOI] [PubMed] [Google Scholar]
- Kikkawa, M., E. P. Sablin, Y. Okada, H. Yajima, R. J. Fletterick, and N. Hirokawa. 2001. Switch-based mechanism of kinesin motors. Nature. 411:439–445. [DOI] [PubMed] [Google Scholar]
- Kojima, H., E. Muto, H. Higuchi, and T. Yanagida. 1997. Mechanics of single kinesin molecules measured by optical trapping nanometry. Biophys. J. 73:2012–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov, S. A., E. A. Vaisberg, N. A. Shanina, N. N. Magretova, Y. Y. Chernyak, and V. I. Gelfand. 1988. The quaternary structure of bovine brain kinesin. EMBO J. 7:353–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, M., H. Higuchi, and T. Yanagida. 2002. Chemomechanical coupling of the forward and backward steps of single kinesin molecules. Nat. Cell Biol. 4:790–797. [DOI] [PubMed] [Google Scholar]
- Nishiyama, M., E. Muto, Y. Inoue, T. Yanagida, and H. Higuchi. 2001. Substeps within the 8-nm step of the ATPase cycle of single kinesin molecules. Nat. Cell Biol. 3:425–428. [DOI] [PubMed] [Google Scholar]
- Okada, Y., H. Higuchi, and N. Hirokawa. 2003. Processivity of the single-headed kinesin KIF1A through biased binding to tubulin. Nature. 424:574–577. [DOI] [PubMed] [Google Scholar]
- Okada, Y., and N. Hirokawa. 1999. A processive single-headed motor: kinesin superfamily protein KIF1A. Science. 283:1152–1157. [DOI] [PubMed] [Google Scholar]
- Okada, Y., and N. Hirokawa. 2000. Mechanism of the single-headed processivity: diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc. Natl. Acad. Sci. USA. 97:640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, S., A. W. Lin, D. Safer, C. L. Hart, N. Naber, B. O. Carragher, S. M. Cain, E. Pechatnikova, E. M. Wilson-Kubalek, M. Whittaker, E. Pate, R. Cooke, and others. 1999. A structural change in the kinesin motor protein that drives motility. Nature. 402:778–784. [DOI] [PubMed] [Google Scholar]
- Svoboda, K., and S. M. Block. 1994. Force and velocity measured for single kinesin molecules. Cell. 77:773–784. [DOI] [PubMed] [Google Scholar]
- Svoboda, K., C. F. Schmidt, B. J. Schnapp, and S. M. Block. 1993. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 365:721–727. [DOI] [PubMed] [Google Scholar]
- Tomishige, M., D. R. Klopfenstein, and R. D. Vale. 2002. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 297:2263–2267. [DOI] [PubMed] [Google Scholar]
- Uemura, S., and S. Ishiwata. 2003. Loading direction regulates the affinity of ADP for kinesin. Nat. Struct. Biol. 10:308–311. [DOI] [PubMed] [Google Scholar]
- Vale, R. D., and R. A. Milligan. 2000. The way things move: looking under the hood of molecular motor proteins. Science. 288:88–95. [DOI] [PubMed] [Google Scholar]
- Veigel, C., L. M. Coluccio, J. J. Jontes, J. C. Sparrow, R. A. Milligan, and J. E. Molloy. 1999. The motor protein myosin-I produces its working stroke in two steps. Nature. 398:530–533. [DOI] [PubMed] [Google Scholar]
- Weber, P. C., D. H. Ohlendorf, J. Wendoloski, and F. R. Salemme. 1989. Structural origins of high-affinity biotin binding to streptavidin. Science. 243:85–88. [DOI] [PubMed] [Google Scholar]
- Yildiz, A., M. Tomishige, R. D. Vale, and P. R. Selvin. 2004. Kinesin walks hand-over-hand. Science. 303:676–678. [DOI] [PubMed] [Google Scholar]
- Young, E. C., H. K. Mahtani, and J. Gelles. 1998. One-headed kinesin derivatives move by a nonprocessive, low-duty ratio mechanism unlike that of two-headed kinesin. Biochemistry. 37:3467–3479. [DOI] [PubMed] [Google Scholar]