Abstract
Folding simulations of polyalanine peptides were carried out using an off-lattice Monte Carlo simulation technique. The peptide was represented as a chain of residues, each of which contains two interaction sites: one corresponding to the Cα atom and the other to the side chain. A statistical potential was used to describe the interaction between these sites. The preferred conformations of the peptide chain on the energy surface, starting from several initial conditions, were searched by perturbations on its generalized coordinates with the Metropolis criterion. We observed that, at low temperatures, the effective energy was low and the helix content high. The calculated helix propagation (s) and nucleation (σ) parameters of the Zimm-Bragg model were in reasonable agreement with the empirical data. Exploration of the energy surface of the alanine-based peptides (AAQAA)3 and AAAAA(AAARA)3A demonstrated that their behavior is similar to that of polyalanine, in regard to their effective energy, helix content, and the temperature-dependence of their helicity. In contrast, stable secondary structures were not observed for (Gly)20 at similar temperatures, which is consistent with the nonfolder nature of this peptide. The fluctuations in the slowest dynamics mode, which describe the elastic behavior of the chain, showed that as the temperature decreases, the polyalanine peptides become stiffer and retain conformations with higher helix content. Clustering of conformations during the folding phase implied that polyalanine folds into a helix through fewer numbers of intermediate conformations as the temperature decreases.
INTRODUCTION
The folding and stability of helices were studied experimentally (Thompson et al., 1997; Scholtz et al., 1991; Huang et al., 2001; Chakrabartty and Baldwin, 1995; Ferguson and Fersht, 2003), theoretically (Lifson and Roig, 1961; Schellman, 1958; Zimm and Bragg, 1959; Doig, 2002; Munoz and Serrano, 1994), and computationally (Bertsch et al., 1998; Chowdhury et al., 2003; Ding et al., 2003; Ferrara et al., 2000; Gilis and Rooman, 2001; Huo and Straub, 1999; Levy and Karplus, 1979; Levy et al., 2001; Smith and Hall, 2001; Young and Brooks, 1996; Yang and Honig, 1995). Most of these studies were based on the Zimm-Bragg model (Zimm and Bragg, 1959) and its generalizations (Lifson and Roig, 1961), which describe thermodynamic properties of the coil-helix transition, in which the biopolymer is modeled as a chain of amino acids that can be in a coil or helical state. The tendency to form helices is determined by the nucleation parameter (σ) and an equilibration constant for helix propagation (s). These parameters provide a convenient means of comparing α-helix propensities as measured by different methods. Alanine-based peptides such as (AAQAA)3, which adopt significant populations of helical structure in aqueous solution, have served as an excellent experimental model system for studies of the helix-coil transition (Huang et al., 2001; Shalongo et al., 1994; Thompson et al., 1997; Chakrabartty et al., 1994)
The Rouse chain model (Rouse, 1953) describes the intrinsic dynamic modes of a flexible chain, taking into account only the local interactions. For globular structures, where tertiary interactions also need to be incorporated, a recent Gaussian network method (GNM) (Bahar et al., 1997a; Haliloglu et al., 1997) can be applied. The GNM assumes that the protein is a three-dimensional elastic network. The nodes are the Cα atoms, and the connectors between the nodes in close proximity are described as springlike interactions that undergo Gaussian-distributed fluctuations. This model serves as an efficient analytical tool to search for the dynamic modes of globular proteins with a given topology.
Although analytical models provide conceptual understanding of α-helix folding and stability, molecular dynamics (Bertsch et al., 1998; Levy et al., 2001; Shirley and Brooks, 1997) can provide us with a detailed atomic understanding, but require computationally intensive simulations. Thus, it is advantageous to use methods that have an intermediate level of complexity between simple analytical models and full dynamic simulations that employ atomistic models. For instance, it is possible to use coarse-grained models, where each residue is represented by a single interaction site. However, such models are deficient in several aspects, including the lack of backbone, side chains, and solvent atoms, as well as the neglect of the effects of the particular amino-acid sequence, e.g., hydrophobicity and polarity. A possible compromise is to use an intermediate-resolution model, which is computationally efficient, yet still provides information about the dynamics and thermodynamics of folding.
In this work we present a novel method to study the folding and stability of short peptides. We present here Monte Carlo (MC) simulations of polyalanine peptides and the alanine-based peptides (AAQAA)3 and AAAAA(AAARA)3A. The latter peptides are commonly used for studies of the helix-coil transition (Huang et al., 2001; Shalongo et al., 1994; Thompson et al., 1997; Chakrabartty et al., 1994), which enables us to compare our results with previous experimental works. A polyglycine peptide was chosen as an example of a nonfolder peptide. We used an off-lattice protein model that, on the one hand, contains enough protein character to capture the peptide's equilibrium and dynamic properties and, on the other hand, is computationally feasible for studies of intact proteins and protein-membrane systems (Bahar et al., 1997b; Haliloglu and Bahar, 1998; Kurt and Haliloglu, 1999, 2001; Kessel et al., 2003). The structure of the peptide was described by a two-bead model, in which each amino acid is represented by two interaction centers; one for the backbone and one for the side chain. Statistical potentials, which have been obtained from the Protein Data Bank of proteins of known structure, were used as an energy function.
THEORETICAL BACKGROUND
Helix-coil theory
According to the Zimm-Bragg model (Zimm and Bragg, 1959), the polypeptide chains are represented as a one-dimensional array of linked residues, where each of the residues can either be in a coil state or in a helix state. The folding is characterized by two parameters: a nucleation parameter σ that corresponds to the statistical weight for nucleating a helical segment; and a propagation parameter s that corresponds to the statistical weight of elongation of a helical segment (a measure of helix propensity). According to this model, at the limit of a long chain, the average number of helical residues 〈n〉 and the average length of the helical segment 〈l〉 are related to σ and s according to Eqs. 1a and 1b (Smith and Hall, 2001; Cantor and Schimmel, 1980),
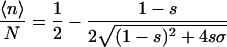 |
(1a) |
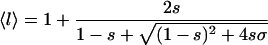 |
(1b) |
where N is the number of residues with well-defined rotational angles in the peptide, as described below. In accordance with the works of others (Mitsutake and Okamoto, 2000; Smith and Hall, 2001; Ferrara et al., 2000), for the purpose of evaluating 〈n〉, a residue i was considered to be in a helical state if it was a part of a segment of at least three residues whose backbone rotational angles were within the helical region. In our reduced model, a residue was defined to be in the helical region if the rotational angles 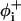 and
and 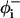 (Fig. 1) of its two adjacent bonds lay within the interval −120° ± 30°. Since we cannot define the rotational angles of the peptide's ends, we neglected the first two residues at the N-terminal and the last two residues at the C-terminal to obtain N = L−4, where L is the chain length.
(Fig. 1) of its two adjacent bonds lay within the interval −120° ± 30°. Since we cannot define the rotational angles of the peptide's ends, we neglected the first two residues at the N-terminal and the last two residues at the C-terminal to obtain N = L−4, where L is the chain length.
FIGURE 1.
A schematic representation of the virtual bond model. A segment between backbone units 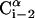 and
and  is shown. The side chain attached to the ith Cα is marked as Si. φi is the rotational angle of the ith virtual bond (connecting
is shown. The side chain attached to the ith Cα is marked as Si. φi is the rotational angle of the ith virtual bond (connecting  and
and 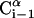 ); it is defined by four consecutive atoms
); it is defined by four consecutive atoms 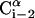 ,
, 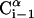 ,
, 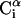 , and
, and 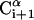 . θi is the bond angle between virtual bonds i and i+1.
. θi is the bond angle between virtual bonds i and i+1. 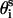 (between li and
(between li and  , where
, where 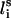 is the side-chain virtual bond vector pointing from
is the side-chain virtual bond vector pointing from 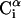 to Si) and
to Si) and 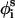 (defined by four consecutive atoms
(defined by four consecutive atoms 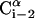 ,
,  ,
,  , and Si) are the side-chain bond and torsional angles, respectively.
, and Si) are the side-chain bond and torsional angles, respectively.
Dynamic modes
The behavior of a flexible chain can be approximated by the Rouse chain model (Rouse, 1953), where intramolecular interactions are limited to the nearest neighbors, and the Brownian motion of each bead along the chain is described by a harmonic oscillator. The slowest mode of the Rouse chain model approximately describes the most cooperative/collective mode of fluctuations expected from a simple flexible chain. In a real protein, however, the interactions may extend to the next neighbors along the sequence and to any residue within the spatial vicinity. The GNM is an analytical tool that incorporates all the interactions within the reach of the first coordination shell to describe the harmonic fluctuations of residues in the structure for a given topology. When only interactions between nearest neighbors are considered, the results of the GNM analysis are reduced to those of the Rouse chain model.
Within the GNM model, the protein structure is modeled as a three-dimensional elastic network, similar to the random network model proposed for polymers (Flory, 1976). The residues, which are represented as interaction sites at their α-carbons, are treated equally regardless of their type. Their mutual interactions depend only on their proximity in three-dimensional space regardless of whether they are covalently bonded to each other or not. In this network representation of the folded protein, the interactions between all residue pairs in close proximity, i.e., separated by less than a preset cutoff distance, are described by a linear spring of a uniform force constant. The residues undergo fluctuations around their mean positions, and the magnitudes of the fluctuations follow a Gaussian distribution. The number of contacts and their spatial distribution in a given topology dictates the dynamic characteristics of the protein, i.e., the spectrum of vibrational modes.
The correlation between the fluctuations ΔRi and ΔRj of residues i and j at equilibrium is given (Bahar et al., 1997a) by
 |
(2) |
where kB is the Boltzmann constant, T is the absolute temperature, and γ is the normalization constant that is the counterpart of the single parameter of the Hookean pairwise potential originally proposed by Tirion (1996). Γ is a symmetric matrix known as the Kirchoff or connectivity matrix of α-carbon atoms, including all residue pairs within the first interaction shell (6.8 Å) (Bahar and Jernigan, 1997; Miyazawa and Jernigan, 1985). The Kirchoff matrix may be viewed as the counterpart of the classical Rouse matrix (Bird et al., 1987; Rouse, 1953; Doi and Edwards, 1986). U is an orthogonal matrix whose columns uk, 1 < k < L, are the eigenvectors of Γ, and Λ is the diagonal matrix of the non-zero eigenvalues (λk) of Γ. The kth eigenvalue represents the frequency of the kth mode of motion, and the kth eigenmode gives the shape of this mode as a function of residue index. Thus, the dynamics of a biomolecular system can be decomposed by GNM into a collection of internal modes of different frequencies, similar to normal mode analysis (Go et al., 1983; Levy and Karplus, 1979), but in a computationally efficient way. GNM provides a detailed description of the dynamics around a local energy minimum for a given topology.
We used GNM to study the fluctuations in the preferred conformations of the peptide during folding and in the folded state. The global/slowest mode describes the elasticity of the whole chain. This analysis by the GNM displays the correlation between the elasticity of the chain, i.e., the distribution of the fluctuations in the most dominant mode, and the sampled low energy conformations during the simulations.
MODEL AND METHODS
Peptide representation
We used a simplified model in which each residue i was represented by two interaction sites, its α-carbon atom (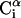 ) and its side-chain interaction center Si (Fig. 1). The side-chain interaction centers were selected on the basis of the specific structure and energy characteristics of the amino acids (Bahar and Jernigan, 1996). The peptide's backbone was represented by the virtual bond model, originally proposed by Flory (1969). Within the model, the actual chemical bonds linking the backbone atoms are replaced by virtual bonds that connect successive Cα atoms; a peptide of L residues has L−1 such bonds. Virtual bonds are highly stiff and were taken here with a fixed equilibrium length li of 3.81 ± 0.03 Å. Thus, the peptide backbone conformation can be defined by the 2L−5 dimensional vector {θ2, θ3, …, θL−1, φ3, φ4, …, φL−1} corresponding to L−2 virtual bond angles (θi) at the ith carbon, and L−3 dihedral angles (φi) at the ith virtual bond. The side-chain interaction centers were selected on the basis of the specific structure and energy characteristics of the amino acids (Bahar and Jernigan, 1996). In this work we modeled four types of amino acids: Ala, Gln, Arg, and Gly. Si of Ala was defined at the Cβ atom. The Si of Gln was defined at the amide group. Si of Arg was defined at the guanidine group. Gly was presented as a single-bead model at the Cα atom. Assuming that the distance between Si and
) and its side-chain interaction center Si (Fig. 1). The side-chain interaction centers were selected on the basis of the specific structure and energy characteristics of the amino acids (Bahar and Jernigan, 1996). The peptide's backbone was represented by the virtual bond model, originally proposed by Flory (1969). Within the model, the actual chemical bonds linking the backbone atoms are replaced by virtual bonds that connect successive Cα atoms; a peptide of L residues has L−1 such bonds. Virtual bonds are highly stiff and were taken here with a fixed equilibrium length li of 3.81 ± 0.03 Å. Thus, the peptide backbone conformation can be defined by the 2L−5 dimensional vector {θ2, θ3, …, θL−1, φ3, φ4, …, φL−1} corresponding to L−2 virtual bond angles (θi) at the ith carbon, and L−3 dihedral angles (φi) at the ith virtual bond. The side-chain interaction centers were selected on the basis of the specific structure and energy characteristics of the amino acids (Bahar and Jernigan, 1996). In this work we modeled four types of amino acids: Ala, Gln, Arg, and Gly. Si of Ala was defined at the Cβ atom. The Si of Gln was defined at the amide group. Si of Arg was defined at the guanidine group. Gly was presented as a single-bead model at the Cα atom. Assuming that the distance between Si and  (
(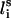 ) is fixed at its equilibrium value and that the angle between li and
) is fixed at its equilibrium value and that the angle between li and 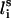 (
(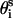 ) is also fixed at its equilibrium value, the conformation of side-chain i can be expressed by the torsion angle (
) is also fixed at its equilibrium value, the conformation of side-chain i can be expressed by the torsion angle (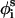 ).
).
Effective energy
The effective energy of any conformation Φ can be decomposed into contributions from short-range (ESR) and long-range (ELR) interactions:
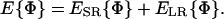 |
(3) |
The short-range conformational energy was calculated as described by Bahar et al. (1997c) using the formulation
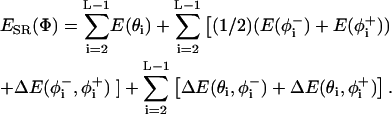 |
(4) |
The first summation refers to the distortion of the virtual bond angles and the second is for the bond rotational angles, in which φ− and φ+ refer to the rotational angles of the virtual bonds preceding and succeeding the ith α-carbon and the coupling between the latter angles. The last term accounts for the coupling between the rotational and bond-angle distortions.
ELR {Φ} in Eq. 3 was calculated based on the statistical potential of Bahar and Jernigan (1996), using the expression
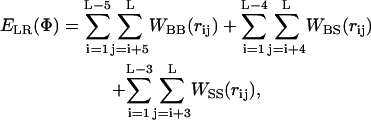 |
(5) |
where rij is the distance between interaction sites i and j in conformation Φ. Equation 5 accounts for backbone-backbone (BB), backbone-side chain (BS), and side-chain/side-chain interactions (SS).
The potentials were derived from structures in the Protein Data Bank and were based on large and tightly packed proteins (Bahar and Jernigan, 1996; Bahar et al., 1997c). These residue-specific potentials differ from a typical 6–12 Lennard Jones atomic potential function by the existence of multiple minima (Bahar and Jernigan, 1997). This is a characteristic property of potentials of mean force in dense systems, and the multiple minima emerge because in a large protein each residue is typically surrounded by two or more layers of residues. The potential of mean force contains information about the supersecondary structures and overall topology. In a typical case, two energy minima appear, reflecting the first and second coordination shells around a residue. In preliminary simulations we found that incorporating all the interactions along the backbone, regardless of the distance between the interacting sites, results in the formation of compact structures such as α-hairpins, even for short peptides of ∼15 residues (data not shown). As a result, the average helix content was much lower and the helical segments shorter. The formation of compact structures for short peptides, which is in conflict with the available experimental data, is attributed to a minimum corresponding to the second coordination shell in the potential of Eq. 5, which overemphasizes the long-range interactions for short peptides. In fact, the residues of parallel α-helices are separated by 8–10 Å, whereas, 5.5–6.5 Å is a typical range for pairs of  and
and 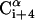 in α-helices (Bahar and Jernigan, 1997). To reduce the contribution of long-range interactions, we introduced into Eq. 5 a cutoff distance of 6.8 Å, which corresponds to the first coordination shell of interactions (Bahar and Jernigan, 1997; Miyazawa and Jernigan, 1985). That is, residue pairs i,j with ri,j > 6.8 Å were excluded from the summation in Eq. 5.
in α-helices (Bahar and Jernigan, 1997). To reduce the contribution of long-range interactions, we introduced into Eq. 5 a cutoff distance of 6.8 Å, which corresponds to the first coordination shell of interactions (Bahar and Jernigan, 1997; Miyazawa and Jernigan, 1985). That is, residue pairs i,j with ri,j > 6.8 Å were excluded from the summation in Eq. 5.
The WBB interactions, which are generic rather than residue-specific, were the only exception to this rule, in that the i/i+5 and i/i+6 terms were included regardless of the cutoff distance (please recall that the interactions between sites i/i+1 through i/i+4 are included anyway as a part of the short-range terms). The incorporation of the i/i+5 and i/i+6 terms induces correlation among the states of three successive residues along the peptide backbone, hence restricting the size of the accessible conformational space of the peptide significantly. This, in turn, promotes chain stiffness, contributes to its stability, and induces protein-like behavior (Skolnick and Kolinski, 1999; Pappu et al., 2000).
The potentials can be found at http://klee.bme.boun.edu.tr/energy.html.
Sampling peptide conformations
New conformations were generated by perturbing the generalized coordinates of φi, 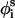 , and θi. Each MC cycle was composed of two steps; in the first, the generalized coordinates φi and
, and θi. Each MC cycle was composed of two steps; in the first, the generalized coordinates φi and 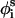 were perturbed randomly, and in the second,
were perturbed randomly, and in the second,  and θi. The random perturbations were of the type
and θi. The random perturbations were of the type
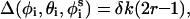 |
(6) |
where r is a random number on the interval [0–1] and δk is the maximum variation for the respective coordinates. The maximal variation in φi was taken as 3°, and a value of 0.5° was used for both θi and 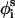 . The sensitivity of the results to the maximal variation of the generalized coordinates was checked. The optimum values were selected based on the criterion that the variations are large enough for effective sampling of the conformational space, i.e., the chain is given a chance to explore minimum energy states, yet small enough to keep the structure in the basin of the minimum energy states. The simulations were not affected by slight changes in the magnitude of δk.
. The sensitivity of the results to the maximal variation of the generalized coordinates was checked. The optimum values were selected based on the criterion that the variations are large enough for effective sampling of the conformational space, i.e., the chain is given a chance to explore minimum energy states, yet small enough to keep the structure in the basin of the minimum energy states. The simulations were not affected by slight changes in the magnitude of δk.
Each move, which creates a new conformation from the present conformation, was accepted according to the Metropolis criterion (Metropolis et al., 1953), based on the value of ΔE/c kT. ΔE is the energy difference between the new and old conformations in units of kT, and c is the reduced temperature, which is a scaling parameter that effectively controls the system's temperature. A large value of c corresponds to a high temperature, thus having an enhanced probability to accept high-energy conformations, whereas a low value of c corresponds to low temperatures.
Peptides
Three types of peptides were used: 1), polyalanine, (Ala)L, of various length L = 10–35; 2), the alanine-based peptides (AAQAA)3 and AAAAA(AAARA)3A; and 3), polyglycine (Gly)20.
Simulations
Between three and five runs of approximately two-million MC cycles each were performed with each peptide. In each MC cycle we randomly perturbed the peptide conformation L times (as the number of residues in the chain). Each time, random perturbations with maximum variation of δk were attempted to the generalized coordinates of φi, θi, and 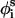 of one residue, as described above.
of one residue, as described above.
Clustering analysis
To find the dominant folds during the simulations and to estimate the conformational diversity, we clustered the conformations based on their root mean-square deviations (RMSD), following the method that was described by Ferrara et al. (2000). We pooled conformations from the simulation trajectories and calculated the RMSD between the Cα atoms of each pair of conformations in the pool. The neighbors of each conformation with RMSD ≤ 2.5 Å were counted and the conformation with the greatest number of neighbors was assigned as the center of the first cluster. All neighbors of this conformation were removed from the pool. The center of the second cluster was determined similarly with the new pool of conformations, and the procedure was repeated until each conformation was assigned to a cluster. To evaluate the robustness of the clustering analysis, we repeated it with similarity measures of 3, 3.5, and 4 Å. The results were qualitatively the same, which implies that the conclusions are not subjective to the magnitude of the similarity measure.
RESULTS
Helix formation and stability
We studied the folding and stability of polyalanine and alanine-based peptides during the simulations, starting from random conformations but excluding conformations with secondary structures. As a counter-example, we studied the folding of the nonfolder peptide (Gly)20.
Effective energy
The value of E{Φ} during a typical simulation of (Ala)20 at a reduced temperature c = 1.4 is presented in Fig. 2 A. The initial conformation was a random coil, where the peptide was unfolded and E{Φ} was high. As the simulation advanced, the peptide folded into more stable conformations and E{Φ} gradually decreased until an equilibrium was reached and the energy fluctuated about its mean value. The observed broad energy fluctuations indicate transitions between different conformations at equilibrium. The corresponding change in the percentage of helix content of the peptide chain during the course of the simulation is shown in Fig. 2 B; it reflects the occurrence of different conformations from a coil to a helix. The peptide oscillated between many conformations, rather than adopting a complete helix or random coil conformation. The average helix content of the chain during four independent runs using the same reduced temperature c = 1.4 was 62 ± 3%.
FIGURE 2.
The effective energy E{φ} (A) and the corresponding helical content (B) as a function of simulation time in MC cycles for (Ala)20 at c = 1.4.
Temperature-dependence
Fig. 3 (circles) shows the average helix content, 〈n〉/N, of (Ala)20 as a function of the reduced temperature c. Low reduced temperatures promote helix formation, whereas high reduced temperatures oppose it. This can also be observed from the clustering of the conformations (on RMSD measure of 2.5 Å) in the trajectories; the conformations were grouped into 2–4 clusters at c = 1, and into ∼10 clusters at c = 1.4. The dependence of the helix content on c, as observed, showed a transition between the completely coiled and the completely helical states occurring at ∼c = 1.4. The corresponding values of the effective energy indicate that the lowest energy conformation for polyalanine in our model is the α-helix structure.
FIGURE 3.
The average helix content of (Ala)10 (squares), (Ala)20 (circles), and (Ala)35 (triangles), calculated as described in Theoretical Background, as a function of c, the reduced temperature.
Overall, the melting curve of Fig. 3 is similar to the experimental curve that was obtained by Scholtz et al. (1991).
Chain-length effect
The helix content of polyalanine peptides of various lengths over a range of reduced temperatures is presented in Fig. 3. In general we observed a sharp transition from helix in low reduced temperature to coil at high reduced temperature for each peptide. The folding of short chains occurred at a lower value of the reduced temperature compared to that of long chains, but the influence of the chain length on the transition-reduced temperature was minor, unlike the strong dependence that was observed in the experiments of Scholtz et al. (1991). For example, at 0°C (Scholtz et al., 1991) observed ∼40%, 65%, and 85% helix content in peptides of 10, 20, and 32 residues, respectively. The helix contents that we observed at the simulations at c = 1.4 were ∼45%, 65%, and 65% in peptides of 10, 20, and 35 residues, respectively. Apparently we achieved a better agreement with experiment when dealing with shorter peptides (Ala(10) and Ala(20)), but observed lower helix content in longer peptides. The reason for the relatively low helicity of long peptides is unclear to us. In this respect it is noteworthy that a similarly weak dependence of the melting curve on the peptide length was also reported in the computational work of Smith and Hall (2001).
Zimm-Bragg's σ and s
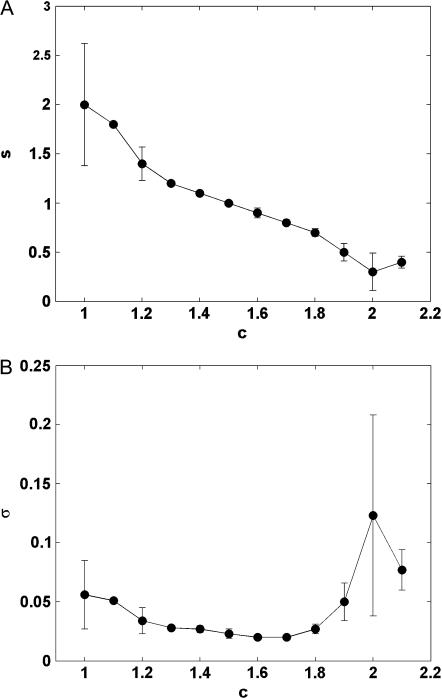
We calculated the Zimm-Bragg parameters from the average helix content (〈n〉/N) and average length of a helical segment (〈l〉) for each peptide length at several reduced temperatures using Eqs. 1a and 1b. The dependence of s and σ on the reduced temperature calculated from simulations of (Ala)35 are plotted in Fig. 4, A and B. Fig. 4 A shows that s, which is related to the helix propensity, decreases with c because thermal fluctuations decrease helix stability. The transition from an unstable to a stable helix (i.e., from s < 1 to s > 1) usually occurred at ∼c = 1.4. Fig. 4 B shows that the value of σ is constant around a level of 0.03 in the reduced temperature interval in which the helix is stable (1.2 < c < 1.8). Its value increases up to 0.13 at higher temperatures in which the helix is unstable, as the nucleation process becomes more difficult.
FIGURE 4.
The Zimm-Bragg parameters σ (A) and s (B) as a function of c calculated from simulations of (Ala)35.
The formalism that was used to estimate the σ and s parameters of the Zimm-Bragg model is valid only for long peptides (Cantor and Schimmel, 1980); therefore we needed to estimate the value of L that can be considered large enough. We estimated the required L indirectly as the value at which σ and s reached their stable levels in our simulations. Fig. 5, A and B, present the dependence of σ and s on L for c = 1.4. As can be seen, the values of σ and s converged to their steady levels (σ = 0.03 and s = 1.1) for L ≥ 20, thus we suggest that L = 20 is large enough.
FIGURE 5.
The Zimm-Bragg parameters σ (A) and s (B) of polyalanine as a function of the chain length at c = 1.4.
Alanine-based peptides
Two alanine-based peptides, (AAQAA)3 and AAAAA(AAARA)3A, were simulated with the present model and potential at various reduced temperatures. The peptides' behavior was similar to polyalanine peptides of equivalent lengths. The peptides went through a clear transition from a coil to a helix around the same reduced temperature (data not shown). The helix formation parameters at c = 1.4 were similar to the values obtained for polyalanine peptides: σ = 0.059 and s = 1.046 for (AAQAA)3, and σ = 0.031 and s = 1.005 for AAAAA(AAARA)3A.
Polyglycine
Polyglycine is known as a nonfolder (Schulz and Schirmer, 1979). We studied the folding of a 20-mer peptide starting from random coil and helix conformations. At c = 1.4, a reduced temperature for which (Ala)20 folded, the (Gly)20 peptide consistently remained unfolded (with a helix content of <1%), regardless of the starting conformation. At a low reduced temperature of c = 1.0 (Gly)20 contained 33 ± 4% helix, whereas that of (Ala)20 was above 90%.
Dynamics
To estimate the difference in the chain's dynamics along the simulation's trajectory, we divided the trajectory chronologically into two phases: the initial phase of the trajectory in which the peptide folded from a coil to a helix (referred to as the folding phase), and the latter phase of the trajectory in which the peptide fluctuated around an equilibrium state (referred to as the equilibrium phase). Fig. 6 depicts the mean-square fluctuations of the Cα atoms of (Ala)20 in the slowest dynamic mode averaged over sampled conformations during the folding phase (A) and in the equilibrium phase (B) from one representative simulation at c = 1.4. In the case of the folding phase, the simulation is not at equilibrium; however, each curve that contributes to the average value in phase A describes a local equilibrium state during the folding process.
FIGURE 6.
The mean-square fluctuations of the Cα atoms in the slowest mode of dynamics calculated by GNM of (Ala)20 conformations from the folding (A) and equilibrium (B) phases of the simulation at c = 1.4. (C) Same as B from simulation at c = 1. Each curve is an average of thousands of conformations randomly chosen from the respective equilibrated trajectory. The error bars indicate the standard deviation between the fluctuations of different conformations in the given state.
In both cases, the middle of the chain was less flexible than the termini, which implies that it was intrinsically confined and unable to explore the conformational space as efficiently as the ends. As expected, the fluctuations (and their deviations) were larger in the folding phase, as the peptide is more flexible in the early stages of the folding, and the conformations are more diverse. In the equilibrium phase, where the peptide contains stable helical segments, it became stiffer, thus the mode's shape was flatter and the deviations were smaller. The mode's shape of conformations from the equilibrium phase of (Ala)20 at c = 1.0 (Fig. 6 C) implies that the chain became more rigid as the temperature decreased because it assumed more stable helix conformations. The increased chain's rigidity reduced the conformational space, which led to a reduction in the number of conformation's clusters (data not shown). It also lowered the deviations between the fluctuations of different conformations manifested by small error bars. The absolute values of the fluctuations are immaterial because the value of the force constant γ in the GNM needs to be scaled with respect to a reference. However, the distribution of fluctuations along the sequence and the relative standing of the mode shapes for different cases reflect the relative behavior of the structures.
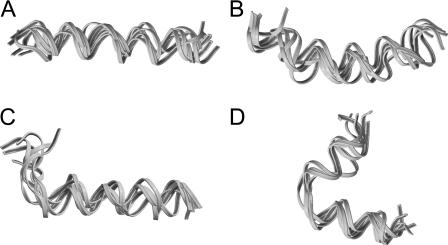
Clustering analysis of the conformations of the equilibrium phase also demonstrated that the chain became more confined after equilibration. Approximately 40% of the conformations were found in the first cluster with a similarity measure of 2.5 Å. These conformations exhibited a helical structure (Fig. 7 A). Approximately 10% assumed a banana-shape helix and were grouped in the second cluster (Fig. 7 B), ∼5% were unwound at the N-terminal region (Fig. 7 C), and ∼5% were in an α-hairpin structure (Fig. 7 D). Approximately 40% of the conformations were distributed over many other clusters, most of which are singletons.
FIGURE 7.
Four main clusters of conformations from simulation of Ala(20) at c = 1.4.
Folding
The clustering of conformations from early stages of the simulations, during peptide's folding, may shed light on the critical stages of helix formation. We clustered the conformations from the folding phase of the simulations on an RMSD measure of 2.5 Å to obtain clusters of similar conformations that were generated during the simulations. We marked the number of clusters that were required to contain half of the conformations from each simulation, and repeated the procedure with conformations from simulations that were carried out at different temperatures. The histograms of Fig. 8 present distinct behaviors at each reduced temperature: At a high reduced-temperature (Fig. 8 C), the conformations constantly changed during the simulations, hence they could not be clustered efficiently—more than 40 clusters were required to contain 50% of the conformations. At these temperatures, the peptide was mostly a random coil and the probability to find it in an energy well, i.e., in an ordered structure, was minor. At a lower reduced-temperature (Fig. 8, A and B), however, we could group the majority of the conformations in only a few clusters. This indicates that, during folding, the peptide visited only a few energy wells in the energy landscape rather than exhaustively searching the conformations.
FIGURE 8.
A histogram of the number of clusters that were required to populate 50% of the conformations of peptides during the folding phase of the simulations. The clustering is based on the RMSD measure of 2.5 Å. The ensemble of conformations contained conformations from simulations of peptides of the length 20–35. (A) Simulations at reduced temperature of 0.8 ≤ c ≤ 1.2, (B) c = 1.4, and (C) 1.8 ≤ c ≤ 2.1.
DISCUSSION
In this work we used an MC model to explore the folding and stability of short peptides. To this end, we used a reduced representation of the peptide and a statistical potential, which was derived from the structure of globular proteins. The potential was successfully used to simulate motions of globular proteins near their folded state, but it was necessary to test its performance in the simulation of folding and stability of short peptides. We checked several measures that characterize the thermodynamics and dynamics of the helix-coil transition of polyalanine and polyalanine-based peptides.
Helix content
Experimental work and simulations demonstrated the relation between peptide length, temperature and helix stability. The common observation was that the helix content of the chain increases with length and decreases with temperature. In this work we observed the same relations between these factors. We saw a decrease in the helicity as the (reduced) temperature was increased, and the shape of the helix-content-versus-temperature curve (Fig. 3) implies that the helix is the most stable conformation.
The helix content of the peptide (AAQAA)3 was measured and calculated by different techniques. Nuclear magnetic resonance (NMR) chemical shift measurements (Shalongo et al., 1994) at 274 K have yielded, on average, 44% helix content for Ace-(AAQAA)3-NHMe. The corresponding value that was reported from a molecular dynamics (MD) simulation study (Ferrara et al., 2000) is much higher, 70% (∼60% at 300 K), and is apparently the result of the overemphasized noncovalent interactions along the backbone in the potentials. On the other hand, some other MD simulations (Sung and Wu, 1996; Shirley and Brooks, 1997) have pointed out that this peptide folds into an α-helix with percent helicity of ∼47% at 273 K. In our simulations, the average helix content was 55%, which is in reasonable agreement with the experimental results considering the arbitrariness in the definition of residue helicity. The NMR work of Shalongo et al. (1994) indicated that the peptide's N-terminal is more stable as a helix than the C-terminal, mostly at the ultimate and penultimate residues of the peptide. The asymmetry in the stability of the helix along the chain is not a trivial outcome of the classic Zimm-Bragg (Zimm and Bragg, 1959) and Roig-Lifson (Lifson and Roig, 1961) models. Shalongo et al. (1994) argued that hydrogen bonds between polar side chains and unpaired amide groups at the N-terminal stabilize this end of the chain as a helix. The MD simulations of Ferrara et al. (2000) supported this behavior. In our simulations, the middle part of the peptide was more helical than the ends, but we did not observe the asymmetric picture that came out of the NMR data of Shalongo et al. (1994). This is mainly because the torsion angles of the ultimate and penultimate residues are not well-defined in the reduced model of the peptide used here as mentioned above.
In folding simulations of the peptide AAAAA-(AAARA)3A we observed a similar helix content (55%). Thompson et al. (1997) used circular dichroism measurements and observed more stable structures with 75% helix content at 0°C. Stable helix is predicted by AGADIR (Munoz and Serrano, 1997) at room temperature (70%) when the ends of the peptides are capped, and there is a much less stable helix without the caps (20%). MD simulations by Hiltpold et al. (2000) displayed peptides with higher helix content than that observed in the present simulations, although the average helix content was not reported explicitly.
The conformational preferences of alanine-based peptides, both (AAQAA)3 and AAAAA(AAARA)3A, appeared to be very close to those of polyalanine, and were in particular demonstrated by the similar helix content and effective energy values at lower temperatures. The dependence of the helix content of (AAQAA)3 on the reduced temperature c closely follows that of a polyalanine containing 15 residues.
The folding simulations of (Gly)20 were carried out as a negative control experiment to demonstrate the capability of the model to deal with non-helix-forming peptides too. As in other computational and experimental works (Gly)20 was recognized here as a nonfolder that folded only at a very low (reduced) temperature. Chakrabartty et al., (1994) calculated from circular dichroism spectroscopy measurements a propagation parameter of s = 0.05 for Gly, which demonstrated the helix-breaker nature of this amino acid. Our results are in agreement with the computational results of Okamoto and Hansmann (1995), who observed 5.2% helix content at high temperature and 8.45% helix content at low temperature, and with other works (Smith and Hall, 2001; Bertsch et al., 1998).
Ends effects
The relatively large flexibility of the chain ends reduces the stability of the α-helical states of the terminal segments. Thus, the shorter the chain, the more likely it is for the ends to unravel, causing the whole helix to dissolve. This behavior was observed in this (Fig. 5) and previous studies (Smith and Hall, 2001; Okamoto and Hansmann, 1995). In our model, this behavior is pronounced because the torsion angles of the residues at the ends are ill-defined. It artificially reduces the helix content and is particularly dominant in short chains. Chakrabartty et al. (1993) demonstrated that the identity of the N-terminal amino acid has a crucial effect on the raveling of the chain ends and hence on the stability of the helix, probably due to specific capping interactions between the residue of the first amino acid and the backbone. They found that a peptide with asparagine at the N-terminal forms a helix that was approximately fivefold more stable than a peptide with alanine at the N-terminal. They also found that N-acetylation of the peptide has a similar effect on helix stability—acetylated peptide with alanine at the N-terminal was sixfold more stable than non-acetylated peptide. Unlike other simulation studies, such as these of Huo and Straub (1999), Ferrara et al. (2000), and Shirley and Brooks (1997), our model lacks the capacity to deal with such modifications and effects, which influence helix stability. However, in these studies the particular effect of the acetylation on helix stability is not reported.
Zimm-Bragg's parameters
Calculation of the Zimm-Bragg's parameters, s and σ, from simulations provides a basis for comparison with experimental observations and other simulations. The corresponding values from the simulations of (Ala)35 at c = 1.4 were σ = ∼0.02 and s = ∼1.1; similar values were obtained for shorter peptides of 20 residues. The experimental values of σ were 8 × 10−4 by Platzer et al. (1972), ∼0.003 by Scholtz et al. (1991) and Rohl et al. (1992), and 0.00117 or 0.025 by Thompson et al. (1997) (depending on the fitting parameters). The values of s that were obtained from experiments are 1.35 by Scholtz et al. (1991), 1.54 by Chakrabartty et al. (1994), and 1.48 or 1.525 by Thompson et al. (1997) (depending on the fitting parameters). Overall the predicted s-values from previous simulation studies have been in the range reported by the experiments. The values of σ have been more diverse and differ from the experimental values by some orders-of-magnitude. Okamoto and Hansmann (Hansmann and Okamoto, 1999; Okamoto and Hansmann, 1995) reported a value of σ = 0.051, Smith and Hall (2001) found σ = 0.095, Sung and Wu (1996) found σ = 0.056, Daggett et al. (1991) found σ = 0.5, and values of 2.12 × 10−22 to 9.6 × 10−7 (based on different solvation models) were reported by Yang and Honig (1995). The value of σ that was obtained from our simulation is among the best obtained by simulation studies, but still an order-of-magnitude larger than the experimental value (Scholtz et al., 1991). In free energy terms, the difference between the calculated ΔGnuc and the measured value is ∼2 kT, which is small but not negligible.
Calibration of the simulation temperature
Since our energy function was generated from crystallographic data that were collected in various non-native temperatures, the correspondence between the reduced temperature in the simulations and the real temperature is unknown. This is a well-known problem of statistical potentials. Moreover, here we modified the potentials that were derived from globular proteins for the peptide simulations. To calibrate the simulation's temperature, we compared the thermodynamic quantities that were calculated to empirical data. Using reduced temperatures in the range of c = 1.3–1.5, we successfully reproduced the experimental propagation parameter s at 0°C. At this range of (reduced) temperature we could obtain a σ-value that is reasonably close to the measured one, albeit an order-of-magnitude larger than the experimental value (as described above). Thus, even though we cannot tell what the simulation temperature is, we can roughly estimate that the average value c = 1.4 is comparable to ∼0°C. In this context it is important to note that we still could not estimate the magnitude (in degrees) of the intervals in the reduced temperature scale.
Folding and dynamics
The simulations imply that a peptide folding from a coil to a helix does not go through an exhaustive search. The clustering analysis of the conformations from the early stages of the simulations show that, right from the onset of the folding process, the peptide tends to prefer specific conformations of an intermediate energy minimum, rather than exploring the available conformation space. This tendency, of course, closely depends on the simulation temperature; at high temperatures the peptide explored many conformations and did not remain in shallow minima. However, in low-temperature simulations, we observed stable intermediates with an α-hairpin conformation. It is interesting to note that intermediate β-hairpins structures were reported by Ding et al., (2003) and Nguyen et al., (2004) in MD simulations of polyalanine peptides. The α-hairpin conformation observed here may be an artifact of the potential that promotes compact structures as it was derived from a database of globular proteins. Otherwise it may be an intrinsic property of folding at low temperatures.
The elastic behavior of the chain was studied by analysis of the fluctuations of residues in the most dominant mode. Along the simulations' course, the difference between the fluctuations of the residues along the chain decreased as the helical conformation became more stable. This implies that the correlation between the fluctuations of the residues along the chain increases with the chain's rigidity or stiffness. The observations from the conformations' clustering and from the GNM analyses support the hypothesis that, during folding, small secondary structures of low energy form first. This restricts the conformations that are accessible to the peptide and directs the peptide toward the equilibrium state, where the energy is low and the structure is most stable.
Analyzing the conformations during the dynamic equilibrium phase using the GNM indicated that the distribution of the residues' fluctuations follows a pattern: The middle residues fluctuated less than the residues that are close to the termini. Indeed, this reflects the intrinsic peculiarity of a flexible chain, where the local interactions are emphasized as also described by the Rouse chain model (Rouse, 1953).
CONCLUSIONS
In this work, we presented a method that provides a reasonable description of the thermodynamic and dynamic properties of helical peptides at equilibrium and during the folding. The method employs statistical potentials and a reduced representation of the peptide that facilitates the energy calculation, along with an MC search algorithm to explore the conformation properties. We demonstrated that the equilibrium properties of the peptide, as well as the dynamic properties, starting from a random conformation, can be observed. The estimated values of the Zimm-Bragg parameters σ and s that were obtained here are in reasonable agreement with experimentally derived values. The content of helical segments in the peptide, the behavior of the effective energy, and the temperature-dependence agreed with those obtained using other approaches. The simulations displayed the correlation between the distribution of the fluctuations along the slowest dynamic mode and the helix content of the structure. The generic behavior of flexible chains, where local interactions are emphasized, may influence the conformations sampled during folding and also in the dynamic equilibrium state.
Acknowledgments
We greatly acknowledge North Atlantic Treaty Organization grants LST-CLG-977836 and LST-CLG-980180. T.H. acknowledges Turkish State Planning Organization grant No. 01K120280 and Bogazici University research grants 02HA501 and 04HA502. N.B.T. acknowledges the support of grant No. 222/04 from the Israel Science Foundation.
References
- Bahar, I., A. R. Atilgan, and B. Erman. 1997a. Direct evaluation of thermal fluctuations in protein using a single parameter harmonic potential. Fold. Des. 2:173–181. [DOI] [PubMed] [Google Scholar]
- Bahar, I., B. Erman, T. Haliloglu, and R. L. Jernigan. 1997b. Efficient characterization of collective motions and inter-residue correlations in proteins by low-resolution simulations. Biochemistry. 36:13512–13523. [DOI] [PubMed] [Google Scholar]
- Bahar, I., and R. L. Jernigan. 1996. Coordination geometry of nonbonded residues in globular proteins. Fold. Des. 1:357–370. [DOI] [PubMed] [Google Scholar]
- Bahar, I., and R. L. Jernigan. 1997. Inter-residue potentials in globular proteins and the dominance of highly specific hydrophilic interactions at close separation. J. Mol. Biol. 266:195–214. [DOI] [PubMed] [Google Scholar]
- Bahar, I., M. Kaplan, and R. L. Jernigan. 1997. Short-range conformational energies, secondary structure propensities, and recognition of correct sequence-structure matches. Proteins. 29:292–308. [DOI] [PubMed] [Google Scholar]
- Bertsch, R. A., N. Vaidehi, S. I. Chan, and W. A. Goddard, III. 1998. Kinetic steps for α-helix formation. Proteins. 33:343–357. [DOI] [PubMed] [Google Scholar]
- Bird, R. B., C. F. Curtiss, R. C. Armstrong, and O. Hassager. 1987. Dynamics of Polymeric Liquids. Kinetic Theory. Wiley and Sons, New York.
- Cantor, C. R., and P. R. Schimmel. 1980. Conformational equilibria of polypeptides and proteins: the helix-coil transition. In Biophysical Chemistry: The Behavior of Biological Macromolecules. W.H. Freeman, New York. 1060–1066.
- Chakrabartty, A., and R. L. Baldwin. 1995. Stability of α-helices. Adv. Protein Chem. 46:141–176. [PubMed] [Google Scholar]
- Chakrabartty, A., A. J. Doig, and R. L. Baldwin. 1993. Helix capping propensities in peptides parallel those in proteins. Proc. Natl. Acad. Sci. USA. 90:11332–11336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabartty, A., T. Kortemme, and R. L. Baldwin. 1994. Helix propensities of the amino acids measured in alanine-based peptides without helix-stabilizing side-chain interactions. Protein Sci. 3:843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, S., W. Zhang, C. Wu, G. Xiong, and Y. Duan. 2003. Breaking non-native hydrophobic clusters is the rate-limiting step in the folding of an alanine-based peptide. Biopolymers. 68:63–75. [DOI] [PubMed] [Google Scholar]
- Daggett, V., P. A. Kollman, and I. D. Kuntz. 1991. A molecular dynamics simulation of polyalanine: an analysis of equilibrium motions and helix-coil transitions. Biopolymers. 31:1115–1134. [DOI] [PubMed] [Google Scholar]
- Ding, F., J. M. Borreguero, S. V. Buldyrey, H. E. Stanley, and N. V. Dokholyan. 2003. Mechanism for the α-helix to β-hairpin transition. Proteins. 53:220–228. [DOI] [PubMed] [Google Scholar]
- Doi, M., and S. Edwards. 1986. The Theory of Polymer Dynamics. Clarendon, Oxford, UK.
- Doig, A. J. 2002. Recent advances in helix-coil theory. Biophys. Chem. 101–102:281–293. [DOI] [PubMed] [Google Scholar]
- Ferguson, N., and A. R. Fersht. 2003. Early events in protein folding. Curr. Opin. Struct. Biol. 13:75–81. [DOI] [PubMed] [Google Scholar]
- Ferrara, P., J. Apostolakis, and A. Caflisch. 2000. Thermodynamics and kinetics of folding of two model peptides investigated by molecular dynamics simulations. J. Phys. Chem. B. 104:5000–5010. [Google Scholar]
- Flory, P. J. 1969. Statistical Mechanics of Chain Molecules. Wiley-Interscience, New York.
- Flory, P. J. 1976. Statistical thermodynamics of random networks. Proc. R. Soc. Lond. A. 351:351–380. [Google Scholar]
- Gilis, D., and M. Rooman. 2001. Identification and ab initio simulations of early folding units in proteins. Proteins. 42:164–176. [DOI] [PubMed] [Google Scholar]
- Go, N., T. Noguti, and T. Nishikawa. 1983. Dynamics of a small globular protein in terms of low-frequency vibrational modes. Proc. Natl. Acad. Sci. USA. 80:3696–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haliloglu, T., and I. Bahar. 1998. Coarse-grained simulations of conformational dynamics of proteins: application to apomyoglobin. Proteins. 31:271–281. [PubMed] [Google Scholar]
- Haliloglu, T., I. Bahar, and B. Erman. 1997. Gaussian dynamics of folded proteins. Phys. Rev. Lett. 79:3090–3093. [Google Scholar]
- Hansmann, U., and Y. Okamoto. 1999. Finite-size scaling of helix-coil transitions in poly-alanine studied by multicanonical simulations. J. Chem. Phys. 110:1267–1276. [Google Scholar]
- Hiltpold, A., P. Ferrara, J. Gsponer, and A. Caflisch. 2000. Free energy surface of the helical peptide Y(MEARA)(6). J. Phys. Chem. B. 104:10080–10086. [Google Scholar]
- Huang, C.-Y., J. W. Klemke, Z. Getahun, W. F. DeGrado, and F. Gai. 2001. Temperature-dependent helix-coil transition of an alanine-based peptide. J. Am. Chem. Soc. 123:9235–9238. [DOI] [PubMed] [Google Scholar]
- Huo, S., and J. E. Straub. 1999. Direct computation of long time processes in peptides and proteins: reaction path study of the coil-to-helix transition in polyalanine. Proteins. 36:249–261. [PubMed] [Google Scholar]
- Kessel, A., D. Shental-Bechor, T. Haliloglu, and N. Ben-Tal. 2003. Interactions of hydrophobic peptides with lipid bilayers: Monte Carlo simulations with M2δ. Biophys. J. 85:3431–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt, N., and T. Haliloglu. 1999. Conformational dynamics of chymotrypsin inhibitor 2 by coarse-grained simulations. Proteins. 37:454–464. [DOI] [PubMed] [Google Scholar]
- Kurt, N., and T. Haliloglu. 2001. Conformational dynamics of subtilisin-chymotrypsin inhibitor 2 complex by coarse-grained simulations. J. Biomol. Struct. Dyn. 18:713–731. [DOI] [PubMed] [Google Scholar]
- Levy, R. M., and M. Karplus. 1979. Vibrational approach to the dynamics of an α-helix. Biopolymers. 18:2465–2495. [Google Scholar]
- Levy, Y., J. Jortner, and O. M. Becker. 2001. Solvent effects on the energy landscapes and folding kinetics of polyalanine. Proc. Natl. Acad. Sci. USA. 98:2188–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifson, S., and A. Roig. 1961. On the theory of helix-coil transition in polypeptides. J. Chem. Phys. 34:1963–1974. [Google Scholar]
- Metropolis, N., A. W. Rosenbluth, M. N. Rosenbluth, A. H. Teller, and E. J. Teller. 1953. Equation of state calculations by fast computing machines. J. Chem. Phys. 21:1087–1092. [Google Scholar]
- Mitsutake, A., and Y. Okamoto. 2000. Helix-coil transitions of amino-acid homo-oligomers in aqueous solution studied by multicanonical simulations. J. Chem. Phys. 112:10638–10647. [Google Scholar]
- Miyazawa, S., and R. Jernigan. 1985. Estimation of effective interresidue contact energies from protein crystal-structures—quasi-chemical approximation. Macromolecules. 18:534–552. [Google Scholar]
- Munoz, V., and L. Serrano. 1994. Elucidating the folding problem of helical peptides using empiric parameters. Nat. Struct. Biol. 1:399–409. [DOI] [PubMed] [Google Scholar]
- Munoz, V., and L. Serrano. 1997. Development of the multiple sequence approximation within the AGADIR model of α-helix formation: comparison with Zimm-Bragg and Lifson-Roig formalisms. Biopolymers. 41:495–509. [DOI] [PubMed] [Google Scholar]
- Nguyen, H. D., A. J. Marchut, and C. K. Hall. 2004. Solvent effects on the conformational transition of a model polyalanine peptide. Protein Sci. 13:2909–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, Y., and U. Hansmann. 1995. Thermodynamics of helix-coil transitions studied by multicanonical algorithms. J. Phys. Chem. 99:11276–11287. [Google Scholar]
- Pappu, R. V., R. Srinivasan, and G. D. Rose. 2000. The Flory isolated-pair hypothesis is not valid for polypeptide chains: implications for protein folding. Proc. Natl. Acad. Sci. USA. 97:12565–12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platzer, K. A. B., H. Scheraga, R. Andreatt, and V. Anathan. 1972. Helix-coil stability constants for naturally occurring amino acids in water. IV. Alanine parameters from random poly(hydroxypropylglutamine-co-L-alanine). Macromolecules. 5:178–187. [Google Scholar]
- Rohl, C. A., J. M. Scholtz, E. J. York, J. M. Stewart, and R. L. Baldwin. 1992. Kinetics of amide proton exchange in helical peptides of varying chain lengths. Interpretation by the Lifson-Roig equation. Biochemistry. 31:1263–1269. [DOI] [PubMed] [Google Scholar]
- Rouse, P. 1953. A theory of the linear viscoelastic properties of dilute solutions of coiling polymers. J. Chem. Phys. 21:1272–1280. [Google Scholar]
- Schellman, J. A. 1958. The factors affecting the stability of hydrogen bonded polypeptide structures in solution. J. Chem. Phys. 62:1485–1494. [Google Scholar]
- Scholtz, J. M., H. Qian, E. J. York, J. M. Stewart, and R. L. Baldwin. 1991. Parameters of helix-coil transition theory for alanine-based peptides of varying chain lengths in water. Biopolymers. 31:1463–1470. [DOI] [PubMed] [Google Scholar]
- Schulz, G. E., and R. H. Schirmer. 1979. Principles of Protein Structure. Springer, New York.
- Shalongo, W., L. Dugad, and E. Stellwagen. 1994. Distribution of helicity within the model peptide Acetyl(AAQAA)3amid. J. Am. Chem. Soc. 116:8288–8293. [Google Scholar]
- Shirley, W. A., and C. L. Brooks, III. 1997. Curious structure in “canonical” alanine-based peptides. Proteins. 28:59–71. [DOI] [PubMed] [Google Scholar]
- Skolnick, J., and A. Kolinski. 1999. Monte Carlo approaches to the protein folding problem. Adv. Chem. Phys. 105:203–242. [Google Scholar]
- Smith, A. V., and C. K. Hall. 2001. Alpha-helix formation: discontinuous molecular dynamics on an intermediate-resolution protein model. Proteins. 44:344–360. [DOI] [PubMed] [Google Scholar]
- Sung, S. S., and X. W. Wu. 1996. Molecular dynamics simulations of synthetic peptide folding. Proteins. 25:202–214. [DOI] [PubMed] [Google Scholar]
- Thompson, P. A., W. A. Eaton, and J. Hofrichter. 1997. Laser temperature jump study of the helix↔coil kinetics of an alanine peptide interpreted with a “kinetic zipper” model. Submillisecond kinetics of protein folding. Biochemistry. 36:9200–9210. [DOI] [PubMed] [Google Scholar]
- Tirion, M. 1996. Large amplitude elastic motions in proteins from a single-parameter, atomic analysis. Phys. Rev. Lett. 77:1905–1908. [DOI] [PubMed] [Google Scholar]
- Yang, A. S., and B. Honig. 1995. Free energy determinants of secondary structure formation. I. Alpha-helices. J. Mol. Biol. 252:351–365. [DOI] [PubMed] [Google Scholar]
- Young, W. S., and C. L. Brooks, III. 1996. A microscopic view of helix propagation: N- and C-terminal helix growth in alanine helices. J. Mol. Biol. 259:560–572. [DOI] [PubMed] [Google Scholar]
- Zimm, B. H., and J. K. Bragg. 1959. Theory of the phase transition between helix and random coil in polypeptide chains. J. Phys. Chem. 31:526–535. [Google Scholar]