Abstract
Current methods for the functional analysis of microarray gene expression data make the implicit assumption that genes with similar expression profiles have similar functions in cells. However, among genes involved in the same biological pathway, not all gene pairs show high expression similarity. Here, we propose that transitive expression similarity among genes can be used as an important attribute to link genes of the same biological pathway. Based on large-scale yeast microarray expression data, we use the shortest-path analysis to identify transitive genes between two given genes from the same biological process. We find that not only functionally related genes with correlated expression profiles are identified but also those without. In the latter case, we compare our method to hierarchical clustering, and show that our method can reveal functional relationships among genes in a more precise manner. Finally, we show that our method can be used to reliably predict the function of unknown genes from known genes lying on the same shortest path. We assigned functions for 146 yeast genes that are considered as unknown by the Saccharomyces Genome Database and by the Yeast Proteome Database. These genes constitute around 5% of the unknown yeast ORFome.
DNA microarrays simultaneously monitor the expression levels of thousands of genes. The massive gene expression data provide us with unique opportunities to analyze the functional and regulatory relationships among genes. One useful approach is to cluster genes with similar expression patterns. The most popular clustering methods include hierarchical clustering (1), K-means clustering (2), and self-organizing maps (3). On the assumption that genes with similar expression profiles have similar biological functions, functions of unknown genes can be predicted from their expression-similarity to known genes (1, 4).
However, do genes with similar functions always have similar expression profiles? The answer is, of course, no. First, genes with similar functions may not have been exposed to sufficient perturbations for their expression similarities to be revealed. Second, for some genes with similar functions, their product concentrations are partially or totally controlled at levels other than transcription. Third, measurements of expression similarity—e.g., Pearson's correlation or Euclidean distance—may not be able to completely capture the relationship between two expression profiles for such reasons as time-shift (5). Therefore, to identify the functional relationships between genes, we need to look beyond clustering methods. In this paper, we propose a method to group genes involved in the same biological process, even those without significant expression similarity.
First, we introduce an important characteristic of biological processes that we call transitive co-expression. Intuitively, this refers to situations where two genes are not strongly correlated in expression, but are both strongly correlated with the same set of other genes. In the simplest case, suppose genes a and b have strong expression correlation, as well as genes b and c. However, genes a and c do not have strong expression correlation, so we say that they are transitively co-expressed, with gene b serving as the transitive gene. It is clear that in a biological pathway, a gene is likely to show strong expression correlations with its neighbor genes, but not with genes that lie far apart in the pathway. This lack of correlation can be caused by various reasons. For example, (i) some biological processes are protracted in time, e.g., the cell cycle, so that expression relationships are revealed at different time points along the process. Using transitive co-expression, we can establish linkages between these genes to unveil the complete pathway. (ii) genetic and biochemical networks of a cell must withstand substantial random perturbations. Based on the principle of efficiency, mechanisms such as negative feedback loops limit the number of genes that fluctuate in their expressions. Thus different sets of experiments perturb different segments of a biological pathway to different extents, so that the expression relationships among genes along the pathway are revealed in a compartmentalized fashion across experiments. Such compartmentalized pathway segments can be associated through overlapping genes by transitive co-expression so that the biological pathway can be revealed as a whole.
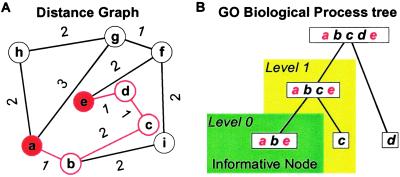
Given two genes known to be involved in the same biological pathway, identifying transitive genes between them may allow us to discover genes involved in the same biological process. Here, we propose a graph-theoretic scheme to identify such transitive genes. In our graph, vertices represent genes, and each gene pair that is highly correlated in expression is connected with an edge, where the edge length is a decreasing function of the expression correlation (Fig. 1A). Given such a graph, there can be multiple expression dependence paths between two genes—e.g., between genes a and e. The shortest of these paths (the shortest path, SP) would then be the most parsimonious explanation of dependence between a and e, given our expression data. If genes a and e are highly correlated, the shortest dependence path between them would just be the edge connecting them; if they are not significantly correlated but are involved in the same pathway, then we may still be able to discover their expression association by constructing the shortest dependence path between them. The transitive genes on the SP are likely to be important intermediate players between the two terminal genes in the same process.
Figure 1.
(A) Application of the shortest-path (SP) algorithm to gene expression data. Nine genes are depicted in the graph. The distance between two genes is a decreasing function of their correlation. For example, there are multiple expression dependence paths leading from gene a to gene e. Among them, the shortest dependence path is a–b–c–d–e, with genes b, c, and d serving as the transitive genes. This is the most parsimonious summary of the expression relationship between the terminal genes a and e. (B) Level 0 (L0) and level 1 (L1) matches of genes on the SP a–b–c–d–e defined according to their relationships in the Gene Ontology (GO) classification tree. With respect to the terminal genes a and e, the transitive gene b is a L0 match because it is annotated in the informative node where a and e are annotated; the transitive gene c is a L1 match because it shares the same direct parent as the two terminal genes; the transitive gene d is neither a L0 nor a L1 match.
In this paper, using yeast expression data, we first validate that the SP method is able to link functionally related genes, even without high expression correlation, and show the results to be highly statistically significant. In addition, we make comparisons to hierarchical clustering to show that our method can more precisely reveal functional relationships among genes. Finally, given that our method can group functionally related genes together, we make predictions for the functions of unknown genes based on those of known genes on the same SP. We made predictions for 146 unknown yeast genes, a significant number of which are supported by evidence other than the data we used.
Methods
Data Processing.
We used the Saccharomyces cerevisiae gene expression profiles from the Rosetta Compendium (6), which includes 300 deletion and drug treatment experiments. Genes were annotated by using the biological process ontology of Gene Ontology (GO) (7) provided by the Saccharomyces Genome Database (SGD) (8). To verify that genes on the same SP are likely to be involved in the same biological process, we applied our method to the Rosetta dataset and checked the results against GO-annotated biological processes in the three major cellular compartments: mitochondria, cytoplasm, and nucleus. Genes are separated according to their subcellular localizations because sometimes a GO process category may actually encompass distinct processes. Although similar in nature, these processes may occur in different cellular compartments and thus are not necessarily tightly controlled in regulation. For example, “protein biosynthesis” can take place in the mitochondria and the cytoplasm, and “membrane transport” processes in different compartments are distinct. We refine the process categories by sorting genes into the three major cellular compartments.
After removing the genes without GO process annotation and the 20 genes for which there are less than 80 experimental measurements in the Rosetta Compendium, we were left with 266 mitochondrial, 398 cytoplasmic, and 659 nuclear GO-annotated genes. For each of the three sets of genes, we calculated the expression similarities of all gene pairs {a, b} using Ca,b, the minimum of the absolute value of leave-one-out Pearson correlation coefficient estimates. This estimate is a measurement robust against single experiment outliers and sensitive to overall similarities in expression patterns.
Graph Construction and SP Computation.
We constructed three graphs, one for each set of the 266 mitochondrial genes, the 398 cytoplasmic genes, and the 659 nuclear genes. In each graph, two genes were assigned an edge if their absolute expression correlation Ca,b was higher than τ = 0.6. This cut-off, while conservative, nonetheless retains a sufficient number of connected gene pairs in the graph. The edge length between vertices a and b is da,b = f(Ca,b) = (1 − Ca,b)k. The powering factor k is used to enhance the differences between low and high correlations. Because the length of a path is the sum of the individual edge lengths, by exaggerating the differences between edge lengths, the SPs will be more likely to cover more transitive genes. Thus by increasing k we gain more power to reveal transitive co-expression. We set k = 6 because for k ≥ 6, the numbers of transitive genes stabilizes (detailed results at www.biostat.harvard.edu/complab/SP/). To ensure the quality of SPs, we consider only SPs with total path lengths <0.008.
We used Dijkstra's algorithm to identify the SPs between a source vertex to all other vertices in a graph. The central idea of Dijkstra's algorithm is the “relaxation” operation, which is based on the fact that every subpath of a SP is also a SP. For instance, in Fig. 1A, suppose that a is the source vertex and that we already have the SP from a to d via b and c. By relaxing each edge leaving d we attempt to see what is the SP from a to all other vertices that can be directly reached from d, in particular vertex e. If the path from a to e going through d is shorter than the current best SP estimate for e, then this estimate is updated to the new path. A detailed introduction to Dijkstra's algorithm can be found in ref. 9 and other algorithm textbooks. Exploiting the sparse nature of our graph, we implemented a priority queue of vertices with a Fibonacci heap to achieve O[n log(n) + m] time complexity, where n is the number of vertices and m is the number of edges in the graph. To determine the SPs starting from v source vertices, we applied Dijkstra's algorithm to each of the v vertices. Thus the overall time complexity is O[nv log(n) + mv].
Analyzing the SP Between Two Genes Involved in the Same Biological Process.
GO is a set of controlled biological vocabularies organized in a rooted directed acyclic graph. For our purposes it can be treated as a tree. Nodes in the tree refer to biological process categories. A parent node refers to a more general annotation than that of its children. SGD annotates each known yeast gene with one or more nodes in the GO tree. From all available annotations, we want to select those process categories that do not include too many genes to guarantee the functional homogeneity of member genes, and that do not include too few genes to provide sufficient numbers of genes for the purpose of validation. Using an approach similar to that proposed by Hvidsten et al. (10), we retrieved such process categories from GO by traversing the tree breadth-first from the root and selecting nodes that satisfy the properties that (i) the node contains more than γ = 30 genes and (ii) each of the node's children contains less than γ genes. We define such GO nodes as informative nodes, and the biological process categories they represent as the informative categories.
To test the validity of the SP method, we need to see whether genes on the same SP share the same GO process annotations. Given any two genes from the same informative process category (terminal genes), we determine whether there is a SP connecting them. If there is a SP including one or more transitive genes, we check the GO process annotations of these genes. A transitive gene is termed a level 0 (L0) match if it is annotated in the informative node in the GO tree from which the terminal genes are selected; it is termed a level 1 (L1) match if it shares the same direct parent node with the terminal genes (Fig. 1B). For all SPs connecting all gene pairs in each informative category, we count the total number of transitive genes as well as the numbers of L0 and L1 matches. For each cellular compartment we sum over the results of its informative categories and calculate the L0 and L1 match ratios relative to the total number of transitive genes.
Using a Permutation Test to Assess the Statistical Significance of the SPs.
For the purpose of validating the SP method, we need to evaluate the numbers of L0 and L1 matches, taking into account the numbers of such matches expected under the null hypothesis. Keeping the graph structure constant, we randomly permute the gene labels over the vertices to decouple gene annotations from their expression profiles. We then perform the SP method over these graphs, and calculate the L0 and L1 match ratios. This is done for 1,000 iterations. The distribution of L0 and L1 match ratios generated under the null hypothesis is compared with the observed quantities. The P values so derived give us an assessment of the ability of the SP method to reveal biological relationships between genes based on microarray data.
Predicting the Functions of Unknown Genes.
We use the SP method to classify previously unannotated yeast genes by adding the 3,255 ORFs unknown to SGD into the graphs of known genes in the mitochondrial, cytoplasmic, and nuclear compartments. As before, an edge is constructed between two genes if their absolute expression correlation is higher than 0.6. For all pairs of known genes, we determine the SPs connecting them. For the purpose of functional prediction, we would like to assign a putative function that is as specific as possible to the gene. Given all known genes on a SP, we achieve this by tracing back their annotations along the GO process tree and finding their lowest common ancestor. If the lowest ancestral node is at least 4 levels below the root of the GO tree, that is, it defines a sufficiently specific gene function, we then assign this function to the unknown genes on the SP. Analogous to the L0 and L1 matches, here the L0 prediction then corresponds to the lowest common ancestor, and the L1 prediction to its direct parent. In this way, the function represented by the lowest common ancestor can be more specific than that defined by the informative nodes. Under two circumstances an unknown gene may be assigned with multiple functions: (i) Because known genes on a SP may each have multiple functions, they may share several lowest common ancestors in the GO tree. (ii) An unknown gene may reside in different SPs with different lowest common ancestors. For each predicted gene function, we provide both the number of support SPs from which the prediction was derived and the number of unique known genes on those support SPs (support genes). The more support genes there are, the more confidence we have in the corresponding prediction. Note that a gene can be assigned putative functions in multiple graphs, because many genes are known to function in multiple cellular compartments.
Results
SP Method Clusters Genes Involved in the Same Biological Process.
We constructed the graphs for all yeast genes with GO process annotations in mitochondria (266 genes), cytoplasm (398 genes), and nucleus (659 genes). Using the procedure defined in Methods, we obtained 4, 8, and 22 informative GO categories for genes in the graphs of mitochondria, cytoplasm, and nucleus, respectively. The numbers of genes in those informative categories range from 31 to 174. In each graph, given any two genes belonging to the same informative GO process category, we identify the SP connecting them. We check the GO process classification of the transitive genes on the SP to identify the L0 and L1 matches, as discussed in Methods.
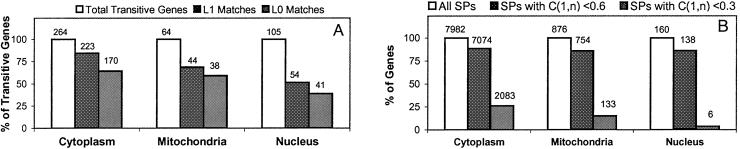
For each compartment, we summarize the results in Fig. 2A. In the cytoplasm and mitochondria graphs the SP method achieved surprisingly accurate results, where it successfully called 84%/64% and 69%/59% of the transitive genes at the L1/L0 levels, respectively. The nuclear graph shows relatively lower match ratios of 51%/39% at the L1/L0 levels. This result can be attributed to regulatory mechanisms exerted at levels other than transcription in the nucleus, for example regulated import/export of nuclear proteins. For the L1/L0 match ratios in all compartments, our method showed better performance than the 1,000 iterations of the permutation test, giving us P values less than 0.001. This demonstrates the significant power of the SP method in revealing biological relationships between genes based on microarray data.
Figure 2.
Summary of the performance of the SP method. (A) The percentages of L0- and L1-matched transitive genes in the three cellular compartments. Values shown above the bars are the numbers of genes. All match ratios at L0 and L1 are statistically significant at P < 0.001 by permutation test. (B) The percentages of SPs with at least one transitive gene in which terminal genes show weak (<0.6) and very weak (<0.3) expression correlations. C(1, n) denotes the expression correlation between the terminal genes. Values shown above the bars are the numbers of SPs.
It should be noted that, first, the match ratios we compute are in fact conservative. Because GO annotation is based only on positive biological evidence and is sparse in SGD, genes that are not classified as L1 or L0 matches may still be functionally related to the terminal genes on the SP. Second, we believe one major source of errors is the heterogeneity of perturbations in the Rosetta Compendium, where experimental conditions overall were not specifically designed for any particular biological process. Thus genes on a SP may be involved in several distinct biological processes that are nonetheless affected simultaneously by nonspecific perturbations. With experiments specifically designed for particular biological processes, we believe our approach will achieve higher accuracy.
SP Method Reveals Functional Relationships Between Genes with Low Expression Similarity.
Among the identified SPs, particularly interesting are those with low expression similarities between the terminal genes. In fact, over all three graphs, for more than 85% of the SPs that include at least one transitive gene, their terminal genes have weak expression correlations (<0.6) (Fig. 2B). This finding means that, in the graph, there is no edge between those terminal gene pairs; their relationships are revealed only by transitive co-expression by transitive genes. In a considerable subset of the SPs, transitive co-expression is even more pronounced. In fact, there are significant numbers of SPs with very weak expression correlations between the terminal gene pairs (<0.3). In the cytoplasm graph, there are 2,083 such SPs, which make up 26% of the total SPs. Even for transitive genes in these SPs, we achieved a L1 match ratio of 74%. This result confirms that our method can successfully reveal the relationship between functionally related gene pairs through transitive co-expression, even if their expression correlation is not significantly high.
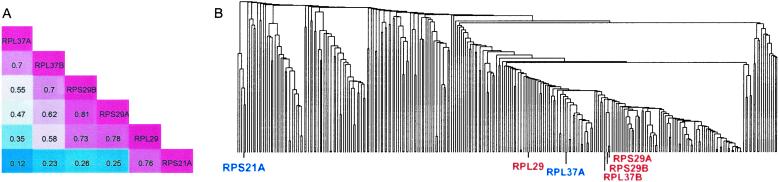
An example is the SP we identified in the graph of cytoplasm, RPL37A–RPL37B–RPS29B–RPS29A–RPL29–RPS21A from the GO process “protein biosynthesis.” The correlation matrix of the 6 genes is shown in Fig. 3A. In this SP, the expression correlation between the terminal genes is only 0.12. In addition, it is apparent that genes farther apart on the SP have lower expression correlations. All 6 genes encode ribosomal proteins and participate in protein biosynthesis. However, it is clear that they are not tightly coregulated as a group. We compare our method to standard hierarchical clustering methods based on expression correlations from the same Rosetta Compendium dataset. Among different linkages of hierarchical clustering, single linkage is the closest to the SP method in its greedy approach to building clusters by incrementally joining the closest, most correlated genes. For the SP mentioned above, using the 398 genes in the cytoplasm graph, the minimum hierarchical clustering subtree covering the 2 SP terminal genes (RPL37A and RPS21A) contains 276 genes (Fig. 3B). Among them, 144 are not ribosomal genes. Even genes involved in carbohydrate metabolism, osmosensory signaling, and fatty acid biosynthesis are included. Such remarkable contrasts between hierarchical clustering and the SP method are common in our results. As another example, the genes on the SP COR1–RIP1–SDH2–SDH4–STF1 in the mitochondria graph are all involved in oxidative phosphorylation. The correlation between the two terminal genes is 0.16. By hierarchical clustering, the minimum subtree covering the 2 SP terminal genes contains 241 genes, which constitutes 91% of the genes in the mitochondria graph. The weakness of the hierarchical clustering method is that once a gene is assigned to a cluster, comparisons with other genes are no longer done at the gene-to-gene level but at the cluster-to-gene or cluster-to-cluster level. Thus it does not offer us as parsimonious a description of the expression dependence as does the SP method. The same drawback also exists in other commonly used clustering methods, such as K-means clustering. Examples of such comparisons are available at our web site (www.biostat.harvard.edu/complab/SP/).
Figure 3.
(A) The correlation matrix for the shortest path RPL37A–RPL37B–RPS29B–RPS29A–RPL29–RPS21A. The gene names and their correlations in the Rosetta dataset are indicated. The magnitudes of the correlations are represented by the colors: red, high; blue, low. (B) The minimum subtree covering the two terminal genes in the SP (RPL37A and RPS21A) contains 276 genes.
Prediction of the GO Process Category for Unknown Genes.
After validating that the SP method can link functionally related genes, we used it to classify previously unannotated yeast genes. We used two rules to predict gene function. (i) A general rule: given all known genes on a SP, if their lowest common ancestor on the GO tree is more than 3 levels below the root, we then assign this function to the unknown genes on the SP. Over all SPs in the graphs of cytoplasm, mitochondria, and nucleus, we have made predictions for 80, 54, 115 genes, respectively, which are considered as unknown by SGD. (ii) Using a conservative rule, we consider those SPs that satisfy the general rule and that contain only one unknown gene. These are predictions for which we have higher levels of confidence, because these unknown genes are each functionally bounded by all of the other genes on the SP. Using the conservative rule, we made predictions for a total of 75 unique genes.
While some genes do not have GO biological process annotations in SGD, the biological processes that they are involved in are annotated in the “Cellular Role” category in the Yeast Proteome Database (YPD; www.incyte.com/sequence/proteome/databases/YPD.shtml). We obtained the YPD cellular role annotations for 24 genes (Table 1) of the total 75 genes predicted with the conservative rule, and we used them as a positive internal control for our prediction. Among the 24 genes, the predictions for 16 matched the experimentally derived annotations in YPD; 3 matched computationally derived annotations in YPD. By “match” we mean that our prediction and the YPD annotation are either identical or closely coupled. In addition, we identified a case (NOC3) in which the YPD cellular role annotation did not correspond to its cited experimental reference, whereas our prediction matched the experimental conclusions perfectly. This amounts to a successful prediction ratio of 83%, again validating our SP method.
Table 1.
Validation of the predictions made by the conservative rule for 24 genes without SGD GO process annotation, against their YPD cellular role annotation (as of March 2002)
| Gene | GO process category prediction (L0) | YPD annotation |
|---|---|---|
| ADH1 | Main pathways of carbohydrate metabolism | Carbohydrate metabolism [E] |
| ADH5 | Amino acid metabolism | Other metabolism [E]; carbohydrate metabolism [E] |
| BMS1 | Nucleobase, nucleoside, nucleotide metabolism | RNA processing/modification [E] |
| BUD28 | Protein biosynthesis | Cell polarity [E] |
| CLC1 | Actin cytoskeleton organization | Vesicular transport [E]; cell polarity [E] |
| COR1 | ATP synthesis coupled proton transport | Energy generation [E]; small molecule transport [P] |
| CPR7 | Cell cycle | Protein folding [P] [protein folding, G1 phase of cell cycle (19)] |
| ELP3 | Transcription, DNA-dependent | Pol II transcription [E, P]; protein modification [P]; chromatin/chromosome structure [P] |
| ERB1 | 35S primary transcript processing | RNA processing/modification [E] [involved in 35S primary transcript processing (11)] |
| GPH1 | Carbohydrate metabolism | Carbohydrate metabolism [E]; cell stress [E] |
| GSP1/Ran | Protein biosynthesis; ribosome biogenesis; 35S primary transcript processing; G2/M transition of mitotic cell cycle | Cell cycle control [E]; nuclear–cytoplasmic transport [E]; RNA processing/modification [E] |
| MAK16 | Ribosome biogenesis; RNA processing | RNA processing/modification [E] |
| NFI1/SIZ2 | Protein metabolism and modification; protein degradation | Cell cycle control [E]; protein modification [E] |
| NOC3 | Ribosome biogenesis; 35S primary transcript processing | Protein synthesis [E] [involved in the biogenesis of the 60S ribosomal subunit (13)] |
| NUG1 | RNA metabolism | Nuclear–cytoplasmic transport [E] [export of ribosomal subunit] (20) |
| PRO2 | Biosynthesis | Amino acid metabolism [E] |
| QCR8 | ATP synthesis coupled proton transport | Energy generation [E]; small molecule transport [P] |
| RIB5 | Amino acid and derivative metabolism | Other metabolism [E] |
| RIP1 | ATP synthesis coupled proton transport | Energy generation [E]; small molecule transport [P] |
| RSM26 | Protein biosynthesis | Energy generation [E]; cell cycle control [E]; protein synthesis [P] [protein of the mitochondrial ribosome small subunit (21)]; cell stress [E] |
| THR1 | RNA processing | Amino acid metabolism [E] |
| UGP1 | Carbohydrate metabolism | Carbohydrate metabolism [E] |
| XKS1 | Protein metabolism and modification | Carbohydrate metabolism [E] |
| YGL068W | Protein biosynthesis | Energy generation [P]; cell cycle control [E]; protein synthesis [P] [homolog of Escherichia coli L7/L12 ribosomal protein (22)] |
The gene with underlined names have YPD annotations that either agree or are closely coupled to our predictions. “[E]” denotes experimental evidence; “[P]” denotes computational prediction. We provide additional references in parentheses if (i) the YPD cellular role annotation is not sufficiently specific (ERB1, NUG1), (ii) YPD has misclassified (NOC3), or (iii) computational cellular role predictions included by YPD match our prediction (CPR7, RSM26, YGL068W).
For example, COR1, RIP1, and QCR8 are all predicted to be involved in “ATP synthesis coupled proton transport.” YPD annotates them as “energy generation” and “small molecule transport.” ERB1 is predicted to be involved in “35S primary transcript processing.” In a recent study it was found to be essential for 35S primary transcript processing (11). The nuclear trafficking protein GSP1/Ran was assigned with a set of diverse functions in both the cytoplasm and the nucleus graphs, including “protein biosynthesis,” “ribosome biogenesis,” and “G2/M transition of mitotic cell cycle.” Evidence in the literature suggests that it plays a central role in a large number of biological processes and is a master regulator of cell cycle and proliferation (12), which explains our putative functional assignments. The fact that we made predictions about GSP1/Ran from both nucleus and cytoplasm graphs agrees with the multiple subcellular localizations of this nuclear trafficking protein.
We predicted NOC3 to be in the GO category “ribosome biogenesis” and “35S primary transcript processing.” While this did not agree with its YPD annotation “protein biosynthesis,” a close inspection of the YPD-cited reference (13) revealed that NOC3 is in fact involved in the biogenesis of the 60S ribosomal subunit, which exactly matched our prediction.
The 51 novel genes predicted with the conservative rule for which no biological process annotation is available in either database are listed in Table 2. The additional 95 novel genes predicted by using the general rule that are unknown to both databases are listed on our web site (www.biostat.harvard.edu/complab/SP/). Together, we have assigned functions to around 5% of the unknown yeast ORFome. While some unknown genes have been assigned three-letter gene symbols, their cellular roles remain uncharacterized. For a significant portion of the genes listed we found partial experimental support, each to a different extent. We present some examples below.
Table 2.
Predictions of GO process category made by the conservative rule for 51 genes with neither SGD GO process annotation nor YPD cellular role annotation
| GO process category prediction (L1 ⇒ L0) | Gene [no. unique support genes, no. support SPs, graph] |
|---|---|
| Amino acid metabolism ⇒ amino acid biosynthesis | YHR029C [9, 14, C] |
| Cell cycle ⇒ DNA replication and chromosome cycle | TOS4 [5, 5, N] |
| Cell cycle ⇒ M phase | YPL267W [2, 1, N] |
| Cell cycle ⇒ mitotic cell cycle | TOS4 [6, 8, N] |
| Cell organization and biogenesis ⇒ cytoplasm organization and biogenesis | YNR046W* [5, 4, N] |
| Cytoplasm organization and biogenesis ⇒ organelle organization and biogenesis | RIM21 [2, 1, M] |
| Cytoplasm organization and biogenesis ⇒ ribosome biogenesis | PUF6 [14, 46, N], RRP12* [3, 2, N], YDR324C* [6, 6, N], YGR128C* [2, 1, C], YIL019W* [2, 1, C], YJL010C* [3, 2, N], YML093W* [2, 1, C], YML093W* [2, 1, M] (23), YML093W* [3, 2, N], YPL146C* [4, 3, N] |
| DNA metabolism ⇒ DNA repair | YBR089W [2, 1, N] |
| Carbohydrate metabolism ⇒ catabolic carbohydrate metabolism | PST2 [2, 1, M] (24) |
| Metabolism of energy reserves ⇒ trehalose metabolism | TFS1 [2, 1, C] |
| Metabolism ⇒ biosynthesis | YDR165W [2, 1, N], YLR132C* [118, 117, C], YPL246C [3, 2, C] (16) |
| Metabolism ⇒ coenzymes and prosthetic group metabolism | YLR356W [2, 1, N] |
| Metabolism ⇒ nucleic acid metabolism | DAT1 [3, 2, N], YBR267W [7, 6, N], YDL063C [5, 4, N], YDR165W [2, 1, N], YGR128C* [4, 4, N], YGR145W [2, 1, M], YLR132C* [3, 2, N], YLR287C [4, 4, N], YNL311C [2, 1, N], YOR004W* [4, 3, N], YOR042W [2, 1, N], YPR045C [5, 8, N] |
| Metabolism ⇒ protein metabolism and modification | RNY1 [3, 3, C], TOS5 [2, 1, C] (16), YDR165W [114, 113, C] (23), YKL053C-A [16, 62, M], YKL195W* [25, 37, M] (16), YLR356W [46, 45, C], YLR434C [119, 124, C] |
| Mitotic cell cycle ⇒ S phase of mitotic cell cycle | YBR089W [2, 1, N] (25) |
| Monosaccharide metabolism ⇒ hexose metabolism | YCR013C* [3, 2, C] (18) |
| Nucleic acid metabolism ⇒ RNA metabolism | YDR324C* [3, 1, N], YML093W* [3, 2, C] |
| Protein biosynthesis ⇒ protein synthesis initiation | LSG1 [3, 2, C] |
| Protein complex assembly ⇒ cytochrome c oxidase biogenesis | YKL053C-A [3, 3, M] |
| Protein metabolism and modification ⇒ protein biosynthesis | RNQ1 [9, 8, M] (16), YGL069C* [24, 44, M] (26), YGL102C* [4, 4, C] (27) |
| Protein-mitochondrial targeting ⇒ mitochondrial translocation | RNQ1 [3, 2, M] |
| Ribosome biogenesis ⇒ ribosomal large subunit assembly | YDR496C [2, 1, M], YML093W* [2, 1, N] |
| Ribosome biogenesis ⇒ rRNA processing | YDR496C [12, 38, N] |
| RNA metabolism ⇒ RNA processing | BCP1* [8, 14, N], TCI1 [2, 1, M], TCI1 [2, 1, N], YGR145W* [5, 5, N] |
| rRNA processing ⇒ 35S primary transcript processing | BCP1* [5, 5, N], PUF6 [2, 1, M], YDR101C [9, 24, N], YHR085W* [4, 5, N], YJL010C* [2, 1, N], YML093W* [2, 1, M] |
| Transcription ⇒ transcription, DNA-dependent | RIO2* [5, 6, N], YNL114C* [4, 3, N] |
| Transcription, DNA-dependent ⇒ transcription, from Pol I promoter | SAS10* [7, 6, N], YDR101C [13, 16, N], YGR210C [3, 2, N], YMR310C [3, 2, N], YNL182C* [8, 9, N] |
The last two levels of the predicted GO processes are shown. Essential genes—i.e., genes with lethal null phenotypes—are marked with
. For each prediction, we show in square brackets the number of support SPs, the number of unique support genes, and the graph in which the prediction is made. References to experimental supports are noted in parentheses. “C” denotes cytoplasm, “M” denotes mitochondria, and “N” denotes nucleus.
One intriguing gene for which we made predictions is RNQ1. It is known to form the [PIN+] prion, and it possesses a glutamine- and asparagine-rich domain characteristic of other yeast prions (14). The function of RNQ1, however, is so far unknown. We predicted it to be involved in both “mitochondrial translocation” and “protein biosynthesis.” Interestingly, the prion responsible for the [PSI+] determinant, SUP35, not only is an important factor for protein translation termination but also is implicated in the system of cotranslational translocation into the mitochondria (15). In addition, a large-scale transcription profiling study found RNQ1 to be clustered with genes involved in protein biosynthesis (16).
There are numerous other interesting cases. BRX1 is predicted to be “ribosome biogenesis.” In fact, a very recent experimental study showed that it is involved in ribosomal large subunit assembly (17). YCR013C was predicted to be involved in “hexose metabolism.” There is evidence based on SAGE data (18) that supports our prediction. In total, for 11 of the 51 genes we have collected suggestive experimental evidences corroborating our predictions (Table 2). Most of the evidence comes from gene expression studies. To determine the biological function and verify our predictions of those genes, further experimental work is required.
Discussion
Systematic approaches to the functional annotation of genes identified in the genome sequencing projects are urgently needed in the postgenomic era. The rapid increase in large-scale gene expression data provides us unique opportunities to meet this need. However, the development of methodology to discover novel gene functions by using expression data has so far been slow. With its 300 deletion and drug-treatment experiments, the Rosetta Compendium (6) is to date the most systematic expression profiling of the yeast genome published. In its original publication, by clustering approaches the authors identified and experimentally confirmed 8 novel gene functions. The Compendium has since remained mostly unexplored. In this paper we have proposed a systematic approach to predict gene functions based on such large-scale data. Applying it to the Rosetta Compendium, we assigned putative functions to 75 unknown yeast genes in SGD by using the conservative rule. We have shown that a positive internal control based on 24 genes gives a successful prediction ratio of 83%. Based on the general prediction rule, we assigned functions to an additional 95 unknown yeast genes. Together these assignments make up 5% of the unknown yeast ORFome. While impressive, the Compendium contains only a tiny fraction of possible perturbations. We believe the method can achieve even better results with more comprehensive expression datasets.
The strength of our method is its utilization of transitive co-expression, an important feature among genes of the same biological process. To link such genes, we applied the SP algorithm. In contrast with traditional clustering approaches, ours is able to group not only functionally related genes with correlated expression profiles but also those without. Moreover, the expression dependence relationships between individual genes are given by the SPs. Using transitive co-expression, we are able to capture the functional relationships between genes beyond “synexpression” (6). While clustering is a useful approach to group genes with similar expression patterns, it is not sensitive to other types of expression relationships such as transitive co-expression. As we have seen, the SP method offers a complementary and informative tool for large-scale expression data analysis.
Another advantage of the SP method over traditional clustering methods is that it transparently and actively uses available biological knowledge as a guide to discover additional relevant genes. Clustering methods first group genes according to their expression profiles, then make inferences on the functions of genes within clusters. In contrast, our method starts with two genes with a known biological function, then uses them as a bound to identify intermediate genes related to them. Such active incorporation of biological annotation into the knowledge discovery process is one of the current challenges in microarray data analyses.
The SP method is scalable to larger graphs. For our prediction based on the nuclear graph with 3,914 vertices (genes) and 20,815 edges, determination of the SP from the 659 known genes as the source vertices took only 7 min on a 700-MHz Pentium III processor running Linux. Because for a given graph the computation needs to be done only once and is easily distributed over multiple processors, the SP method is applicable to the transcriptomes of higher eukaryotes such as the human and mouse.
Acknowledgments
The work of X.Z. and W.H.W. is supported by National Institutes of Health Grant 1R01HG02341. The work of M.-C.J.K. is supported by a Howard Hughes Medical Institute Predoctoral Fellowship.
Abbreviations
- SP
shortest path
- L0
level 0
- L1
level 1
- GO
Gene Ontology
- SGD
Saccharomyces Genome Database
- YPD
Yeast Proteome Database
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 12509.
References
- 1.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tavazoie S, Hughes J D, Campbell M J, Cho R J, Church G M. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 3.Golub T R, Slonim D K, Tamayo P, Huard C, Gaasenbeek M, Mesirov J P, Coller H, Loh M L, Downing J R, Caligiuri M A, et al. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 4.Niehrs C, Pollet N. Nature (London) 1999;402:483–487. doi: 10.1038/990025. [DOI] [PubMed] [Google Scholar]
- 5.Qian J, Dolled-Filhart M, Lin J, Yu H, Gerstein M. J Mol Biol. 2001;314:1053–1066. doi: 10.1006/jmbi.2000.5219. [DOI] [PubMed] [Google Scholar]
- 6.Hughes T R, Marton M J, Jones A R, Roberts C J, Stoughton R, Armour C D, Bennett H A, Coffey E, Dai H, He Y D, et al. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 7.Ashburner M, Ball C A, Blake J A, Botstein D, Butler H, Cherry J M, Davis A P, Dolinski K, Dwight S S, Eppig J T, et al. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dwight S S, Harris M A, Dolinski K, Ball C A, Binkley G, Christie K R, Fisk D G, Issel-Tarver L, Schroeder M, Sherlock G, et al. Nucleic Acids Res. 2002;30:69–72. doi: 10.1093/nar/30.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cormen T H. Introduction to Algorithms. Cambridge, MA: MIT Press; 2001. [Google Scholar]
- 10. Hvidsten, T. R., Komorowski, J., Sandvik, A. K. & Laegreid, A. (2001) Pac. Symp. Biocomput., 299–310. [DOI] [PubMed]
- 11.Pestov D G, Stockelman M G, Strezoska Z, Lau L F. Nucleic Acids Res. 2001;29:3621–3630. doi: 10.1093/nar/29.17.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke P R, Zhang C. Trends Cell Biol. 2001;11:366–371. doi: 10.1016/s0962-8924(01)02071-2. [DOI] [PubMed] [Google Scholar]
- 13.Milkereit P, Gadal O, Podtelejnikov A, Trumtel S, Gas N, Petfalski E, Tollervey D, Mann M, Hurt E, Tschochner H. Cell. 2001;105:499–509. doi: 10.1016/s0092-8674(01)00358-0. [DOI] [PubMed] [Google Scholar]
- 14.Osherovich L Z, Weissman J S. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 15.Shumov N N, Volkov K V, Mironova L N. Genetika. 2000;36:644–650. [PubMed] [Google Scholar]
- 16.Jelinsky S A, Estep P, Church G M, Samson L D. Mol Cell Biol. 2000;20:8157–8167. doi: 10.1128/mcb.20.21.8157-8167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaser A, Bogengruber E, Hallegger M, Doppler E, Lepperdinger G, Jantsch M, Breitenbach M, Kreil G. Biol Chem. 2001;382:1637–1647. doi: 10.1515/BC.2001.199. [DOI] [PubMed] [Google Scholar]
- 18.Velculescu V E, Zhang L, Zhou W, Vogelstein J, Basrai M A, Bassett D E, Jr, Hieter P, Vogelstein B, Kinzler K W. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 19.Fujimori F, Gunji W, Kikuchi J, Mogi T, Ohashi Y, Makino T, Oyama A, Okuhara K, Uchida T, Murakami Y. Biochem Biophys Res Commun. 2001;289:181–190. doi: 10.1006/bbrc.2001.5925. [DOI] [PubMed] [Google Scholar]
- 20.Bassler J, Grandi P, Gadal O, Lessmann T, Petfalski E, Tollervey D, Lechner J, Hurt E. Mol Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 21.Saveanu C, Fromont-Racine M, Harington A, Ricard F, Namane A, Jacquier A. J Biol Chem. 2001;276:15861–15867. doi: 10.1074/jbc.M010864200. [DOI] [PubMed] [Google Scholar]
- 22.Mewes H W, Frishman D, Guldener U, Mannhaupt G, Mayer K, Mokrejs M, Morgenstern B, Munsterkotter M, Rudd S, Weil B. Nucleic Acids Res. 2002;30:31–34. doi: 10.1093/nar/30.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Causton H C, Ren B, Koh S S, Harbison C T, Kanin E, Jennings E G, Lee T I, True H L, Lander E S, Young R A. Mol Biol Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J, Godon C, Lagniel G, Spector D, Garin J, Labarre J, Toledano M B. J Biol Chem. 1999;274:16040–16046. doi: 10.1074/jbc.274.23.16040. [DOI] [PubMed] [Google Scholar]
- 25.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Traven A, Wong J M, Xu D, Sopta M, Ingles C J. J Biol Chem. 2001;276:4020–4027. doi: 10.1074/jbc.M006807200. [DOI] [PubMed] [Google Scholar]
- 27.Rep M, Krantz M, Thevelein J M, Hohmann S. J Biol Chem. 2000;275:8290–8300. doi: 10.1074/jbc.275.12.8290. [DOI] [PubMed] [Google Scholar]