Abstract
A self-replicating molecule directs the covalent assembly of component molecules to form a product that is of identical composition to the parent. When the newly formed product also is able to direct the assembly of product molecules, the self-replicating system can be termed autocatalytic. A self-replicating system was developed based on a ribozyme that catalyzes the assembly of additional copies of itself through an RNA-catalyzed RNA ligation reaction. The R3C ligase ribozyme was redesigned so that it would ligate two substrates to generate an exact copy of itself, which then would behave in a similar manner. This self-replicating system depends on the catalytic nature of the RNA for the generation of copies. A linear dependence was observed between the initial rate of formation of new copies and the starting concentration of ribozyme, consistent with exponential growth. The autocatalytic rate constant was 0.011 min−1, whereas the initial rate of reaction in the absence of pre-existing ribozyme was only 3.3 × 10−11 M⋅min−1. Exponential growth was limited, however, because newly formed ribozyme molecules had greater difficulty forming a productive complex with the two substrates. Further optimization of the system may lead to the sustained exponential growth of ribozymes that undergo self-replication.
In living systems, replicative processes transfer genetic information from template nucleic acid molecules to newly synthesized, complementary products. Several nonenzymatic template-dependent ligation systems have been devised to study the role of a template in binding and positioning complementary substrates for covalent bond formation (1–5). These have included simple self-replicating systems of the form A + B → T, where A and B are substrates that bind to a complementary template, T, and become joined to form a product molecule that is identical to the template (6–9). The unique aspect of self-replicating systems is that the reaction product has the potential to direct additional reactions. The system is termed autocatalytic when the product is an efficient template, and each covalent bond that is formed generates additional template molecules that can direct further joining reactions. The realization of autocatalytic behavior in a self-replicating system implies that sustained exponential growth may be possible.
The self-replicating systems that have been studied to date use template molecules composed of nucleic acids (6, 7, 10), peptides (11–14), or small organic compounds (15–17). The nucleic acid-based systems are the most straightforward and rely on simple Watson–Crick pairing interactions between a short oligonucleotide template and two complementary oligonucleotide substrates (6, 7, 10). The substrates are bound at adjacent positions along the template and are joined through a reaction involving chemical groups at their opposed ends. Peptide-based self-replicating systems are similar, except that the components are oligopeptides that have the capacity to form α-helices. The template is the hydrophobic face of an α-helix that interacts with the corresponding face of two peptide substrates (11–14). Unlike nucleic acid systems, where the templating interactions involve all of the nucleotide subunits, peptide replication systems involve only a few amino acid residues in the helix–helix interactions. The remaining residues are responsible for maintaining the overall fold of the helix, which demonstrates that the templating properties of a self-replicating system do not necessarily involve interactions with every residue in the polymer.
Self-replicating systems based on organic compounds further generalize the notion of a template, as these systems are not based on a polymer, but rather on a low molecular weight compound that presents a templating surface to bind the two substrates (15–17). With this broader notion of templating in mind, it was of interest to explore more complex nucleic acid polymers that would carry out self-replication mediated by secondary and tertiary interactions, rather than simple Watson–Crick pairing. In particular, it was of interest to examine a ribozyme-based system in which the structured nucleic acid components would add enzymatic activity to the self-replicating system.
Nucleic acid polymers are attractive candidates for self-replication because they can encode genetic information, recognize nucleic acid components with high specificity, and catalyze nucleic acid joining reactions (18–22). A system based on a ligase ribozyme was used to investigate the ability of catalytic RNA molecules to promote their own synthesis from component oligonucleotides. A self-replicating ribozyme was constructed by using a rational design approach, relying on RNA-catalyzed RNA ligation to bring about a reaction of the form A + B → T. An important goal of the design was to have the template and substrates interact with minimal base pairing so that the ligated product could easily dissociate from the template and thus be available to catalyze subsequent ligation reactions. An added consideration was to optimize the construct so that the presence of the template molecule would greatly enhance the rate of ligation of the substrates compared with their ligation in the absence of the template. The R3C RNA ligase ribozyme was chosen for this study because of its simple secondary structure, reasonably fast ligation rate, and ability to function in an intermolecular reaction format (23).
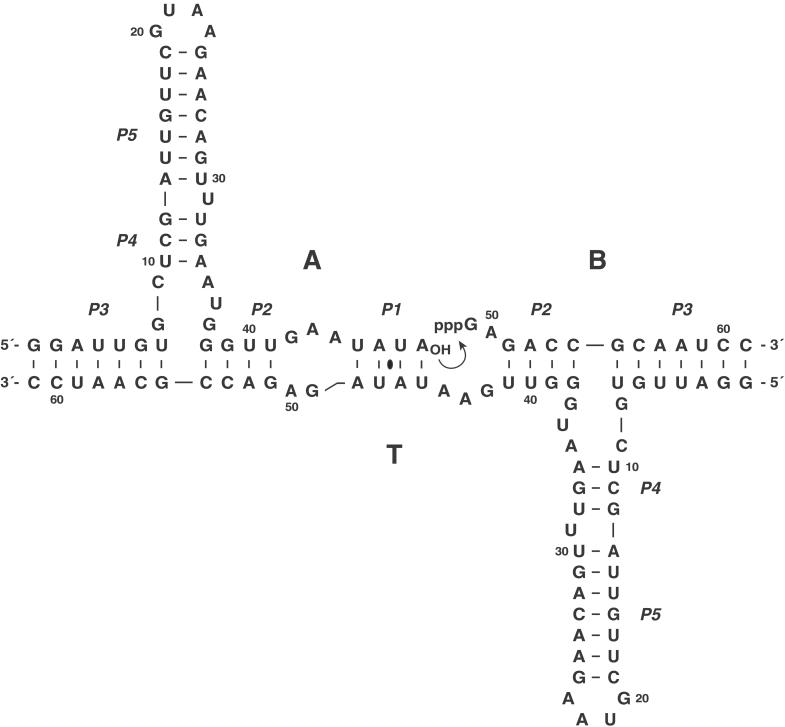
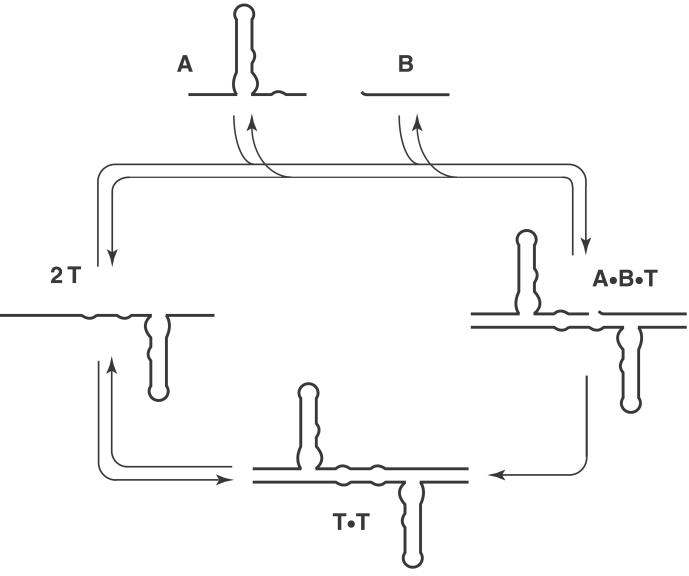
The R3C ribozyme is an RNA-dependent RNA ligase that catalyzes the formation of a 3′,5′-phosphodiester linkage between two RNA molecules, with a catalytic rate of 0.32 min−1 (23). The ribozyme, which was obtained by in vitro evolution, catalyzes attack of the 3′-hydroxyl of the substrate on the α-phosphate of the 5′-triphosphate of the ribozyme, with concomitant release of inorganic pyrophosphate. The overall structure of the ribozyme is centered about a three-way junction, whose arms are defined by the P2, P3, and P4 stems. The ribozyme has been engineered to function in an intermolecular reaction format, with one substrate containing the attacking 3′-hydroxyl and a second substrate containing the 5′-triphosphate (23). In the present study, the R3C ligase was restructured as a symmetrical dimer, such that the ligated product would be identical to the template (Fig. 1). This process resulted in an RNA molecule that underwent RNA-catalyzed self-replication with the potential for exponential growth (Fig. 2).
Figure 1.
Secondary structure of the self-replicating ligase ribozyme. The template molecule T binds the two substrate molecules A and B, which then undergo RNA-catalyzed ligation (arrow) to generate another copy of T. Nucleotides are numbered at every 10th position. The location of the 2-fold axis of symmetry is denoted by the black oval.
Figure 2.
Replication cycle of the R3C ligase ribozyme.
Materials and Methods
Enzymes, Nucleotides, and Oligonucleotides.
Histidine-tagged T7 RNA polymerase was purified from Escherichia coli strain BL21 containing plasmid pBH161 (kindly provided by William McAllister, State University of New York, Brooklyn). T4 polynucleotide kinase was purchased from New England Biolabs. Nucleoside 5′-triphosphates were purchased from Amersham Pharmacia, and [γ-32P]ATP (7 μCi/pmol) was from ICN. Synthetic oligodeoxynucleotides were prepared by using an Expedite Nucleic Acid Synthesis System (Applied Biosystems), with phosphoramidites purchased from either Glen Research (Sterling, VA) or Applied Biosystems. Synthetic RNA molecules were prepared by Dharmacon Research, Boulder, CO. All oligonucleotides were purified by denaturing PAGE and desalted by using a C18 SEP-Pak Cartridge (Waters).
In Vitro Transcription.
RNA templates and substrates were prepared by in vitro transcription unless otherwise stated. The transcription reactions used 15 mM MgCl2, 2 mM spermidine, 50 mM Tris (pH 7.5), 5 mM DTT, 400 nM DNA template, 800 nM synthetic oligodeoxynucleotide having the sequence 5′-GGACTAATACGACTCACTATA-3′ (T7 promoter sequence underlined), 2 mM each of the four NTPs, and 25 units/μl T7 RNA polymerase. The reaction mixture was incubated at 37°C for 2 h, then quenched with an equal volume of gel-loading buffer containing 15 mM Na2EDTA and 18 M urea. The transcription products were purified by denaturing PAGE, eluted from the gel, and desalted by using a Waters C18 SEP-Pak Cartridge.
RNA Substrates and Templates.
The RNA molecules A, B, and T (Fig. 1) were prepared either by automated solid-phase synthesis or in vitro transcription. B and T were obtained by transcription of the corresponding DNA template to yield 5′-pppGAGACCGCAAUCC-3′ and 5′-pppGGAUUGUGCUCGAUUGUUCGUAAGAACAGUUUGAAUGGGUUGAAUAUAGAGACCGCAAUCC-3′, respectively. Due to poor yields obtained for the transcription of A, this oligonucleotide, having the sequence 5′-GGAUUGUGCUCGAUUGUUCGUAAGAACAGUUUGAAUGGGUUGAAUAUA-3′, was prepared synthetically. The mutant forms of B and T, with insertion of a single A residue after position 54, were obtained by in vitro transcription.
Gel-Shift Analysis.
Gel-shift assays were used to examine the ability of the reactant molecules to associate into higher-order complexes. A mixture containing B and/or T was equilibrated separately from a mixture containing A at 23°C for 30 min in the presence of 10 mM MgCl2 and 50 mM N-[2-hydroxyethyl]piperazine-N′-[3-propane-sulfonic acid] (EPPS, pH 8.5), then combined and allowed to stand at 23°C for an additional 3–4 min. Aliquots from the combined mixture were analyzed in a nondenaturing polyacrylamide gel containing 12 mM MgCl2, 2 mM Na2EDTA, and 90 mM Tris-borate buffer (pH 8.5). All gels were run at a constant temperature of 23°C and imaged by using a PhosphorImager (Molecular Dynamics).
Analysis of Catalytic Activity.
Ligation assays were carried out in the presence of 25 mM MgCl2 and 50 mM EPPS (pH 8.5) at 23°C. Before initiating the reaction, B plus or minus T were preincubated in reaction buffer at 23°C for 10 min. The reactions were initiated by the addition of 5′-32P-labeled A, which had been preincubated at four times the final concentration in reaction buffer at 23°C for 10 min. The concentrations of A and B were varied from 0.5 to 12 μM, and the starting concentration of T was varied from 0 to 2 equivalents relative to the amount of A and B. Aliquots were removed from the reaction mixture at various times and quenched by adding an equal volume of gel-loading buffer that contained 25 mM Na2EDTA and 18 M urea. The unreacted substrates were separated from the ligated products by denaturing PAGE and quantitated with a PhosphorImager. The fraction reacted at each time point was determined, and the initial phase of the reaction was fit to a plot of ln (1 − fraction reacted) versus time to obtain values for kobs. Similar values for kobs were obtained when the data were fit to a burst kinetic equation: fraction reacted = a (1 − e−kt) + bt, where k is the exponential rate, a is the amplitude of the burst, and b is the rate of the second phase (kobs = ak).
Results
Design of the Construct.
A 2-fold symmetrical ribozyme-based construct was generated, in which each half contained the catalytic core of the R3C ligase as well as the two substrate domains. The construct was designed such that the product of the R3C-catalyzed reaction between substrates A and B would be identical to the template T, thus generating new template-catalyst molecules with each ligation event. The nucleotide sequences of the P2, P4, and P5 stems were fixed, whereas the sequences of the P1 and P3 stems were changed to form a self-complementary dimeric construct centered about the symmetry axis within the palindromic P1 stem (Fig. 1). The resulting trimolecular complex consisted of a template molecule, T, that could bind the two substrates, A and B, through base-pairing interactions involving the P1, P2, and P3 stems.
The P1 stem was shortened to reduce the rate of ligation within the A⋅B⋅A⋅B tetramolecular complex, while maintaining efficient ligation within the A⋅B⋅T trimolecular complex. When the P1 stem was shortened, it was important to avoid steric hindrance between the adjacent catalytic cores and to maintain proper geometry at the ligation site. The self-complementary P3 stems were lengthened relative to the P1 stem to favor formation of the A⋅B⋅T complex over the A⋅B⋅A⋅B complex. If the P3 stems were made too long, however, the reaction would be limited by dissociation of the T⋅T product complex or inhibited by competing self-structure within the template.
With these criteria in mind, various lengths and sequences of the P1 and P3 stems were tested (Table 1). Constructs that contained a 4-bp P3 stem and a palindromic P1 stem of 6–10 bp all were capable of catalyzing ligation at an appreciable rate even in the absence of a starting amount of template. When the P3 stem was lengthened to 7 bp and the P1 stem contained 6 bp, the template-dependent rate of reaction was comparable to that involving the substrates alone. With a P3 stem of 7 bp and a P1 stem of only 4 bp, the rate of reaction of the substrates alone was greatly diminished relative to the template-dependent reaction. The significant enhancement of ligation efficiency attributable to the template prompted further characterization of this construct.
Table 1.
Rate of template formation for various self-replicative complexes in either the absence or presence of a starting amount of template
| P1 stem
|
P3 stem
|
Reaction rate
|
|||
|---|---|---|---|---|---|
| Length | Sequence | Length | Sequence | − Template | + Template |
| 10 | 5′-UAUAGCUAUA-3′ | 4 | 5′-GGAUUGU-3′ | +++ | ND |
| 3′-AUAUCGAUAU-5′ | 3′-AACG-5′ | ||||
| 8 | 5′-UAUGCAUA-3′ | 4 | 5′-GGAUUGU-3′ | +++ | ++ |
| 3′-AUACGUAU-5′ | 3′-AACG-5′ | ||||
| 6 | 5′-UAGCUA-3′ | 4 | 5′-GGAUUGU-3′ | +++ | +++ |
| 3′-AUCGAU-5′ | 3′-AACG-5′ | ||||
| 6 | 5′-UAUAUA-3′ | 4 | 5′-GGAUUGU-3′ | ++ | ++ |
| 3′-AUAUAU-5′ | 3′-AACG-5′ | ||||
| 6 | 5′-UAGCUA-3′ | 7 | 5′-GGAUUGU-3′ | +++ | +++ |
| 3′-AUCGAU-5′ | 3′-CCUAACG-5′ | ||||
| 6 | 5′-UAUAUA-3′ | 7 | 5′-GGAUUGU-3′ | ++ | ++ |
| 3′-AUAUAU-5′ | 3′-CCUAACG-5′ | ||||
| 4 | 5′-UAUA-3′ | 7 | 5′-GGAUUGU-3′ | + | ++ |
| 3′-AUAU-5′ | 3′-CCUAACG-5′ | ||||
ND., not determined.
Characterization of the Construct.
The behavior of the system in the presence of the template was found to be sensitive to the order of addition of the individual components, A, B, and T. The trimolecular reaction could be initiated by the addition of either T or B to the other two components, but initiation by the addition of A to a mixture containing B and T resulted in a significantly faster initial rate of ligation. Thus, the standard protocol for initiating the reaction involved addition of A to a preincubated mixture containing B and T.
The behavior of the substrate molecule A and the template molecule T was examined by gel-shift analysis. There was concern that a high degree of A⋅A dimerization would result in a high background reaction via an A⋅B⋅A⋅B quaternary complex. Accordingly, it was important to assess the strength of base pairing between the P1 stems of two A molecules. Only a small amount of A⋅A complex was formed for concentrations of A ranging from 0.5 to 20 μM (data not shown). There also was concern that if the T⋅T complex was too stable, newly formed copies of T would not readily dissociate, and thus not be able to enter another replication cycle. However, over a concentration range for T of 0.1–100 μM, the majority of molecules were present as a T monomer, with far lesser amounts of T⋅T dimers and higher-order complexes of T. The prevalence of the T monomer over such a large concentration range suggests that T is stabilized by intramolecular interactions. Secondary structure prediction using mfold (version 3.1; refs. 24 and 25) indicates that this structure involves base pairing between the P2 and P3 stems located at both ends of the molecule.
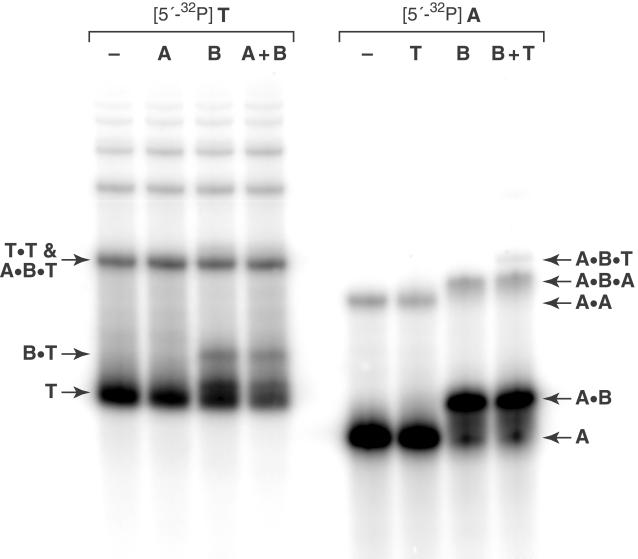
Gel-shift studies were carried out to examine the formation of multimeric complexes involving A, B, and T. In the presence of 2 μM each of these three RNAs, there was no detectable binding between A and T and only weak binding between B and T (Fig. 3). One possible explanation for the low affinity of A for T and of B for T may be competing intramolecular structure of T involving the P2 and P3 stems. Although T did not readily form dimeric complexes or bind with either of the two substrates, the two substrates were found to bind readily with each other. A mixture of 2 μM each of A and B was present mostly as an A⋅B complex, accompanied by a small amount of what appeared to be an A⋅B⋅A complex. The desired A⋅B⋅T complex was observed in a mixture containing 2 μM each of A, B, and T, but was dominated by the A⋅B dimer and other species.
Figure 3.
Gel-shift analysis of mixtures of A, B, and T, each present at 2 μM concentration. Complex formation was detected in a nondenaturing 12% polyacrylamide gel based on reduced mobility of either 5′-32P-labeled T (Left) or 5′-32P-labeled A (Right).
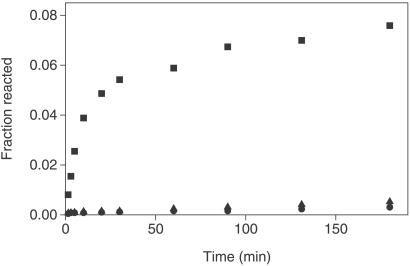
Initial assessment of the ribozyme-catalyzed reaction set out to characterize its template dependence (Fig. 4). The ligation reaction involving 2 μM each of A and B in the absence of T was very slow, with less than 1% product formed after 3 h. When 0.5 equivalents of T were added to a reaction mixture that contained 2 μM each of A and B, a 400-fold increase in the initial rate of ligation was observed. In contrast, addition of another 0.5 equivalents of both A and B resulted in only a 2-fold increase in the initial rate of ligation. This small rate enhancement may be attributable to a slightly increased reaction resulting from interaction of the P1 region of two A molecules. The template dependence of the reaction between A and B was deemed to be significant because the rate enhancement achieved by the introduction of T could not be achieved by the use of more substrates alone.
Figure 4.
Time course of template production in reaction mixtures containing either the A and B substrates alone or the two substrates in the presence of T. ●, 2 μM A and 2 μM B; ■, 2 μM A, 2 μM B, and 1 μM T; ▴, 3 μM A and 3 μM B.
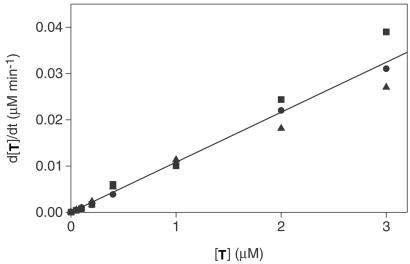
The relationship between the initial rate of reaction and the starting concentration of T was investigated to determine whether the template-dependent reaction fit to a model of self-replication (Fig. 5). The kinetic behavior of a self-replicating system can be characterized by the equation: [d[T]/dt]initial = ka[T0]p + kb, where the initial rate of reaction is proportional to the starting template concentration, [T0], raised to the reaction order p (8, 9, 26, 27). A plot of [d[T]/dt]initial versus [T0]p has a y-intercept, kb, that is equal to the rate of reaction in the absence of template, and a slope, ka, that is equal to the autocatalytic rate enhancement due to the template. In the presence of 2 μM each of A and B, a linear relationship was evident between the initial reaction rate and the starting concentration of template, with p = 1.0, ka = 0.011 ± 0.002 min−1, and kb = 3.3 ± 0.2 × 10−11 M⋅min−1. A similar linear relationship was observed for concentrations of A and B of either 0.5 or 8 μM each (data not shown). These results are indicative of a first-order reaction and suggest that the newly formed product molecules are able to dissociate from T⋅T complexes (8, 9, 26, 27), and thus become available to catalyze subsequent ligation reactions (Fig. 2).
Figure 5.
Initial rate of template formation as a function of the starting concentration of template. Reactions used: 2 μM A, 2 μM B, and 0–3 μM T. The line corresponds to a linear fit of three replicates (●, ■, and ▴), with slope, ka, of 0.011 min−1 and y-intercept, kb, of 3.3 × 10−11 M⋅min−1.
Insertion of a single adenosine residue between the P2 and P3 stems of the R3C ribozyme diminishes its catalytic rate by about 100-fold (23). This insertion does not interfere with base-pairing involving the P1, P2, or P3 stems. When the corresponding mutation was introduced in the replicative complex (after position 54), there was a 25-fold decrease in the rate of ligation for the reaction involving 2 μM each of A and B in the absence of T. In the presence of 0.2 equivalents of template, the reaction rate for the mutated complex was diminished by 1,550-fold. The significant decrease in ligation activity resulting from the single-nucleotide insertion demonstrates that the self-replicative behavior of the system derives from its intrinsic activity as a ribozyme.
The self-replication reaction involving A, B, and T exhibited an initial burst, followed by a second, slower phase of reaction (Fig. 4). Attempts to improve the amplitude of the burst phase of the reaction involved testing varying concentrations of A and B, ranging from 0.5 to 16 μM. The reaction between A and B in the absence of T was significantly slower than the reaction in the presence of T, except when the concentration of both A and B approached 16 μM. Using equimolar concentrations of A and B, the amplitude of the initial burst corresponded to 10–15% of the starting concentration of template. When the concentration of B exceeded that of A, the initial rate and burst amplitude were decreased, whereas at concentrations of A greater than B, the initial rate and burst amplitude were increased. The greatest burst amplitude was obtained when the concentration of A was less than 16 μM and the concentration of B was 2- to 3-fold lower than that of A.
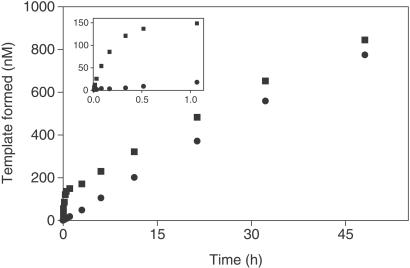
The behavior of the system was further examined in the presence of 12 μM A and 4 μM B and either the presence or absence of 0.4 μM T (Fig. 6). The initial burst generated product molecules corresponding to about one-third of the input concentration of template molecules. The slower second phase of the reaction continued to generate product molecules over a 48-h period. After approximately 17 h, the amount of newly synthesized template exceeded the starting amount of template. The reaction in the absence of template did not exhibit an initial burst, but proceeded at a rate comparable to the slow phase of the template-dependent reaction, generating 0.4 μM product after about 24 h. Although the initial burst of the template-dependent reaction could be enhanced by increasing the concentration of A relative to B, the second phase of the reaction remained about the same in the presence or absence of T for varying concentrations of the two substrates.
Figure 6.
Time course demonstrating the production of newly synthesized template that exceeds the starting amount of template. Reactions used either (●) 12 μM A and 4 μM B, or (■) 12 μM A, 4 μM B, and 0.4 μM T. (Inset) The course of the reaction over the first hour.
Discussion
Self-Replication of a Ligase Ribozyme.
The R3C ligase ribozyme, which catalyzes the formation of a 3′,5′-phosphodiester linkage between two RNA molecules, was engineered as a 2-fold symmetrical construct that brought about the template-dependent ligation of two substrate molecules to form another copy of the template (Fig. 1). It was designed to function as a self-replicating system of the form A + B → T, whereby the newly formed template molecules could direct subsequent ligation reactions (Fig. 2). This behavior is unique among reported self-replicating systems because the template-substrate complex is a ribozyme that adds more catalytic potential to the system with each template-forming reaction. Base-pairing interactions between the template and substrate are required for the formation of the A⋅B⋅T ternary complex, but the overall secondary and tertiary structure of the complex also are essential for the observed autocatalytic behavior.
The presence of the template results in a significantly accelerated initial rate of reaction that cannot be mimicked by adding comparable amounts of the two substrates (Fig. 4). The template-catalyzed reaction is biphasic, with a rapid initial burst followed by a slower second phase. The initial burst appears to reflect the fraction of A⋅B⋅T complexes that form as a result of binding of A to preformed B⋅T complexes. The burst amplitude is substantially greater when A is added to a mixture of B and T, compared with when B is added to a mixture of A and T. When A is added last, the amplitude of the burst can be improved by increasing the concentration of A relative to B (Fig. 6). One explanation for this behavior is that the excess A, beyond what is needed to saturate binding with B to form A⋅B complexes, remains available to saturate binding with B⋅T to form fast-reacting A⋅B⋅T complexes. The slower second phase appears to reflect the reaction of A and B independent of the template, as indicated by the similar rate of this phase and the rate of reaction of A and B in the absence of preformed T (Fig. 6). Template molecules that are present at the start of the reaction have a pronounced effect on the initial rate of replication, but newly formed template molecules do not appear to be capable of acting in a similar manner.
Most self-replicating systems that have been described previously exhibit an initial rate of reaction that is proportional to the square root of the starting template concentration, reflecting the rate-limiting dissociation of the T⋅T complex (8, 9, 26, 27). If T⋅T dissociation is not rate limiting, then one would expect to see a first-order, linear relationship between the initial rate of reaction and the starting concentration of template. There is a recent example of a self-replicating system that exhibits a reaction order of 0.91 (14). That system used short peptides of the minimum length necessary to form coiled–coil complexes, which allowed the reaction products to dissociate at a rate that was not strictly rate limiting. The time course of the reaction exhibited sigmoidal behavior, consistent with exponential growth and product turnover (27). Another noteworthy system, composed of 3-aminobenzamides and 2-formylphenoxyacetic acids, exhibited a reaction order of 1.0, although in that case the reaction product was not identical to the template (16).
The R3C ligase self-replicating system is a symmetrical system that exhibits a first-order, linear relationship between the initial rate of reaction and the starting concentration of template (Fig. 5). The reaction order of 1.0 indicates that the availability of template at the start of the reaction is not limited by dissociation of T⋅T complexes, consistent with the weak self-binding of T that was observed by gel-shift analysis (Fig. 3). This finding implies that the T⋅T product complex also dissociates readily, which would allow product turnover. However, sigmoidal growth was not observed in this system. The time course of the reaction was modeled by using the experimentally determined values for autocatalytic and nonautocatalytic growth (ka and kb, respectively; Fig. 5). Even allowing for product turnover, the simulated time course exhibited very little sigmoidal character when the concentration of both A and B was 2 μM and the concentration of T was 0.2–2 μM. This is because the ribozyme is a very efficient self-replicator, with an efficiency (ɛ = ka/kb; ref. 10) of 3.3 × 108 M−1. Under the reaction conditions that were used, the substrates are consumed very quickly and newly produced template molecules do not greatly increase the total concentration of template. The initial rate of reaction predicted by modeling, allowing product turnover, does not differ significantly from what was observed experimentally. After the initial burst, however, the experimental data deviates from the model as the reaction enters the slower second phase.
It would be desirable to extend the burst phase beyond the point at which the amount of newly formed template exceeds the starting amount of template. This would allow exponential growth to continue indefinitely, either in the context of a continuous flow reactor or by a serial transfer procedure. Serial transfer involves taking an aliquot from a completed reaction mixture and using it to seed a new reaction mixture that contains a fresh supply of substrates. The chief obstacle to realizing this possibility is that newly formed template molecules, after their dissociation from the T⋅T complex, will encounter A and B simultaneously. This is disadvantageous for formation of the fast-reacting A⋅B⋅T complex because A and B already will exist mostly as the tightly bound A⋅B dimer, which is not easily displaced by T. Further optimization of the system might lead to improved formation of the catalytic A⋅B⋅T complex relative to the A⋅B complex. Perhaps this could be achieved by using in vitro evolution methods, which thus far have not been applied to self-replicating ribozymes.
The self-replicating R3C ligase is amenable to a variety of sequence modifications within each of the component stem regions (23). One could maintain the sequences of the P1, P2, and P3 stems, while changing the sequences of the P4 and P5 stems in search of variants with more desirable self-replicative properties. These variants might be allowed to compete for a common substrate, leading to selective enrichment of the most efficient replicators. Another approach would be to change the sequences of the P1, P2, and P3 stems, while maintaining the sequences of the P4 and P5 stems. A set of symmetrical self-replicating ribozymes could be constructed, each operating exclusively on its corresponding substrates to generate additional copies of itself. If that exclusivity was relaxed, for example, by using substrates that differ by only 1 or 2 nt, then one template might direct the ligation of several different pairs of substrates to generate several different products. One or more of these products might direct the synthesis of the starting template. Cross-catalytic systems of this type have been demonstrated for nucleic acids, peptides, and small organic molecules (13, 26, 28–34).
A more challenging task would be to generate a pair of self-replicating molecules that are exclusively cross-catalytic. Nucleic acid replication in biology behaves in this manner, with a plus-strand template directing the synthesis of a minus-strand product, and vice versa. An exclusively cross-catalytic system based on the self-replicating ribozyme might be developed by breaking the symmetry of the A⋅B⋅T complex. This would require two pairs of substrates (A and B; A′ and B′) that bind to their respective template (T′ and T), resulting in reactions of the form A + B → T and A′ + B′ → T′. Assuming that dissociation of the T⋅T′ complex continues not to be rate limiting, the lack of self-complementarity between the 5′ and 3′ ends of T might allow it to displace the undesirable A′⋅B dimer, leading to the fast-reacting A′⋅B′⋅T complex. Similarly, T′ might be able to displace the A⋅B′ dimer to form the A⋅B⋅T′ complex. If this were the case, self-replication with exponential growth could proceed indefinitely.
Reconciling Chemical Self-Replication and Darwinian Evolution.
Self-replication is of special interest to both chemists and biologists because it is a fundamental property of living systems. Self-replication, broadly defined, gives rise to new copies of an organism and results in the transfer of genetic information from a parent to its progeny. Self-replication alone, however, is not sufficient for life unless it allows for the possibility of heritable mutations. Variation is needed in a population of self-replicators if that population is to withstand and adapt to changing environmental conditions. Competition for finite resources among a heterogeneous population of self-replicators leads to Darwinian behavior, resulting in survival of the fittest.
All of the chemical self-replication systems that have been described, including the ribozyme-based system of the present study, are not capable of undergoing Darwinian evolution. These systems usually offer no choice other than to form a particular product molecule. Even in those cases where one template can direct the synthesis of multiple products (13, 31, 35), the products do not “breed true” such that each product species only gives rise to additional copies of itself. Furthermore, product formation typically involves only a single joining reaction. Thus the information content of a particular self-replicating species can be no more than log2(NA·NB), where NA is the number of different A substrates and NB is the number of different B substrates that can be joined and faithfully replicated.
Laboratory systems have been devised for the Darwinian evolution of nucleic acid or protein molecules (36–38). These systems use large heterogeneous populations of compounds, each of which is assembled from many subunits. The maximum possible information content of a nucleic acid species of length n is log2(4n), whereas that of a protein species is log2(20n). An evolutionary search of this vast number of possibilities is an open-ended process that can lead to the development of molecules with complex structural and functional properties. However, all of the laboratory evolution systems that have been described do not involve self-replication; replication is instead carried out by polymerase proteins that are not part of the evolving system.
It is intriguing to contemplate the possibility of a chemical system that would be capable of undergoing Darwinian evolution in a self-sustained manner, that is, without the assistance of evolved molecules that are not part of the evolving system. By some measures, this would constitute a working example of life in the laboratory (39). One can consider two general approaches toward this goal. First, starting from a chemical self-replicating system, one might expand the process to involve several joining reactions, each requiring a choice from among two or more different combinations of substrates. Once a particular combination had been assembled, it would breed true, subject to an occasional mutation, which also would be maintained in a heritable fashion. A heterogeneous population of such self-replicating entities could be made to compete for limited resources, for example, a fixed supply of common substrate building blocks.
A second approach to a self-sustained evolving system might involve the in vitro evolution of a nucleic acid (or protein) enzyme that catalyzes the replication of many different nucleic acid (or protein) molecules, including copies of itself. The evolution process that would be used to obtain such an enzyme would not be self-sustained, but the product might be able to evolve in a self-sustained manner. Substantial progress has been made along these lines, although the goal of a self-replicating, evolving enzyme has not been achieved. Starting from a ribozyme that catalyzes the template-directed joining of two RNA molecules (40), an RNA polymerase ribozyme was evolved that catalyzes the polymerization of up to 14 NTPs on an external RNA template, operating with high fidelity and generality for almost any template sequence (22). The ribozyme itself contains approximately 200 nt, so it is not nearly capable of catalyzing the synthesis of additional copies of itself. Perhaps a smaller and/or more active form of the RNA polymerase ribozyme might be developed.
Reconciling self-replication and Darwinian evolution requires either bringing the fundamental biological principle of heritable genetic information to a chemical self-replicating system or instilling the chemistry of autocatalysis in a Darwinian system. Such an accommodation would bring history to chemistry and material continuity to biology. Rather than focus on the extreme versions of the two approaches outlined above, it may be fruitful to explore a middle ground in which self-replicating molecules direct the assembly of new copies of themselves from a modest assortment of component modules. Competition for utilization of these components might provide the basis for Darwinian evolution, while the threshold for achieving self-replication would be greatly lowered compared with that required for residue-by-residue copying of a long polymer.
Acknowledgments
We thank Julius Rebek and Günter von Kiedrowski for helpful discussions and Dong-Eun Kim for assistance with kinetics simulations. This work was supported by Research Grant NAG5-9386 from the National Aeronautics and Space Administration and by the Skaggs Institute for Chemical Biology.
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on May 1, 2001.
References
- 1.Naylor R, Gilham P T. Biochemistry. 1966;5:2722–2728. doi: 10.1021/bi00872a032. [DOI] [PubMed] [Google Scholar]
- 2.Lohrmann R, Orgel L E. J Mol Evol. 1979;12:237–257. doi: 10.1007/BF01732341. [DOI] [PubMed] [Google Scholar]
- 3.Inoue T, Orgel L E. Science. 1983;219:859–862. doi: 10.1126/science.6186026. [DOI] [PubMed] [Google Scholar]
- 4.Rohatgi R, Bartel D P, Szostak J W. J Am Chem Soc. 1996;118:3332–3339. doi: 10.1021/ja953712b. [DOI] [PubMed] [Google Scholar]
- 5.Zhan Z J, Lynn D G. J Am Chem Soc. 1997;119:12420–12421. [Google Scholar]
- 6.von Kiedrowski G. Angew Chem. 1986;25:932–935. [Google Scholar]
- 7.Zielinski W S, Orgel L E. Nature (London) 1987;327:346–347. doi: 10.1038/327346a0. [DOI] [PubMed] [Google Scholar]
- 8.Bag B G, von Kiedrowski G. Pure Appl Chem. 1996;68:2145–2152. [Google Scholar]
- 9.Robertson A, Sinclair A J, Philp D. Chem Soc Rev. 2000;29:141–152. [Google Scholar]
- 10.von Kiedrowski G, Wlotzka B, Helbing J, Mazten M, Jordan S. Angew Chem. 1991;30:423–426. [Google Scholar]
- 11.Lee D H, Granja J R, Martinez J A, Severin K, Ghadiri M R. Nature (London) 1996;382:525–528. doi: 10.1038/382525a0. [DOI] [PubMed] [Google Scholar]
- 12.Yao S, Ghosh I, Zutshi R, Chmielewski J. J Am Chem Soc. 1997;119:10559–10560. [Google Scholar]
- 13.Lee D H, Severin K, Yokobayashi Y, Ghadiri M R. Nature (London) 1997;390:591–594. doi: 10.1038/37569. [DOI] [PubMed] [Google Scholar]
- 14.Issac R, Chmielewski J. J Am Chem Soc. 2002;124:6808–6809. doi: 10.1021/ja026024i. [DOI] [PubMed] [Google Scholar]
- 15.Tijvikua T, Ballester P, Rebek J., Jr J Am Chem Soc. 1990;112:1249–1250. [Google Scholar]
- 16.Terfort A, von Kiedrowski G. Angew Chem. 1992;31:654–656. [Google Scholar]
- 17.Wintner E A, Conn M M, Rebek J., Jr J Am Chem Soc. 1994;116:8877–8884. [Google Scholar]
- 18.Doudna J A, Couture S, Szostak J W. Science. 1991;251:1605–1608. doi: 10.1126/science.1707185. [DOI] [PubMed] [Google Scholar]
- 19.Green R, Szostak J W. Science. 1992;258:1910–1915. doi: 10.1126/science.1470913. [DOI] [PubMed] [Google Scholar]
- 20.Doudna J A, Usman N, Szostak J W. Biochemistry. 1993;32:2111–2115. doi: 10.1021/bi00059a032. [DOI] [PubMed] [Google Scholar]
- 21.Cuenoud B, Szostak J W. Nature (London) 1995;375:611–614. doi: 10.1038/375611a0. [DOI] [PubMed] [Google Scholar]
- 22.Johnston W K, Unrau P J, Lawrence M S, Glasner M E, Bartel D P. Science. 2001;292:1319–1325. doi: 10.1126/science.1060786. [DOI] [PubMed] [Google Scholar]
- 23.Rogers J, Joyce G F. RNA. 2001;7:395–404. doi: 10.1017/s135583820100228x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuker M, Mathews D H, Turner D H. In: RNA Biochemistry and Biotechnology. Barciszewski J, Clark B F C, editors. Dordrecht, The Netherlands: Kluwer; 1999. pp. 11–43. [Google Scholar]
- 25.Mathews D H, Sabina J, Zuker M. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 26.Sievers D, Achilles T, Burmeister S, Jordon A, Terfort A, von Kiedrowski G. In: Self-Production of Supramolecular Structures. Fleischaker G R, Colonna S, Luisi P L, editors. Dordrecht, The Netherlands: Kluwer; 1994. pp. 45–62. [Google Scholar]
- 27.von Kiedrowski G. In: Bioorganic Frontiers. Dugas H, editor. Vol. 3. New York: Springer; 1993. pp. 113–146. [Google Scholar]
- 28.Pieters R J, Huc I, Rebek J., Jr Angew Chem. 1994;33:1579–1581. [Google Scholar]
- 29.Achilles T, von Kiedrowski G. Angew Chem. 1993;32:1198–1201. [Google Scholar]
- 30.Sievers D, von Kiedrowski G. Nature (London) 1994;369:221–224. doi: 10.1038/369221a0. [DOI] [PubMed] [Google Scholar]
- 31.Sievers D, von Kiedrowski G. Chem Eur J. 1998;4:629–641. [Google Scholar]
- 32.Severin K, Lee D H, Martinez J A, Vieth M, Ghadiri M R. Angew Chem. 1998;37:126–128. [Google Scholar]
- 33.Yao S, Ghosh I, Zutshi R, Chmielewski J. Nature (London) 1998;396:447–450. doi: 10.1038/24814. [DOI] [PubMed] [Google Scholar]
- 34.Zhan Z J, Ye J, Li X, Lynn D G. Curr Org Chem. 2001;5:885–902. [Google Scholar]
- 35.Hong J-I, Feng Q, Rotello V, Rebek J., Jr Science. 1992;255:848–850. doi: 10.1126/science.255.5046.848. [DOI] [PubMed] [Google Scholar]
- 36.Wilson D S, Szostak J W. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 37.Taylor S V, Kast P, Hilvert D. Angew Chem. 2001;40:3310–3335. doi: 10.1002/1521-3773(20010917)40:18<3310::aid-anie3310>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 38.Powell K A, Ramer S W, del Cardayré S B, Stemmer W P C, Tobin M B, Longchamp P F, Huisman G W. Angew Chem. 2001;40:3948–3959. doi: 10.1002/1521-3773(20011105)40:21<3948::aid-anie3948>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 39.Chyba C F, McDonald G D. Annu Rev Earth Planet Sci. 1995;23:215–249. doi: 10.1146/annurev.ea.23.050195.001243. [DOI] [PubMed] [Google Scholar]
- 40.Bartel D P, Szostak J W. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]