Abstract
Heme-based oxygen sensors are part of ligand-specific two-component regulatory systems, which have both a relatively low oxygen affinity and a low oxygen-binding rate. To get insight into the dynamical aspects underlying these features and the ligand specificity of the signal transduction from the heme sensor domain, we used femtosecond spectroscopy to study ligand dynamics in the heme domains of the oxygen sensors FixL from Bradyrhizobium japonicum (FixLH) and Dos from Escherichia coli (DosH). The heme coordination with different ligands and the corresponding ground-state heme spectra of FixLH are similar to myoglobin (Mb). After photodissociation, the excited-state properties and ligand-rebinding kinetics are qualitatively similar for FixLH and Mb for CO and NO as ligands. In contrast to Mb, the transient spectra of FixLH after photodissociation of ligands are distorted compared with the ground-state difference spectra, indicating differences in the heme environment with respect to the unliganded state. This distortion is particularly marked for O2. Strikingly, heme–O2 recombination occurs with efficiency unprecedented for heme proteins, in ≈5 ps for ≈90% of the dissociated O2. For DosH–O2, which shows 60% sequence similarity to FixLH, but where signal detection and transmission presumably are quite different, a similarly fast recombination was found with an even higher yield. Altogether these results indicate that in these sensors the heme pocket acts as a ligand-specific trap. The general implications for the functioning of heme-based ligand sensors are discussed in the light of recent studies on heme-based NO and CO sensors.
Heme-based sensors carry out crucial roles in biological signaling in prokaryotic and eukaryotic organisms by responding to the gaseous ligands nitric oxide (NO), carbon monoxide (CO), and oxygen (O2). Binding of these physiological messenger molecules initiates chemistry that eventually results in the organism's response to changes in ligand availability. Apart from the best-studied NO-activated mammalian enzyme guanylate cyclase (GC), bacterial sensors of CO [CooA from Rhodospirillum rubrum (1, 2)] and O2 [FixL in nitrogen-fixing bacteria (3) and Dos in Escherichia coli (4)] have been discovered in recent years. In these regulatory systems, the ligand-specific binding to a heme domain leads to either activation or inhibition of a regulated (catalytic) domain. Thus, a local perturbation is transduced over a long distance within the protein. Structural and spectroscopic studies indicate that the mechanisms of this transduction vary considerably within these systems, already at the level of the heme pocket. In CooA and Dos, the sensed ligand, CO or O2, respectively, replaces one of the two protein residues bound to the (six-coordinate) heme. In GC, the unliganded heme is five-coordinate, and NO binding to the heme leads to dissociation of the proximal histidine. In FixL, binding of O2 induces a spin change of the heme (five- to six-coordinate; ref. 5).
The molecular mechanism and the origin of ligand specificity of the signal transduction of the heme-based sensors are open questions. One way to address these issues is to study the effect of photodissociation of ligands from the heme, thus triggering the inverse of the ligand-binding reaction. Recent studies on GC (6) and CooA (7) have revealed extremely fast geminate recombination of a large fraction of dissociated NO and CO, respectively. Here we study photodissociation and rebinding of O2 from the heme sensor domain of FixL in comparison with other ligands and with O2 in the heme domain of Dos. We report that, in FixL, O2 rebinds to the heme even faster than NO, an unprecedented feature for heme proteins, indicating that very fast rebinding of the sensor-specific physiological ligand to the heme is a general feature of heme-based sensors. Moreover, spectroscopic evidence is obtained for ligand-specific alterations of the heme pocket.
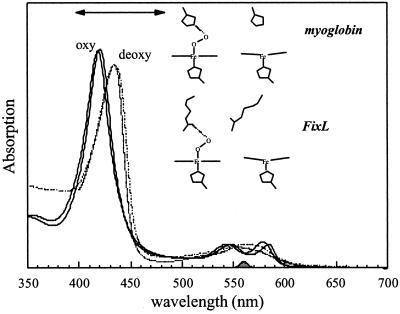
FixL, a rhizobial oxygen-sensor protein (for reviews see refs. 8–10), contains a heme sensor domain and a histidine kinase domain (3). The kinase activity strongly diminishes after binding of molecular oxygen [and to a much lesser extent of other diatomic ligands (11)] to the heme, and therewith the expression of genes involved in nitrogen fixation. As in the oxygen-storage protein myoglobin (Mb), the unliganded and oxygenated form of the heme are (high-spin) five-coordinate and (low-spin) six-coordinate, respectively (5), and the corresponding absorption spectra (as well as those for the NO and CO adducts) also are very similar to those of Mb (Fig. 1). Structural studies indicate similarities in the heme geometry of the oxycomplexes of both proteins (12), and also the bond strength between the heme iron and O2 appears similar (13). However, the ligand-binding properties are very different (8), and whereas the oxygen dissociation rate is similar, in particular the oxygen affinity and oxygen-binding rates are much lower in FixL (14), possibly adapting its sensitivity to changes in the environmental oxygen concentration. Crystal structures have been obtained for the heme domains of FixL (FixLH) from Bradyrhizobium japonicum (12, 15) and Rhizobium meliloti (16). Based on these structures, for B. japonicum FixL the mechanism of signal transduction has been suggested to involve the heme propionates and two arginine residues, one of which is hydrogen-bonded to heme-bound oxygen (12); for R. meliloti FixL, a somewhat different mechanism has been proposed (16). In this work we will show that whereas the two stable (oxy and deoxy) states of FixLH may show similarities with the Mb “model” system, the intermediates are very different and may provide keys for understanding the regulatory mechanism of this oxygen sensor at the level of the heme sensor domain.
Figure 1.
Ground-state absorption spectra of the oxy (solid line) and deoxy (dashed line) forms of FixLH (thick lines) compared with the corresponding spectra for horse-heart Mb (thin lines). The represented cartoons of the heme environments are based on the crystal structures. The spectra are normalized to the same concentration. The spectral profile of the pump pulse and the probe region in the transient absorption experiments are indicated by the shaded area at the bottom and the two-sided arrow, respectively.
Materials and Methods
Bacterial Strains and Growth Conditions.
B. japonicum wild-type strain was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH DSM no. 30131 (American Type Culture Collection 10324) and grown aerobically at 30°C in peptone/salts/yeast extract (PSY) medium (17). All E. coli strains were grown in LB medium (18) at 37°C.
DNA Manipulations.
Isolation of chromosomal DNA from B. japonicum was performed as described (19). Chromosomal DNA from E. coli HB101 was isolated following standard protocols. To amplify the gene fragment between codons 142–270, encoding the FixL heme domain (FixLH), two oligonucleotide primers were designed (forward, 5′-GCC ATG GAG ACC CAC CTC CGC TCG-3′, and reverse, 5′-CAG GCG CGT CGA CAG GAA CTG CAA-3′), introducing an NcoI and a SalI site at the 5′ and 3′ ends, respectively. The amplified product was cloned into the expression vector pET-28a+ (Novagen) as an NcoI–SalI fragment. The DNA fragment corresponding to codons 1–152 from E. coli yddU (renamed dos; ref. 4), coding for the oxygen-sensing PAS domain (DosH), was amplified by using the primers 5′-ATG AAG CTA ACC GAT GCG GAT A-3′ (forward) and 5′-C GAC CGA CCG GTG ATT GTC CTC-3′ (reverse) and cloned into a pBAD TOPO TA cloning vector (Invitrogen) under control of an arabinose-inducible promoter.
Gene Expression and Protein Purification.
Expression of fixlH was induced by the addition of 0.4 mM isopropyl β-D-thiogalactoside to BL21 DE3 cultures in the exponential growth phase. After 16 h of induction at 37°C, the cells were harvested at 4°C. Expression of dosH was induced by adding 0.2% arabinose to exponential TOP10 cultures, and cells were harvested after 5 h of induction at 37°C. Both proteins were purified following instructions of the Xpress System protein purification (Invitrogen) on a nickel-resin column, washed, and stored in 50 mM Tris⋅HCl, pH 7.4. The sizes of purified FixLH and DosH were verified on SDS/PAGE, and protein concentrations were measured by using the BCA protein assay (Pierce).
Sample Preparation.
For the spectroscopic measurements, samples were prepared to a heme concentration of ≈60 μΜ in gas-tight optical cells with an optical path length of 1 mm. For FixLH, the degassed as-prepared (ferric) sample was reduced with 1 mM sodium dithionite to obtain the reduced nonliganded form (deoxy). For the carbonmonoxy form, deoxy FixLH was equilibrated with 1 atm (1 atm = 101.3 kPa) CO. To obtain the oxy and nitrosyl forms, ferric–FixLH was reduced in ≈10-min time with 10 mM sodium ascorbate and subsequently equilibrated with 1 atm O2 or 0.01 atm NO, respectively.
Analysis of the second derivative of the absorption spectrum indicated that the thus-reduced sample was in the O2-bound form, ≈95% in air and ≈99% in 1 atm O2, corresponding to a Kd of ≈0.015 mM, which is somewhat lower than that reported previously for the full-length protein (14).† In a similar way, >99% of the NO-bound form was found to be in the six-coordinated form. This contrasts with the mixtures of five- and six-coordinate forms reported for the dithionite-reduced NO-bound form of R. meliloti FixL (20, 21) but is in agreement with a very recent characterization of B. japonicum FixL–NO (22).
To obtain oxy–DosH, as-prepared DosH was degassed, reduced with 25 mM DTT, and subsequently equilibrated with 1 atm O2. Horse-heart Mb was purchased from Sigma and prepared at a concentration of ≈100 μM in 50 mM Tris buffer, pH 7.4. The reduced unliganded and liganded forms were prepared in a similar way as for FixLH.
Spectroscopy.
For all samples, ground-state spectra, recorded with a Shimadzu UV-Vis 1601 spectrophotometer, were unchanged after the femtosecond experiments. Multicolor femtosecond absorption spectroscopy (23) was performed at a repetition rate of 30 Hz by using pump pulses centered at 563 nm (30 fs, full width at half-maximum) and compressed white-light continuum probe pulses. All experiments were performed at room temperature. It was verified that for MbCO, MbNO, and MbO2 the photodissociation spectra were similar to the steady-state difference spectra.
Global exponential fitting of the data in terms of decay-associated spectra (DAS) was performed with a procedure described previously (24). To avoid artifacts due to the chirp in the probe pulse, this analysis was performed over a limited wavelength region and started at t ≈ 500 fs. Therefore the very fast phases, of a few hundred femtoseconds or less, although within the time resolution of the instrument, were not recovered by this analysis.
Results
Unliganded Species.
As a control, transient spectroscopy was performed on unliganded FixLH in two redox forms (data not shown). Photoexcitation of ferric FixLH resulted in a transient bleaching of the ground state Soret band and an induced absorption on the red side, which decays with time constants of ≈0.5 ps (major component) and ≈5 ps. After 20 ps the ground state is recovered completely. This response, presumably reflecting formation of excited states and their decay, is similar to that found in other oxidized heme proteins (cf. ref. 25) and is consistent with ferric FixLH being five-coordinate unliganded (14, 26). Transient spectra of unliganded ferrous FixLH are similar to those of reduced unliganded Mb (27) and GC (6) and mainly reflect the heme photophysics, which can be understood in the framework of two excited states cascading back to the ground state (28, 29).
FixLH–CO.
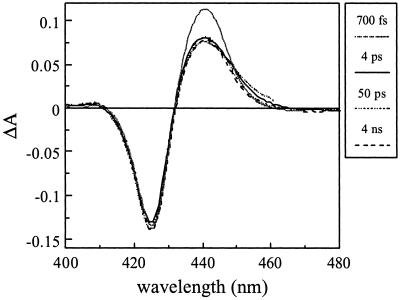
As in all other heme proteins studied by flash photolysis, photoexcitation of CO-liganded FixLH leads to dissociation of CO from the heme. A small spectral evolution is observed with a time constant of ≈200 fs (not shown), presumably reflecting excited-state decay as in MbCO (28). Virtually no further spectral changes are observed up to our longest delay time of 4 ns (Fig. 2), showing that recombination does not occur on this time scale. The transient spectra have the same general shape as the equilibrium difference spectrum but, in contrast to MbCO, are somewhat distorted in that the relative amplitude of the induced absorption is ≈30% smaller. This distortion is not observed in the bimolecular recombination phases on the millisecond time scale (30). The origin of this distortion will be discussed later.
Figure 2.
Transient absorption spectra of FixLH–CO at various delay times. The solid gray line represents the shape of the steady-state unliganded minus CO-liganded difference spectrum.
FixLH–NO.
Photodissociation of NO from FixLH leads to geminate recombination of the FixLH–NO pair in a multiphasic way on the picosecond time scale (Fig. 3). After 1 ns ≈95% of the initial (≈1-ps) signal has decayed; no significant further spectral evolution was observed up to 4 ns (not shown). Analysis of the transient spectra in terms of a multiexponential decay model yields, apart from a small ≈400-fs phase assigned to excited-state decay (cf. ref. 28), picosecond decay phases of 5.3, 20, and 220 ps as well as an asymptotic phase (>4 ns). The spectral characteristics of the last three decay phases are very similar, and these phases can be assigned to recombination of NO with ground-state heme. The bleaching part of the transient spectrum associated with the 5.3-ps decay (5.3-ps DAS) is very similar to that of the other phases; the induced absorption part is slightly more extended to the red side (not shown). As after NO dissociation from heme proteins, red-absorbing (≈450 nm) heme excited states with a lifetime of a few picoseconds have been observed (28), which indicates that at least part of the NO recombination in FixLH corresponding to this decay component occurs with the heme in the excited state. Overall, the geminate recombination of NO with heme is highly multiphasic, similar to what has been observed in other heme proteins (31–35) [with the notable exception of the NO receptor GC (6)], and often is interpreted in terms of relaxation of the protein on the time scale of the recombination.
Figure 3.
Recombination of NO with FixLH. Transient absorption spectra at various delay times are shown. The solid gray line represents the steady-state unliganded minus NO-liganded difference spectrum, normalized on the bleaching part of the 10-ps transient spectrum. (Inset) Kinetics at 440 nm. The solid lines represent a fit with decay phases of 5.3 (62%), 20 (24%), and 220 ps (8%) and a constant (6%). The dashed line represents the recombination of NO with Mb (data taken from ref. 35). Note the different time scales.
Fig. 3 Inset shows that the kinetics are qualitatively similar to those observed in wild-type Mb, with the fastest decay phases having a somewhat higher relative amplitude. However, in contrast to Mb (MbNO, data not shown), the shape of the transient spectra differs considerably from the equilibrium difference spectrum in that the amplitude of the induced absorption is ≈2 times lower (as shown for the 10-ps transient spectrum in Fig. 3). This effect is qualitatively similar to the effect observed after CO dissociation but is much stronger for the case of NO dissociation (see Discussion).
FixLH–O2.
Measurements with oxy–FixL are potentially complicated by the fact that equilibration with air leads to incomplete oxygen binding to FixL (14). To minimize this effect, the samples were equilibrated with 1 atm O2. In this case, only ≈1% of the oxygenated sample is unliganded (see Materials and Methods).
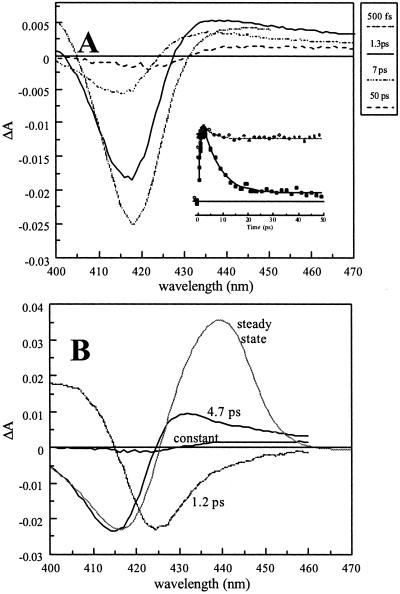
Fig. 4A shows the transient spectra observed after excitation of oxygenated FixLH. The spectra are characterized by a strong initial bleaching around 420 nm and a much weaker induced absorption to the red. The amplitude of the spectra strongly diminishes on the picosecond time scale and remains unchanged from ≈20 ps to >1 ns.
Figure 4.
Recombination of O2 with FixLH. (A) Transient absorption spectra at various delay times. (Inset) Kinetics (>200 fs) at 433 nm (the dashed line represents a fit of the kinetics for MbO2 at the same wavelength). (B) DAS (dashed lines) of the decay components > 1 ps. The solid gray line represents the steady-state unliganded minus O2-liganded difference spectrum, normalized on the bleaching part of the 4.7-ps component.
In a global analysis three main phases with a time constant > 1 ps were found (Fig. 4B).‡ The 1.2-ps phase presumably reflects excited-state decay. Consistently, considerable induced absorption decaying on this time scale was observed around 470 nm (not shown; cf. ref. 28). The other decay phase is the 4.7-ps phase and can be attributed to recombination of photodissociated O2 with the heme. The 4.7-ps DAS-induced absorption maximum appears more to the blue than the steady-state difference spectrum. Because it is expected to be more red-shifted for an excited state (see NO results above and ref. 28), this indicates that most recombination occurs with the heme in the ground state. Decay phases on a similar time scale after excitation of oxy hemes have been observed in Mb and hemoglobin (28, 34), but these (ascribed at least in part to heme excited states) have a much lower relative amplitude (compare Fig. 4A Inset) than in FixLH, where they extend to ≈90% of the signal, as estimated from the ratio of the (integrated) DAS spectra of the 4.5-ps phase and the constant phase.
As for CO and NO, for O2 the amplitude of the induced absorption part of the DAS of the component ascribed to heme–ligand recombination is low compared with the steady-state spectrum. The relative decrease is ≈3–4-fold, and the effect thus seems stronger in the order CO < NO < O2. The remaining low-amplitude signal (constant DAS) has a shape that differs both from the 4.7-ps recombination phase and the steady-state difference spectrum in that the bleaching is more to the red (the induced absorption maximum is at ≈440 nm, as in the steady-state difference spectrum). We note that such distortion is not observed for Mb (data not shown) but that a small distortion is observed for the allosteric protein hemoglobin (28).
DosH–O2.
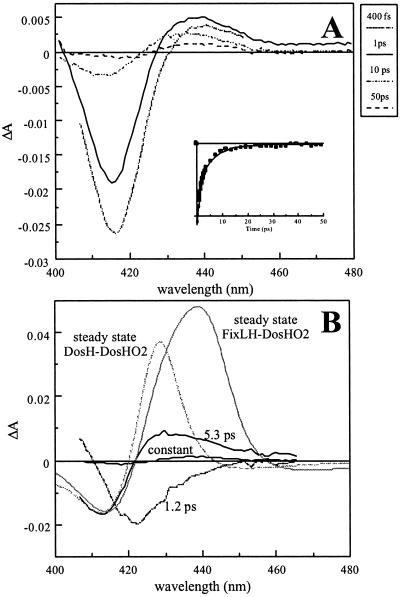
To investigate whether the observed ligand-rebinding properties are specific for oxygen sensors, similar experiments were performed on DosH (Fig. 5), which binds O2 stronger than FixL (4). The spectral evolution is qualitatively similar for DosH and FixLH (Figs. 4 and 5). In particular, there is a strong 5.3-ps recombination component with a relatively weak induced absorption lobe (see below) and a constant phase with red-shifted characteristics with respect to both the 4.5-ps recombination phase and the steady-state difference spectra. The relative amplitude of the constant phase is very small (3–5% of the 5.3-ps phase; for FixLH, ≈10% for the corresponding amplitude), showing that the oxygen-escape probability from the heme domain is very small. The spectra do not change significantly between 50 ps and 1 ns (not shown).
Figure 5.
Recombination of O2 with DosH. (A) Transient absorption spectra at various delay times. (Inset) Kinetics (>200 fs) at 415 nm. (B) DAS (black lines) of the decay components > 1 ps. The solid gray lines represent the steady-state DosH unliganded minus DosH O2-liganded (dashed) and FixLH unliganded minus DosH O2-liganded normalized at ≈415 nm with the 5.3-ps component DAS.
In the unliganded form of DosH, the heme is six-coordinate; the sixth (oxygen-binding) position is occupied by an internal ligand, which has been shown to be a methionine (35). Therefore, the steady-state difference spectrum (Fig. 5B, dashed gray line) differs from that of FixL. At short times after O2 dissociation, the heme presumably is five-coordinate, and therefore comparison of the early transient spectra with a five-coordinate unliganded minus O2-liganded spectrum seems more appropriate. Such a spectrum is constructed (Fig. 5B, solid gray line) by using the ground-state unliganded FixLH spectrum as a model for the five-coordinate product and published extinction coefficients for unliganded FixL and DosH-O2 (4, 14). Using this model, the 5.3-ps DAS appears very different from the expected steady-state difference spectrum and qualitatively similar to the transient spectra for FixLH–O2 (Fig. 4). Similar experiments performed with unliganded and CO- and NO-liganded DosH will be published elsewhere.
Discussion
The present studies on ultrafast ligand dynamics in the heme domains of two oxygen sensors have resulted in two main findings. First, dissociation of oxygen in these proteins leads to heme–O2 recombination described by a very fast (≈5-ps) and nearly complete phase, on a time scale where no long-range transmission can take place. The recombination with O2 is much more efficient than heme–NO recombination in the same protein and unprecedented for heme proteins. A near-zero yield of O2 dissociation from a heme protein has only been observed in the high oxygen-affinity quinol oxidase bd (25), but in this case it was ascribed to a low yield of photodissociation rather than a high rate of recombination. Thus, it would seem that in FixL, once oxygen is bound, its probability to escape from the heme pocket is very low.
Second, the spectra obtained preceding recombination are perturbed with respect to the ground-state spectra for all studied ligands and in a particularly strong way for O2. This observation suggests that intermediate conformations in the switching pathway are generated in this way and that femtosecond techniques can be used to transiently obtain sizeable populations of these intermediates. In the following we will discuss the implications of these findings in more detail.
Significance of Transient Spectra.
For all three ligands tested, CO, NO, and O2, the transient spectra on the picosecond and nanosecond time scale differ from the steady-state (unliganded minus liganded) difference spectra. This difference implies that on this time scale, the heme and its environment have not relaxed yet to the equilibrium unliganded conformation. Because the ligands presumably have not left the protein on this time scale, this might be explained by an influence of the dissociated ligand on the heme environment. However, two arguments indicate that this is not the case. First, the multiphasic dynamics of NO rebinding can be understood as a progressive relaxation of the heme–NO interaction (23, 28, 36). The fact that the shape of the transient spectra does not change for the different recombination phases (Fig. 3) indicates that, at least for NO, the position of the dissociated ligand does not influence the heme spectrum strongly. Second, in the oxygen-storage protein Mb the transient spectra on the picosecond time scale are much more similar to the steady-state difference spectra for MbCO (6, 28, 37), MbNO, and MbO2 (36), whereas, at least for CO dissociation, the ligand is located close to the heme up to the nanosecond time scale (38). By contrast, significant spectral perturbations have been observed after CO and O2 dissociation from hemoglobin (28), which were interpreted in terms of allosteric interactions between the heme and the protein backbone. In view of these comparisons, and in particular because the spectral distortion is strongest for the physiological effector O2, we suggest that these distortions reflect an intermediate where, after ligand dissociation, the heme configuration is restrained initially by the long-distance interactions associated with ligand sensing.
Using resonance Raman spectroscopy with nanosecond pulses, Rodgers et al. have attributed differences between the unliganded and CO-photodissociated forms of R. meliloti FixL to an intermediate after CO dissociation (39). As in our work, this intermediate also showed somewhat different characteristics, because the deoxy species and in particular the Fe–His bond was found to be shorter [a correlation between Fe–His bond length and kinase activity has been suggested (16)]. The observation of a CO-dissociated intermediate species (39) is consistent with the lack of recombination of CO with the heme on the time scale up to a few nanoseconds that we observe (Fig. 2). Our transient absorption spectra of FixLH–CO presumably correspond to the same intermediate, and our time resolution also permits observation of the corresponding intermediates for NO and O2 dissociation, which, in view of the fast recombination processes, are hardly accessible on the nanosecond time scale.
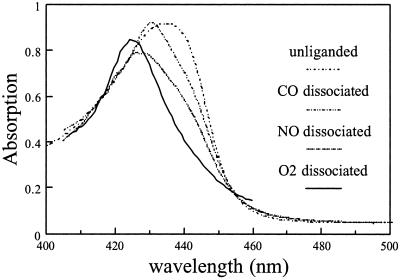
To characterize these intermediate states, we have attempted to construct their absolute absorption spectra in the Soret region (Fig. 6) for normalization, making use of the fact that the bleaching lobes of the transient spectra overlap well with those of the equilibrium spectra. With respect to the equilibrium nonliganded spectrum (maximum 435 nm), the intermediate spectra, with maxima at 431 (CO), 427 (NO), and 424 nm (O2) all are shifted in the direction of the initial bound form. As noted above, the deviation is strongest for O2, the physiological ligand. Comparison of the structures of the O2 and NO-bound forms of B. japonicum FixLH (ref. 12; a structure for the CO-bound form is not available) may help to understand this feature. The (heme-bound) O2 is stabilized on the distal side by a hydrogen bond to Arg-220, whereas this residue is turned away from the heme pocket when NO is bound [and presumably in the reduced unliganded form, which is available only for R. meliloti FixLH (16)]. This hydrogen bond may keep O2 close to the heme after dissociation, thus limiting heme conformational changes associated with the spin change and, in addition, favoring the recombination of oxygen with the heme, which proceeds with unprecedented efficiency (see below). Whereas the signal mechanism may be somewhat different for B. japonicum FixL and R. meliloti FixL (16), our observations on B. japonicum FixLH are in general agreement with the modeling result on the deoxy–R. meliloti FixLH structure, indicating that O2 cannot be accommodated in the heme pocket without serious conformational changes in the protein (16).
Figure 6.
Absolute absorption spectra of the dissociated species of FixLH, obtained by normalizing the bleaching part of the DAS of the recombination phases (nondecaying, 20-ps, and 4.7-ps phase for CO, NO, and O2, respectively) on those of the equilibrium difference spectra and adding the DAS to the corresponding absolute spectrum of the liganded form.
The above reasoning may explain the strong spectral effect on O2 as compared with NO and may be relevant for the sensing mechanism. However, we note that in MbO2, where severe spectral distortions are not observed in the time-resolved spectra, O2 is stabilized also by a hydrogen bond (40, 41) to a distal histidine. Therefore, and because distortions in FixLH are observed also for the other ligands, additional constraints on the heme, not present in Mb, must be present after dissociation of all three ligands. These constraints may be imposed by the propionates, which have been proposed to play an important role in the signal-transduction mechanism (12, 16, 39). From the changes in the Soret band alone one cannot directly deduce the associated structural changes, but our results demonstrate that they are present on the picosecond time scale for all ligands and suggest strongly that they are much stronger for O2, and to a lesser extent for NO, than for CO. Structural characterization of these intermediates will be addressed by vibrational spectroscopic studies with picosecond time resolution.
We note that similar perturbations were observed for the DosH–O2 complex. This indicates that, whereas the signal-transduction pathway has been proposed to be very different in Dos (4) and involves a distal methionine, constraints on the heme also play a regulatory role in the primary signal-transduction interactions.
Fast Geminate Recombination and Sensors.
Geminate recombination of ligands has now been studied for heme-based sensors specific for three different physiological ligands: NO [mammalian GC (6)], CO [CooA from R. rubrum (7)], and O2 (FixL and Dos, present work). The suggested mechanisms for signal transduction from the ligand-binding site to the regulated domain vary strongly between the different sensor proteins (see the introduction). Table 1 summarizes the main picosecond kinetic phases for these studies and compares them with Mb. A striking feature is that for all sensors, the rates and yields of geminate recombination are particularly high for the physiological ligand. In these sensors, the geminate recombination takes place from a heme environment very different from the nonliganded state. Thus it seems that as a general feature, the “off” (ligand-bound) configuration favors fast recombination once the iron–ligand bond is broken.
Table 1.
Picosecond decay phases of geminate recombination of three ligand sensors and Mb with different ligands
| CO
|
NO
|
O2
|
||||
|---|---|---|---|---|---|---|
| Decay, ps | Asymptote | Decay, ps | Asymptote | Decay, ps | Asymptote | |
| CooA* | 78 (0.6) | 0.1 | — | — | — | — |
| 386 (0.3) | ||||||
| Guanylate cyclase† | (0.0) | 1.0 | 7.5 (0.97) | 0.03 | — | — |
| FixLH‡ | (0.0) | 1.0 | 5.3 (0.62) | 0.06 | 4.7 (0.90) | 0.10 |
| 20 (0.24) | ||||||
| 220 (0.08) | ||||||
| DosH‡ | — | — | — | — | 5.3 (0.96) | 0.04 |
| Mbठ| (0.0) | 1.0 | 5 (0.53) | 0.04 | 6.5 (0.15) | 0.85 |
| 50 (0.22) | ||||||
| 500 (0.21) | ||||||
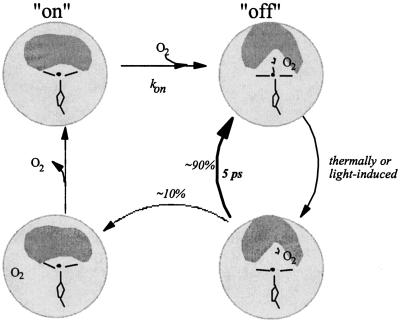
On the other hand, the overall ligand-binding rates kon are low for FixL and Dos and presumably also for CooA,§ giving rise to a relatively low affinity. Thus, for these heme sensors, a picture emerges where a high barrier of ligand binding to the heme in the “on” (unliganded) state is combined with a high initial barrier of ligand escape after dissociation from the off state. For those dissociated ligands that have overcome this initial escape barrier (≈10% for FixLH), the recombination probability is low (we observed no further rebinding up to the nanosecond time scale), suggesting that the binding barrier determining the low kon is in place and that some conformational change in the heme pocket toward the on state already occurs on the picosecond time scale. Our observation of different transient spectral shapes associated with the ≈5-ps recombination phase and the remaining phase is in agreement with this suggestion. These features are summarized in Fig. 7 for FixL. Altogether, it appears that in these sensor proteins, the heme pocket acts as a bi-stable switch that is protected from rapid fluctuations in both the on and off states.
Figure 7.
Schematic view of O2 binding and dissociation in FixL. The hatched circle represents the protein, and the inner gray area represents the heme environment involved in signal transduction. The right-hand conformations correspond to the off state, and the left-hand conformations correspond to the on state. There is a high barrier for ligand binding to the heme in the on state and a high initial barrier for ligand escape after dissociation from the off state, a feature that seems common for heme-based sensors.
The above general reasoning has many points in common for the environmental ligand-concentration sensors FixL and Dos (O2) and CooA (CO). The NO receptor GC also displays fast and efficient rebinding of NO, but the NO binding rate (kon) is very high (43), and in the absence of external effectors, the koff is very low (6, 44). This finding is in agreement with the fact that NO is a locally produced messenger molecule that is highly reactive in solution, thus requiring a very high-affinity receptor.
Outlook.
The possibility to synchronize ligand dissociation events by photolysis on a femtosecond time scale makes ultrafast spectroscopy a unique tool to study the ligand on → off regulation in heme-based sensors. The low yield of photodissociation of O2 from FixLH and DosH after a few picoseconds will present a major challenge for attempts to resolve the dynamics of the signal through the heme domain by using time resolved vibrational spectroscopy methods. Therefore, such studies will have to be combined with those using other ligands and with the full-length catalytic domain containing protein, which may exert strain on the heme domain.
Abbreviations
- GC
guanylate cyclase
- Mb
myoglobin
- DAS
decay-associated spectra
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Millisecond ligand-binding studies indicate that this lower Kd is predominantly the result of a higher kon (L. Kiger, M. C. Marden, U.L., and M.H.V., unpublished results).
A somewhat better fit was obtained with an additional decay phase, where the 4.7-ps phase was separated in two phases, with best fits for the time constants of ≈4-ps phase and (smaller phase) 7 ps. The closeness of the two time constants implies significant uncertainty in the fit. For simplicity we treat these phases here as a single one.
The kon for CO binding to CooA has been measured only by determining the bimolecular recombination after flash photolysis under conditions where the heme remained five-coordinated and was found to be in the range of 1–30 × 106 M−1⋅s−1 (42). Presumably, the binding rate will be much lower when the heme is six-coordinate, as in equilibrium nonliganded CooA.
References
- 1.Aano S, Nakajima H, Saito K, Okada M. Biochem Biophys Res Commun. 1996;228:752–756. doi: 10.1006/bbrc.1996.1727. [DOI] [PubMed] [Google Scholar]
- 2.Shelver D, Kerby R L, He Y, Roberts G P. Proc Natl Acad Sci USA. 1997;94:11216–11220. doi: 10.1073/pnas.94.21.11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilles-Gonzales M A, Ditta G S, Helinski D R. Nature (London) 1991;350:170–172. doi: 10.1038/350170a0. [DOI] [PubMed] [Google Scholar]
- 4.Delgado-Nixon V M, Gonzales G, Gilles-Gonzales M A. Biochemistry. 2000;39:2685–2691. doi: 10.1021/bi991911s. [DOI] [PubMed] [Google Scholar]
- 5.Gilles-Gonzales M A, Gonzales G, Perutz M F. Biochemistry. 1995;34:232–236. doi: 10.1021/bi00001a027. [DOI] [PubMed] [Google Scholar]
- 6.Négrerie M, Bouzhir-Sima L, Martin J-L, Liebl U. J Biol Chem. 2001;276:46815–46821. doi: 10.1074/jbc.M102224200. [DOI] [PubMed] [Google Scholar]
- 7.Kumazaki S, Nakajima H, Sakaguchi T, Nakagawa E, Shinahara H, Yoshihara K, Aano S. J Biol Chem. 2000;275:38378–38383. doi: 10.1074/jbc.M005533200. [DOI] [PubMed] [Google Scholar]
- 8.Perutz M F, Paoli M, Lesk A M. Chem Biol. 1999;6:R291–R297. doi: 10.1016/s1074-5521(99)80121-5. [DOI] [PubMed] [Google Scholar]
- 9.Rodgers K R. Curr Opin Chem Biol. 1999;3:158–167. doi: 10.1016/S1367-5931(99)80028-3. [DOI] [PubMed] [Google Scholar]
- 10.Pellequer J-L, Brudler R, Getzoff E D. Curr Biol. 1999;9:R416–R418. doi: 10.1016/s0960-9822(99)80257-7. [DOI] [PubMed] [Google Scholar]
- 11.Tuckerman J R, Gonzales G, Dioum E M, Gilles-Gonzales M A. Biochemistry. 2002;41:6170–6177. doi: 10.1021/bi025628w. [DOI] [PubMed] [Google Scholar]
- 12.Gong W, Hao B, Chan M K. Biochemistry. 2000;39:3955–3962. doi: 10.1021/bi992346w. [DOI] [PubMed] [Google Scholar]
- 13.Tamura K, Nakamura H, Tanaka Y, Oue S, Tsukamoto K, Nomura M, Tsuchiya T, Adachi S, Takahashi S, Iizuka T, Shiro Y. J Am Chem Soc. 1996;118:9434–9435. [Google Scholar]
- 14.Gilles-Gonzales M A, Gonzales G, Perutz M F, Kiger L, Marden M C, Poyart C. Biochemistry. 1994;33:8067–8073. doi: 10.1021/bi00192a011. [DOI] [PubMed] [Google Scholar]
- 15.Gong W, Hao B, Mansy S S, Gonzales G, Gilles-Gonzales M A, Chan M K. Proc Natl Acad Sci USA. 1998;95:15177–15182. doi: 10.1073/pnas.95.26.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyatake H, Mukai M, Park S-Y, Adachi S, Tamura K, Nakamura H, Nakamura K, Tsuchiya T, Iizuka T, Shiro Y. J Mol Biol. 2000;301:415–431. doi: 10.1006/jmbi.2000.3954. [DOI] [PubMed] [Google Scholar]
- 17.Regensburger B, Hennecke H. Arch Microbiol. 1983;135:103–109. doi: 10.1007/BF00408017. [DOI] [PubMed] [Google Scholar]
- 18.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 19.Hahn M, Hennecke H. Mol Gen Genet. 1984;193:46–52. [Google Scholar]
- 20.Lukat-Rodgers G S, Rodgers K R. Biochemistry. 1997;36:4178–4187. doi: 10.1021/bi9628230. [DOI] [PubMed] [Google Scholar]
- 21.Rodgers K R, Lukat-Rodgers G S, Tang L. J Biol Inorg Chem. 2000;5:642–654. doi: 10.1007/s007750000150. [DOI] [PubMed] [Google Scholar]
- 22.Tomita T, Gonzales G, Chang A L, Ikeda-Saito M, Gilles-Gonzales M A. Biochemistry. 2002;41:4819–4826. doi: 10.1021/bi0158831. [DOI] [PubMed] [Google Scholar]
- 23.Martin J-L, Vos M H. Methods Enzymol. 1994;232:416–430. doi: 10.1016/0076-6879(94)32057-8. [DOI] [PubMed] [Google Scholar]
- 24.Liebl U, Lambry J-C, Leibl W, Breton J, Martin J-L, Vos M H. Biochemistry. 1996;35:9925–9934. doi: 10.1021/bi960462i. [DOI] [PubMed] [Google Scholar]
- 25.Borisov V B, Liebl U, Rappaport F, Martin J-L, Zhang J, Gennis R B, Konstantinov A A, Vos M H. Biochemistry. 2002;41:1654–1662. doi: 10.1021/bi0158019. [DOI] [PubMed] [Google Scholar]
- 26.Winkler W C, Gonzales G, Wittenberg J B, Hille R, Dakappagari N, Jacob A, Gonzales L A, Gilles-Gonzales M A. Chem Biol. 1996;3:841–850. doi: 10.1016/s1074-5521(96)90070-8. [DOI] [PubMed] [Google Scholar]
- 27.Vos M H, Lambry J-C, Martin J-L. J Chin Chem Soc (Taipei) 2000;47:765–768. [Google Scholar]
- 28.Petrich J W, Poyart C, Martin J-L. Biochemistry. 1988;27:4049–4060. doi: 10.1021/bi00411a022. [DOI] [PubMed] [Google Scholar]
- 29.Franzen S, Kiger L, Poyart C, Martin J-L. Biophys J. 2001;80:2372–2385. doi: 10.1016/S0006-3495(01)76207-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodgers K R, Tang L, Lukat-Rodgers G S, Wengenack N L. Biochemistry. 2001;40:12932–12942. doi: 10.1021/bi011237q. [DOI] [PubMed] [Google Scholar]
- 31.Petrich J W, Lambry J-C, Kuczera K, Karplus M, Poyart C, Martin J-L. Biochemistry. 1991;30:3975–3978. doi: 10.1021/bi00230a025. [DOI] [PubMed] [Google Scholar]
- 32.Petrich J W, Lambry J-C, Balasubramanian S, Lambright D G, Boxer S G, Martin J-L. Biochemistry. 1994;238:437–444. doi: 10.1006/jmbi.1994.1302. [DOI] [PubMed] [Google Scholar]
- 33.Carlson M L, Regan R, Elber R, Li H, Phillips G N, Jr, Olson J S, Gibson Q H. Biochemistry. 1994;33:10597–10606. doi: 10.1021/bi00201a005. [DOI] [PubMed] [Google Scholar]
- 34.Walda K N, Liu X Y, Sharma V S, Magde D. Biochemistry. 1994;33:2198–2209. doi: 10.1021/bi00174a029. [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez G, Dioum E M, Bertolucci C M, Tomita T, Ikeda-Saito M, Cheesman M R, Watmough N J, Gilles-Gonzalez M A. Biochemistry. 2002;41:8414–8421. doi: 10.1021/bi025845x. [DOI] [PubMed] [Google Scholar]
- 36.Négrerie M, Berka V, Vos M H, Liebl U, Lambry J-C, Tsai A-L, Martin J-L. J Biol Chem. 1999;274:24694–24702. doi: 10.1074/jbc.274.35.24694. [DOI] [PubMed] [Google Scholar]
- 37.Reynolds A H, Rentzepis P M. Biophys J. 1982;38:15–18. doi: 10.1016/S0006-3495(82)84525-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Šrajer V, Teng T, Ursby T, Pradervand C, Ren Z, Adachi S, Schildkamp W, Bourgeois D, Wulff M, Moffat K. Science. 1996;274:1726–1729. doi: 10.1126/science.274.5293.1726. [DOI] [PubMed] [Google Scholar]
- 39.Rodgers K R, Lukat-Rodgers G S, Tang L. J Am Chem Soc. 1999;121:11241–11242. [Google Scholar]
- 40.Phillips S E V. J Mol Biol. 1980;42:531–554. doi: 10.1016/0022-2836(80)90262-4. [DOI] [PubMed] [Google Scholar]
- 41.Springer B A, Sligar S G, Olson J S, Phillips G N., Jr Chem Rev. 1994;94:699–714. [Google Scholar]
- 42.Uchida T, Ishikawa H, Takahashi S, Ishimori K, Morishima I, Ohkubo K, Nakajima H, Aano S. J Biol Chem. 1998;32:19988–19992. doi: 10.1074/jbc.273.32.19988. [DOI] [PubMed] [Google Scholar]
- 43.Zhao Y, Brandish P E, Ballou D P, Marletta M A. Proc Natl Acad Sci USA. 1999;96:14753–14758. doi: 10.1073/pnas.96.26.14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brandish P E, Buechler W, Marletta M A. Biochemistry. 1998;37:16989–16907. doi: 10.1021/bi9814989. [DOI] [PubMed] [Google Scholar]