Abstract
Overproduction of the Her2 oncoprotein has been found in ≈30% of breast tumors, and patients who have Her2 excesses typically have more aggressive disease. Here we show that the expression of the Her2 gene can be decreased by inhibiting the interaction of the two cancer-linked proteins, DRIP130/CRSP130/Sur-2 (a Ras-linked subunit of human mediator complexes) and ESX (an epithelial-restricted transcription factor). Disruption of the interaction by a short cell-permeable peptide reduced the expression of the Her2 gene and specifically impaired the growth and viability of Her2-overexpressing breast cancer cells. The association of ESX with DRIP130 is mediated by a small hydrophobic face of an 8-aa helix in ESX, suggesting a therapeutic approach to incapacitating the Her2 gene by small organic molecules.
Overexpression of the oncogene Her2 (neu/ErbB2) has been found in ≈30% of breast tumors (1, 2). It is unclear why overexpression of this transmembrane receptor occurs in breast cancers, but patients who have Her2 excesses typically have more aggressive disease with enhanced metastasis and increased resistance to chemotherapy (1–6). Monoclonal antibodies against the Her2 protein have been successful in treating these Her2-positive patients (7–10). A humanized antibody called Herceptin has demonstrated tumor-inhibitory and chemosensitizing effects in clinical studies and is the only drug that the Food and Drug Administration has approved for treatment of Her2-overexpressing breast tumors (9, 10). The clinical success of the Her2-antibody therapy was an excellent example of the translation of basic cancer biology into clinical cancer treatment. However, the antibody therapy alone may not be ideal for therapeutic intervention of Her2-overexpressing breast cancers. In theory, down-regulation of Her2 may be accomplished efficiently by inhibiting the expression of the Her2 gene rather than targeting elevated levels of the Her2 proteins that are already overexpressed. By analogy with treatments for AIDS, cocktails of drugs, each with different mechanisms of action, might also be more effective at achieving complete remission of breast tumors. Thus, discovering a means to provide external control over Her2 expression, particularly through small organic molecules, remains appealing.
One of the critical transcription factors that activate the Her2 gene in breast cancers is ESX (ESE-1/ELF3/ERT/Jen), an Ets factor that is expressed specifically in epithelial cells including mammary glands (11–13). ESX binds and strongly activates the Her2 promoter (11), and the ESX-binding site in the Her2 promoter is absolutely required for the high-level expression of Her2 in breast cancer cells (14). The expression of ESX is boosted in Her2-overexpressing breast cancers (11); a simple correlation seems to exist between the expression levels of ESX and Her2 in breast cancer cells. Here we isolate DRIP130/CRSP130/Sur-2, a Ras-linked metazoan-specific subunit of human mediator complexes, as a nuclear cofactor that binds specifically to the transactivation domain of ESX. Disruption of the interaction between these two cancer-linked proteins decreases the expression of the Her2 gene and down-regulates the proliferation and viability of Her2-expressing breast cancer cells. Our findings suggest a therapeutic approach to incapacitating the Her2 gene in breast cancers and provide a broad implication that pharmacological manipulation of transcriptional coactivators may be a useful approach in the treatment of human disease.
Materials and Methods
Materials.
HeLa S3 and MCF7 cells were purchased from American Type Culture Collection. MCF7, MCF7-HER18, SK-BR-3, MDA-MB-453, and MDA-MB-468 cells were grown in DMEM/F-12 medium with 10% FBS. Z. Songyang (Baylor College of Medicine) kindly supplied 293Tag cells. The cDNA plasmid construct of DRIP130 (pcDNA3-FLAG-DRIP130) was a kind gift from L. P. Freedman (Memorial Sloan-Kettering Cancer Center). Peptides were synthesized by the fluorenylmethoxycarbonyl chemistry by using Rink amide methylbenzhydrylamine resins, purified by HPLC, and characterized by electrospray ionization mass spectroscopy and/or NMR.
Isolation of DRIP130.
HeLa cell nuclear extracts were prepared as described (15, 16). GST fusion proteins of the ESX activation domain or its activation-deficient mutant were obtained as described (17). The glutathione beads binding GST fusion protein or GST only were incubated with HeLa cell nuclear extracts and then extensively washed by buffer containing 20 mM Tris⋅Cl (pH 7.5), 150 mM NaCl, and 0.1% Nonidet P-40. The bound proteins were separated by SDS/PAGE and stained with Coomassie brilliant blue R-250. The protein bands stained with Coomassie brilliant blue R-250 were excised from the gel and microsequenced by mass spectrometry as described (18).
In Vitro Protein–Protein Interaction.
35S-Labeled DRIP130 proteins were prepared by TNT T7 Quick Coupled Transcription/translation system (Promega). The glutathione beads binding to GST fusion protein or GST only were incubated with the in vitro translated proteins in 500 μl of binding buffer containing 100 mM NaCl, 0.4% Nonidet P-40, 10% glycerol, 20 mM Tris⋅HCl (pH 7.5), 10 mM MgCl2, and 2 mM DTT for 1 h at 4°C. After extensive washing with the same buffer, the bound proteins were analyzed by SDS/PAGE.
NMR Studies.
ESX129–145 was dissolved in 95% H2O plus 5% 2H2O containing 50 mM KCl, 20 mM perdeuterated Tris-AcOH (pH 6.2), and 10 μM EDTA, and then the pH of the solution was adjusted to ≈6.1 by adding dilute KOH. The final peptide concentration was determined by UV absorption to be 3.6 mM. NMR experiments were performed on a Bruker AMX600 spectrometer. The sequential assignment of the peptide signals was obtained by using a combination of total correlation spectroscopy, double-quantum-filtered COSY, and NOESY data sets. In the NOESY spectra, 512 free induction decays were recorded at 295 K with mixing times of 200 and 350 ms.
Transcription Assay.
We used the mammalian expression vectors each encoding the activation domains of ESX, VP16, ALL1, C/EBPβ, or NF-κB p65 fused with the GAL4 DNA-binding domain (amino acids 1–94) as described (17). These vectors were cotransfected to 293Tag cells with a secreted alkaline phosphatase (SEAP) reporter gene (pG5IL2SX) and the expression vectors encoding full-length DRIP130 or its fragments. After 40-h incubation, an aliquot of the culture was assayed for SEAP activity as described (17). The total amounts of transfected DNA were normalized.
Immunoblotting.
The primary antibody used was the monoclonal antibody c-neu-Ab-3 (Oncogene Science). The blots were then incubated with horseradish peroxidase-conjugated goat anti-mouse Ig. Protein signals were detected with ECL chemiluminescence Western blotting detection reagents (Amersham Pharmacia).
Cell Nuclear Staining.
Cells were cultured on coverslips for 16 h in the presence or absence of peptides and fixed with acetone for 10 min at −20°C. After washing with PBS, cells were incubated with 0.5 μg/ml Hoechst 33342 and photographed with a Zeiss fluorescence microscope.
Results and Discussion
Isolation of DRIP130.
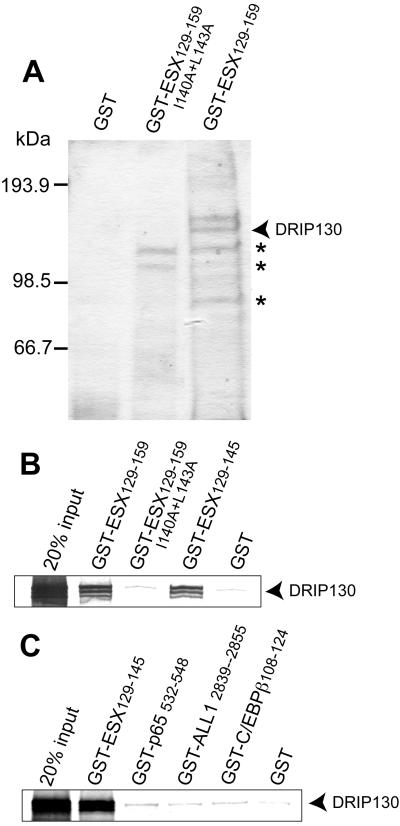
ESX has a strong activation domain whose activation potency is similar to that of the viral transcription factor VP16 (19). Although this 31-aa module has a binding site for hTAFII31, a general coactivator of transcription (17), its high potency cannot be explained by the presence of the single general coactivator. We isolated human proteins that bound to the activation domain of ESX from HeLa nuclear extracts (Fig. 1A). SDS/PAGE analyses of the isolated proteins showed that a 130-kDa protein bound tightly to the ESX activation domain but not to its activation-deficient mutant protein. Microsequencing data of a tryptic peptide matched to DRIP130 (CRSP130/Sur-2), a metazoan-specific component of the human mediator complexes known as DRIP (vitamin D receptor coactivator complex), CRSP (coactivator complex for Sp1), and ARC (activator-recruited cofactor) complexes (20–24). Other proteins that bound to the ESX activation domain (Fig. 1A) were abundant house-keeping proteins such as ATP-citrate lyase and 6-phosphofructokinase, and had affinity, although weaker, even to the activation-deficient mutant as revealed by silver staining of gels. We thus focused on DRIP130/Sur-2 for further analysis.
Figure 1.
(A) Purification of DRIP130 (CRSP130/Sur-2). A 130-kDa protein that binds specifically to the ESX activation domain was purified from HeLa nuclear extracts and determined to be human DRIP130 by microsequencing. Nonspecific proteins are indicated by stars. (B) In vitro translated DRIP130 binds the ESX activation domain but not its activation-deficient mutant. It is evident that a short peptide of ESX (ESX129–145) is sufficient for the interaction with DRIP130. (C) DRIP130 binds specifically to the ESX activation domain. Analogous amino acid sequences from the activation domains of human NF-κB p65, ALL1, C/EBPβ, and HSF-1 had no detectable affinity to DRIP130.
In vitro translated DRIP130 bound to the ESX activation domain but not its activation-deficient mutant protein (Fig. 1B). The 17-aa segment of the ESX activation domain (ESX129–145) was sufficient for mediating the interaction with DRIP130. DRIP130 seemed to recognize this short segment of ESX in a specific manner because DRIP130 had no detectable affinity to analogous the 17-aa peptides from the activation domains of NF-κB p65, ALL1, and C/EBPβ (Fig. 1C). DRIP130 is likely to be a nuclear protein that binds specifically to the activation domain of ESX.
Ectopic Expression of a Small DRIP130 Domain Impairs ESX.
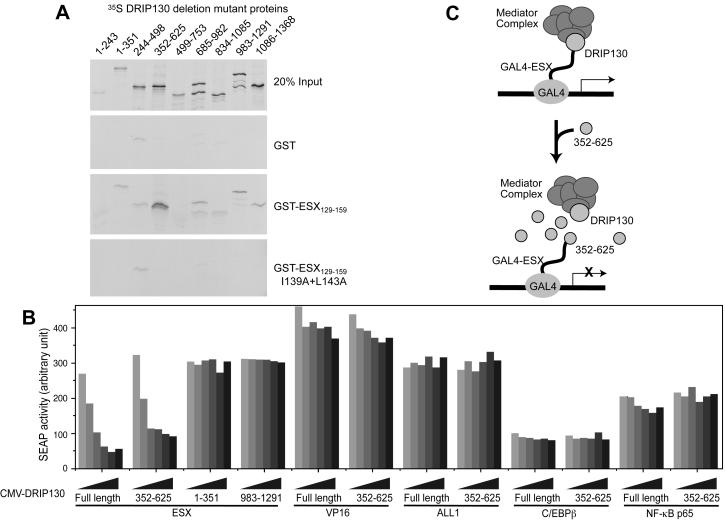
To map the minimal domain of DRIP130 that binds the ESX activation domain, a series of 35S-labeled deletion mutant proteins of DRIP130 were incubated with the GST fusion of the ESX activation domain, and the bound proteins were analyzed by SDS/PAGE. The DRIP130 fragment of amino acids 352–625 bound most selectively to the ESX activation domain, and its affinity was as high as that of full-length DRIP130 (Fig. 2A). The domain important for the interaction thus lies in the segment that emcompasses residues 352–625. Ectopic expression of this 274-aa fragment or full-length DRIP130 impaired the ability of the ESX activation domain to activate transcription in HEK293T cells, presumably by squelching the ESX activation domain from binding to endogenous DRIP130-containing mediator complexes (Fig. 2B). In contrast, overexpression of other DRIP130 fragments that had little affinity to the ESX activation domain, DRIP1301–351 and DRIP130983–1291, had no detectable impacts on the ability of the ESX activation domain to activate transcription. The inhibitory effect seems to be specific to the ESX activation domain because the forced expression of DRIP130 or its fragment had little effect on the activity of the activation domains of NF-κB p65, ALL1, or C/EBPβ. These results suggest that DRIP130 acts, by means of the DRIP/CRSP/ARC complex, as an ESX-selective coactivator.
Figure 2.
(A) Identification of a small DRIP130 domain that binds the ESX activation domain. A series of DRIP130 protein fragments were assayed for the ability to bind the GST fusion of the ESX activation domain (GST-ESX129–159). The amino acids 352–625 are sufficient for the interaction with the ESX activation domain. (B) Overexpression of the small DRIP130 domain specifically inhibits transcriptional activation by the ESX activation domain. A secreted alkaline phosphatase reporter gene driven by five GAL4-binding sites was transfected into 293Tag cells either with expression vectors of GAL4-ESX or GAL4-VP16. The effects of the cotransfection of DRIP130 expression vectors were examined. (C) Model for inhibition of GAL4-ESX activation by overexpression of DRIP130352–625.
DRIP130-Binding Peptide Reduces Her2 Expression in Breast Cancer Cells.
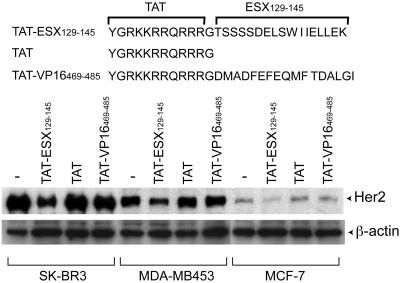
DRIP130 is a human homolog of Clostridium elegans Sur-2, a protein of unknown function whose mutations suppress the phenotype of an activated ras mutation (25), and it has been linked to the transcription-activating function of the adenovirus E1A protein and the Elk-1 transcription factor (22, 26). DRIP130 may act late in a Ras-MAP kinase signaling pathway as a unique coactivator for growth-promoting transcription factors including ESX. The functional association of DRIP130/Sur-2 both with the Ras-mediated tumor growth and the Her2 overexpression suggests that the DRIP130-ESX interaction might serve as a potential target for breast cancer therapy. To test this, we designed a cell-permeable peptide that binds DRIP130 (referred to as TAT-ESX129–145). The design takes advantage of the binding affinity of ESX129–145 and the cell-internalization property of the 11-aa peptide derived from the HIV TAT protein (27, 28). We first examined whether TAT-ESX129–145 reduces endogenous expression of the Her2 gene in three Her2-overexpressing breast cancer cell lines, SK-BR-3, MDA-MB-453, and MCF7 (ref. 29; Fig. 3). Cells were incubated in the presence or absence of TAT-ESX129–145, and the protein levels of Her2 were examined by Western blot analyses of normalized amounts of cell lysates. Repeated experiments showed that TAT-ESX129–145 reduced the protein levels of Her2 in all three Her2-overexpressing cell lines we tested, whereas the TAT sequence alone or TAT-VP16469–485 had no detectable effects. The expression levels of actin were unchanged by TAT-ESX129–145, suggesting its selective inhibition of Her2 expression.
Figure 3.
Effects of TAT-ESX129–145 on the protein levels of Her2 in Her2-expressing breast cancer cell lines. TAT-ESX129–145 (12.5 μM) significantly reduced the protein levels of Her2 in all of the three cell lines tested (SK-BR-3, MDA-MB-453, and MCF7), whereas TAT alone or TAT-VP16469–485 had little effects. The loaded amounts of cell lysates were normalized by Bradford assays and Coomassie blue staining. The Her2 protein and β-actin were detected by Western blots.
Inhibition of DRIP130 Induces Cell Death in Her2-Overexpressing Breast Cancer Cells.
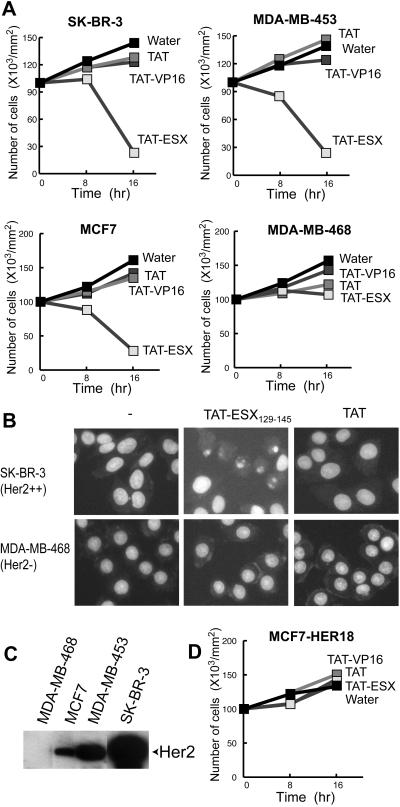
We next examined the effects of TAT-ESX129–145 on the growth of Her2-overexpressing breast cancer cells. The growth of SK-BR-3, MDA-MB-453, and MCF7 cells was completely blocked in the presence of TAT-ESX129–145, whereas TAT alone and TAT-VP16469–485 had little effects on the growth of these cell lines (Fig. 4A). Prolonged incubation with TAT-ESX129–145 reduced the number of living cells and resulted in apoptotic cell death as judged by the nuclear condensation (Fig. 4B). This observation is consistent with the notion that Her2 plays a role in antiapoptosis pathways (30); for example, Her2 antisense oligonucleotides have been shown to induce apoptosis in Her2-overexpressing cells, just as we observed in the present study (31, 32).
Figure 4.
(A and D) Effects of TAT-ESX129–145 on the growth of breast cancer cells. Five breast cancer cell lines were incubated with TAT-ESX129–145. TAT-ESX129–145 impaired the growth and viability of Her2-expressing breast cancer cells (SK-BR-3, MDA-MB-453, and MCF7), whereas it had much less effect on MDA-MB-468 cells with no detectable levels of Her2. MCF7-HER18, an MCF7-derived cell line in which Her2 is overexpressed on a heterologous promoter, was resistant to TAT-ESX129–145. The concentration of TAT-ESX129–145 is 25 μM. (B) Nuclear condensation induced by TAT-ESX129–145. SK-BR-3 and MDA-MB-468 cells were incubated with TAT-ESX129–145 (25 μM) or TAT (25 μM) for 8 h. The cells were then fixed and stained with Hoechst 33342 (10 μg/ml) for 10 min. A typical apoptotic phenotype of the nuclei was observed only when the Her2-overexpressing SK-BR-3 cells were incubated with TAT-ESX129–145. (C) Western blots of Her2. It is evident that MDA-MB-468 has no detectable levels of Her2.
In contrast to the Her2-overexpressing cells, MDA-MB-468, a breast cancer cell line with no detectable levels of Her2 (Fig. 4C), was resistant to TAT-ESX129–145 (Fig. 4 A and B). The limited response of MDA-MB-468 to TAT-ESX129–145 demonstrates the specificity of TAT-ESX129–145 in down-regulating the proliferation and viability of Her2-expressing breast cancer cells. These results collectively suggest that disruption of the interaction between ESX and DRIP130 decreases Her2 expression and causes cell death in Her2-expressing cancer cells.
Ectopic Expression of Her2 on a Heterologous Promoter Rescues Cells from the TAT-ESX129–145-Mediated Cell Death.
DRIP130 and ESX are almost certainly involved in the control of several growth-linked genes in addition to Her2. It is possible to imagine that the apoptotic cell death observed with Her2-positive breast cancer cells is mediated by some other genes that are regulated by ESX or DRIP130. To address this issue, we examined the effects of TAT-ESX129–145 on an MCF7 cell line stably transfected with the Her2 gene. The ectopic expression of Her2 on a heterologous promoter effectively rescued MCF7 cells from the TAT-ESX129–145-mediated cell death (Fig. 4D). Although the expression of other genes may be influenced by TAT-ESX129–145, the expression of Her2 is likely to be the primary target for the TAT-ESX129–145 peptide to induce apoptosis specifically in Her2-positive cells.
Characterization of the Interaction of ESX with DRIP130.
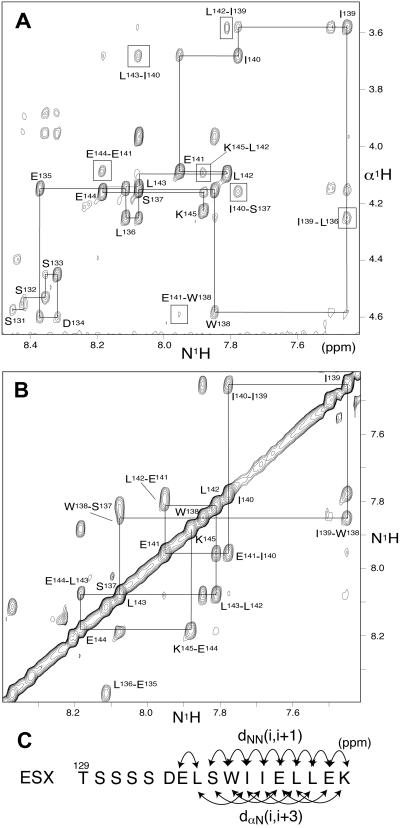
Although TAT-ESX129–145 specifically killed Her2-positive cells by inhibiting Her2 expression, the peptide is not practical to use clinically because of its rapid degradation and poor cellular uptake. Nonpeptidylation of a lead peptide usually requires its detailed characterization. We examined the structural characteristics of isolated ESX129–145 by NMR spectroscopy. Despite the low molecular weight of ESX129–145 and the standard experimental condition we used (in a 5% D2O/95% H2O solution buffered at pH 6.1 at 295 K), ESX129–145 exhibited a substantial number of interresidue nuclear Overhauser effects (NOEs) including those between successive amide protons in the main chain and those arising from dαN(i, i + 3) (Fig. 5 A and B). The pattern of the NOE connectivities is characteristic of the pattern observed for α-helical secondary structures, especially in the region from Ser-137 to Lys-145 (Fig. 5C). Thus, the residues from Ser-137 to Lys-145 have strong helical propensity and perhaps form a transient α-helical structure that may play a role in the association with DRIP130.
Figure 5.
NMR studies. (A) Expansion of the NH-αH region of an NOESY spectrum of ESX129–145. The sequential connectivities and long-range interresidue NOEs are indicated by solid lines and boxes, respectively. (B) Expansion of the NH-NH region of a NOESY spectrum of ESX129–145. Clear NH-NH connectivities were observed in a COOH-terminal segment of the peptide as indicated by solid lines. (C) Summary of the interresidue NOE data. The NOE connectivities in the region from S137 to E144 are characteristic of that observed for an α-helical structure.
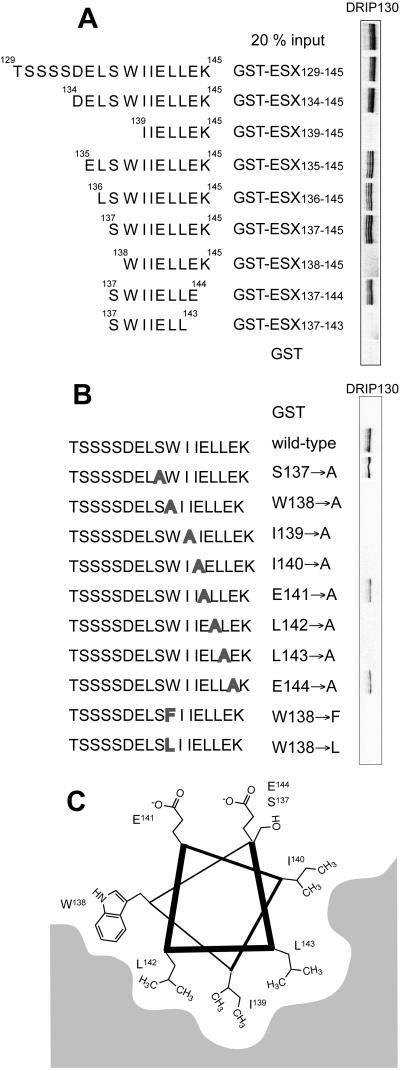
To support this model independently, we constructed deletion and point mutant proteins of ESX129–145 and examined their ability to bind DRIP130 in vitro (Fig. 6A). The results of the deletion mutants indicate that the residues from Ser-137 to Glu-144 are necessary for the interaction with DRIP130; this 8-aa segment corresponds almost precisely to the region that was detected to be helical by NMR. Substitutions of bulky hydrophobic residues in the short peptide abolished the interaction with DRIP130, whereas mutations of the hydrophilic residues had no detectable effects (Fig. 6B). The projection of residues Ser-137 to Glu-144 onto a helical wheel reveals that these hydrophobic residues all lie along one face of the short helix, perhaps making direct contact with the chemically complementary surface in DRIP130 (Fig. 6C).
Figure 6.
(A) Identification of the minimal ESX peptide that binds DRIP130 in vitro. It is evident that 35S-labeled DRIP130 binds an 8-aa segment of the ESX activation domain (ESX137–144). (B) Summary of mutational studies. Point mutants of ESX129–145 were tested for their ability to bind DRIP130. Although the Ala substitution of hydrophilic residues had no effect, those of hydrophobic residues in the putative α-helical region abolished the interaction with DRIP130. The indole structure of Trp-138 seems to be important for the interaction because either substitution with Ala, Phe, or Leu abolished the interaction with DRIP130. (C) Projection of the minimal binding peptide, ESX137–144, onto a helical wheel.
Toward the Discovery of Small-Molecule Inhibitors.
Despite the recent success of Her2 receptor-targeted therapeutics, emerging resistance mechanisms now point to the clinical need for additional therapeutic alternatives and combinatorial approaches. Strategies that transcriptionally inhibit Her2 expression by small molecules would be desirable; however, discovery of simple organic molecules that modulate transcription of specific genes has always been a challenge. In an early effort, an antieheumatic gold salt, which inhibits DNA binding of AP2 and other zinc finger transcription factors, was shown to down-regulate Her2 promoter activity and endogenous Her2 expression in cultured cells (33). Triple-helix-forming oligonucleotides that target the Her2 promoter have also been tested by several groups (34–37). Other strategies include design of pyrrole-imidazole polyamides that bind the ESX-binding site in the Her2 promoter (38). None of these approaches that address DNA–protein interactions have yet reached clinical trial.
Our results suggest that the DRIP130-ESX interaction serves as a pharmaceutical target for Her2-overexpressing breast cancers. A combination of NMR and biochemical experiments indicated that the interaction of ESX with DRIP130 is mediated primarily by five hydrophobic residues clustered onto one face of the helix: Trp-138, Ile-140, Leu-142, Ile-139, and Leu-143. The relatively small size of the interface suggests the feasibility of developing organic molecule inhibitors. Among those five residues that are clustered onto one face of the helix, Trp-138 may make a unique contribution to the interaction: substitutions of Trp-138 either with Leu or Phe greatly impaired the affinity to DRIP130, and thus it is not only the simple hydrophobic or aromatic character of Trp-138 but its indole structure that plays an essential role in binding specifically to DRIP130 (Fig. 6A). Taken together, it seems reasonable to foresee the discovery of low-molecular-weight inhibitors from chemical libraries enriched in the structural families of indole, carbazole, and benzodiazepin, indole-mimicking π-electron-rich pharmacores that are found in many bioavailable drugs. Successful isolation of small molecule inhibitors from a chemical library would demonstrate the “druggability” of the ESX-DRIP130 interaction and supply promising leads toward the clinical application.
Current drug therapy addresses only about 500 molecular targets. Cell membrane receptors and enzymes account for ≈80% of all current drug targets, and a few drug targets are in the nucleus except for nuclear receptors (and DNA in the case of cancer chemotherapy) (39). Discovering new “druggable” targets in the nucleus would extend the scope of drug targets; for instance, recent discovery of histone deacetylases as a potential target for cancer therapy had tremendous impact on the drug-discovery research (40–43). In this report we propose DRIP130, another type of nuclear protein involved in transcription, as a pharmaceutical target for cancer therapy. The potentiality of transcriptional coactivators as a drug target has never been explored because of the lack of expected therapeutic phenotypes. Our findings suggest that a subset of coactivators may serve as a target for pharmaceutical intervention. Drug-discovery efforts for ESX-DRIP130 inhibitors may provide a unique case study.
Acknowledgments
We thank X. Gao for assistance in using the NMR instruments at the University of Houston, L. P. Freedman for a plasmid construct of DRIP130, Z. Songyang for 293Tag cells, and Y. Torii for technical assistance. We are also grateful to S. Yoshikawa, Y. Watanabe, and the members of the Uesugi laboratory for encouragement and discussion. This work was supported in part by grants from Concern Foundation and Welch Foundation. Y.C. is a predoctoral fellow of the U.S. Department of Defense.
Abbreviations
- SEAP
secreted alkaline phosphatase
- DRIP
vitamin D receptor coactivator cofactor
- NOE
nuclear Overhauser effect
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Slamon D J, Clark G M, Wong S G, Levin W J, Ullrich A, McGuire W L. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 2.Slamon D J, Godolphin W, Jones L A, Holt J A, Wong S G, Keith D E, Levin W J, Stuart S G, Udove J, Ullrich A, et al. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 3.Guy C T, Webster M A, Schaller M, Parsons T J, Cardiff R D, Muller W J. Proc Natl Acad Sci USA. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu D H, Hung M C. Oncogene. 1991;6:1991–1996. [PubMed] [Google Scholar]
- 5.Gusterson B A, Gelber R D, Goldhirsch A, Price K N, Save-Soderborgh J, Anbazhagan R, Styles J, Rudenstam C M, Golouh R, Reed R, et al. J Clin Oncol. 1992;10:1049–1056. doi: 10.1200/JCO.1992.10.7.1049. [DOI] [PubMed] [Google Scholar]
- 6.Yu D, Liu B, Jing T, Sun D, Price J E, Singletary S E, Ibrahim N, Hortobagyi G N, Hung M C. Oncogene. 1998;16:2087–2094. doi: 10.1038/sj.onc.1201729. [DOI] [PubMed] [Google Scholar]
- 7.Drebin J A, Link V C, Stern D F, Weinberg R A, Greene M I. Cell. 1985;41:695–706. doi: 10.1016/s0092-8674(85)80050-7. [DOI] [PubMed] [Google Scholar]
- 8.Drebin J A, Link V C, Weinberg R A, Greene M I. Proc Natl Acad Sci USA. 1986;83:9129–9133. doi: 10.1073/pnas.83.23.9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stebbing J, Copson E, O'Reilly S. Cancer Treat Rev. 2000;26:287–290. doi: 10.1053/ctrv.2000.0182. [DOI] [PubMed] [Google Scholar]
- 10.Dillman R O. Cancer Biother Radiopharmacol. 1999;14:5–10. doi: 10.1089/cbr.1999.14.5. [DOI] [PubMed] [Google Scholar]
- 11.Chang C H, Scott G K, Kuo W L, Xiong X, Suzdaltseva Y, Park J W, Sayre P, Erny K, Collins C, Gray J W, Benz C C. Oncogene. 1997;14:1617–1622. doi: 10.1038/sj.onc.1200978. [DOI] [PubMed] [Google Scholar]
- 12.Tymms M J, Ng A Y, Thomas R S, Schutte B C, Zhou J, Eyre H J, Sutherland G R, Seth A, Rosenberg M, Papas T, et al. Oncogene. 1997;15:2449–2462. doi: 10.1038/sj.onc.1201427. [DOI] [PubMed] [Google Scholar]
- 13.Andreoli J M, Jang S I, Chung E, Coticchia C M, Steinert P M, Markova N G. Nucleic Acids Res. 1997;25:4287–4295. doi: 10.1093/nar/25.21.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott G K, Daniel J C, Xiong X, Maki R A, Kabat D, Benz C C. J Biol Chem. 1994;269:19848–19858. [PubMed] [Google Scholar]
- 15.Dignam J D, Martin P L, Shastry B S, Roeder R G. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 16.Okumura K, Sakaguchi G, Takagi S, Naito K, Mimori T, Igarashi H. J Biol Chem. 1996;271:12944–12950. doi: 10.1074/jbc.271.22.12944. [DOI] [PubMed] [Google Scholar]
- 17.Choi Y, Asada S, Uesugi M. J Biol Chem. 2000;275:15912–15916. doi: 10.1074/jbc.275.21.15912. [DOI] [PubMed] [Google Scholar]
- 18.Ogryzko V V, Kotani T, Zhang X, Schlitz R L, Howard T, Yang X J, Howard B H, Qin J, Nakatani Y. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 19.Chang C H, Scott G K, Baldwin M A, Benz C C. Oncogene. 1999;18:3682–3695. doi: 10.1038/sj.onc.1202674. [DOI] [PubMed] [Google Scholar]
- 20.Naar A M, Beaurang P A, Zhou S, Abraham S, Solomon W, Tjian R. Nature (London) 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 21.Rachez C, Lemon B D, Suldan Z, Bromleigh V, Gamble M, Naar A M, Erdjument-Bromage H, Tempst P, Freedman L P. Nature (London) 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 22.Boyer T G, Martin M E, Lees E, Ricciardi R P, Berk A J. Nature (London) 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 23.Wang G, Cantin G T, Stevens J L, Berk A J. Mol Cell Biol. 2001;21:4604–4613. doi: 10.1128/MCB.21.14.4604-4613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryu S, Zhou S, Ladurner A G, Tjian R. Nature (London) 1999;397:446–450. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- 25.Singh N, Han M. Genes Dev. 1995;9:2251–2265. doi: 10.1101/gad.9.18.2251. [DOI] [PubMed] [Google Scholar]
- 26.Stevens J L, Cantin G T, Wang G, Shevchenko A, Shcevchenko A, Berk A J. Science. 2002;296:755–758. doi: 10.1126/science.1068943. [DOI] [PubMed] [Google Scholar]
- 27.Schwarze S R, Ho A, Vocero-Akbani A, Dowdy S F. Science. 1999;285:1569–1572. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 28.Nagahara H, Vocero-Akbani A M, Snyder E L, Ho A, Latham D G, Lissy N A, Becker-Hapak M, Ezhevsky S A, Dowdy S F. Nat Med. 1998;4:1449–1452. doi: 10.1038/4042. [DOI] [PubMed] [Google Scholar]
- 29.Perou C M, Sorlie T, Eisen M B, van de Rijn M, Jeffrey S S, Rees C A, Pollack J R, Ross D T, Johnsen H, Akslen L A, et al. Nature (London) 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 30.Zhou B P, Hu M C, Miller S A, Yu Z, Xia W, Lin S Y, Hung M C. J Biol Chem. 2000;275:8027–8031. doi: 10.1074/jbc.275.11.8027. [DOI] [PubMed] [Google Scholar]
- 31.Roh H, Pippin J A, Green D W, Boswell C B, Hirose C T, Mokadam N, Drebin J A. Oncogene. 2000;19:6138–6143. doi: 10.1038/sj.onc.1204001. [DOI] [PubMed] [Google Scholar]
- 32.Roh H, Pippin J, Drebin J A. Cancer Res. 2000;60:560–565. [PubMed] [Google Scholar]
- 33.Hollywood D P, Hurst H C. Br J Cancer. 1995;71:753–757. doi: 10.1038/bjc.1995.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ebbinghaus S W, Gee J E, Rodu B, Mayfield C A, Sanders G, Miller D M. J Clin Invest. 1993;92:2433–2439. doi: 10.1172/JCI116850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ebbinghaus S W, Fortinberry H, Gamper H B., Jr Biochemistry. 1999;38:619–628. doi: 10.1021/bi980981g. [DOI] [PubMed] [Google Scholar]
- 36.Noonberg S B, Scott G K, Hunt C A, Hogan M E, Benz C C. Gene. 1994;149:123–126. doi: 10.1016/0378-1119(94)90420-0. [DOI] [PubMed] [Google Scholar]
- 37.Porumb H, Gousset H, Letellier R, Salle V, Briane D, Vassy J, Amor-Gueret M, Israel L, Taillandier E. Cancer Res. 1996;56:515–522. [PubMed] [Google Scholar]
- 38.Chiang S Y, Burli R W, Benz C C, Gawron L, Scott G K, Dervan P B, Beerman T A. J Biol Chem. 2000;275:24246–24254. doi: 10.1074/jbc.M000820200. [DOI] [PubMed] [Google Scholar]
- 39.Drews J. Science. 2000;287:1960–1964. doi: 10.1126/science.287.5460.1960. [DOI] [PubMed] [Google Scholar]
- 40.Kwon H J, Owa T, Hassig C A, Shimada J, Schreiber S L. Proc Natl Acad Sci USA. 1998;95:3356–3361. doi: 10.1073/pnas.95.7.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hassig C A, Tong J K, Schreiber S L. Chem Biol. 1997;4:783–789. doi: 10.1016/s1074-5521(97)90111-3. [DOI] [PubMed] [Google Scholar]
- 42.Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 43.Lin R J, Nagy L, Inoue S, Shao W, Miller W H, Jr, Evans R M. Nature (London) 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]