Abstract
We observed fast aggregation of partially ordered oligomers in an earlier simulation study of an amyloidogenic hexapeptide NFGAIL. In this work, the nucleation of highly ordered oligomers was further investigated by a combined total of 960 ns molecular dynamics simulations with explicit solvent on NFGAIL and its nonamyloidogenic mutant NAGAIL. In these simulations, four dimer subunits that each was constrained by harmonic forces as a two-strand β-sheet were used to enhance the rate of formation. It was found that a critical role played by the aromatic residue Phe was to direct the stacking of β-sheets to form ordered multilayer aggregates. We also found that many molecular arrangements of the peptide satisfied the “cross-β-structure”, a hallmark of amyloid fibrils. The tendency for the peptide to form either parallel or antiparallel β-sheet was comparable, as was the tendency for the β-sheets to stack either in parallel or antiparallel orientation. Overall, ∼85% of the native hexapeptide formed octamers. The fact that only 8% of the octamers were well-ordered species suggests that the dissociation of the disordered oligomers be the rate-limiting step in the formation of highly ordered oligomers. Among the well-ordered subunit pairs, about half was formed by the β-sheet extension along the main-chain hydrogen-bond direction, whereas the other half was formed by the β-sheet stacking. Hence, a delicate balance between intersheet and intrasheet interactions appeared to be crucial in the formation of a highly ordered nucleus of amyloid fibrils. The disordered oligomers were mainly stabilized by nonspecific hydrophobic interactions, whereas the well-ordered oligomers were further stabilized by cross-strand hydrogen bonds and favorable side-chain stacking.
INTRODUCTION
Amyloidogenesis is a common pathogenic process associated with many debilitating human diseases (Dobson, 1999; Kayed et al., 2003; Kelly, 1998) in which proteins and peptides of diverse sequences and structures form toxic soluble oligomers and insoluble fibrils. The resulting oligomers and fibrils share highly similar “cross-β” structures (Kayed et al., 2003), suggesting a common mechanism. Studies have further suggested a nucleation process followed by a conformational change into a predominantly β-sheet secondary structure (Dobson, 1999; Kelly, 1998; Rochet and Lansbury, 2000; Thirumalai et al., 2003). Recent progress included the advent of antiamyloid agents (Mason et al., 2003). Despite these efforts, many important questions remain unanswered. Of particular significance is the mechanism by which early stage ordered oligomers start to form. An improved knowledge on these early stage processes can enhance our ability to design more effective therapeutic agents because the nucleation state is believed to be one of the rate-limiting steps.
Lattice and other simplified models have been used to study protein aggregation (Dima and Thirumalai, 2002), to search possible aggregating conformations of SH3 domain (Ding et al., 2002), to study the competition between protein folding and aggregation of a tetrameric β-sheet complex (Jang et al., 2004), and to investigate spontaneous fibril formation by random-coil peptides (Nguyen and Hall, 2004). All-atom molecular dynamics (MD) simulations have been applied to study amlyoid fibril stability (Li et al., 1999; Zanuy et al., 2003; Zanuy and Nussinov, 2003). Early stage aggregation has also been studied with the assistance of interstrand harmonic restraining forces (Gsponer et al., 2003; Klimov and Thirumalai, 2003). This was necessary because aggregation is an extremely slow process in comparison to the timescales accessible to all-atom molecular dynamics simulations.
The human islet amyloid polypeptide (IAPP) is a 37-amino-acid hormone and is the main constituent of the islet amyloid fibrils found in 95% of type II diabetes mellitus (Hoppener et al., 2000; Westermark et al., 1987). IAPP forms cytotoxic amyloid fibrils in vitro by inducing islet cell apoptosis (Lorenzo et al., 1994). The hexapeptide NFGAIL is a fragment truncated from the human IAPP (residues 22–27). The short NFGAIL fragment is a good model system because it is one of the shortest fragments that can form amyloid fibrils similar to those formed by the full-length polypeptide (Sunde et al., 1997; Tenidis et al., 2000) and the fibrils are also cytotoxic to the pancreatic cell line. Alanine-scanning mutagenesis revealed that the Phe played a critical role in the formation of the fibrils (Azriel and Gazit, 2001). The F23A mutation prohibited the mutant from forming amyloid fibril organization.
In our previous work, formation of tetrameric aggregates of the peptide NFGAIL was studied. The analyses indicated that dimer assembly could be a major pathway to the formation of larger aggregates (Wu et al., 2004). Here, we take a step further and study the formation of octamers by all-atom MD simulations with explicit solvent. The novelty of our approach in this study is that the association of well-ordered oligomers is enhanced by the use of two-strand β-sheets as subunits to allow the association process to take place within affordable computing time. We further examine the two assembling modes: β-sheet stacking and β-sheet extension, and investigate the roles of intersheet (hydrophobic) and intrasheet interactions (hydrogen bonding) in the formation of highly ordered β-sheet complex. The role of Phe in the formation of ordered oligomers is investigated by comparing the simulation results on NFGAIL and its nonamyloidogenic mutant NAGAIL. In addition, the stability of the well-ordered oligomers was studied by simulations at a high temperature with less or no conformational restraints.
METHOD
Systems
A subunit is a two-strand β-sheet in either parallel or antiparallel configuration. The two strands in each subunit were held together by restraining harmonic forces along the main-chain hydrogen bonds (2.8 Å between O and H). The main-chain torsion angles were also restrained to (Φ = −110°, Ψ = 135). The side-chain atoms were allowed to move without any restraints. Four subunits of a given sequence (NFGAIL or NAGAIL) were immersed into a triclinic box of water (∼3553 water molecules), equivalent to a truncated-octahedral box, with the final box dimensions of a = b = c = 52.77 Å, α = β = γ = 109.47°. The effective peptide concentration was ∼117 mM.
MD simulation
The AMBER simulation package was used in both MD simulations and trajectory analysis (Case et al., 2002). The Duan et al. force field (Duan et al., 2003) was chosen to represent the peptides and the N- and C-termini were blocked, respectively, by acetyl and amine groups. The four subunits were initially placed 24 Å away from each other in parallel. The solvent was explicitly represented by the TIP3P water model. The solvated peptide systems were subjected to periodic boundary conditions via both minimum image convention and fast Fourier transformation implemented as part of the particle-mesh Ewald (PME) method (Essmann et al., 1995).
A set of 10 simulations was carried out for each system. The orientations and positions of the four subunits were randomized at 500 K and 1.0 atm pressure for 1.0 ns followed by 1.0 ns simulations at 278 K and 1.0 atm pressure to adjust the system size and density and to fully solvate the peptides. Production simulation was carried out at 278 K and constant volume for 21.0 ns. The PME method (Essmann et al., 1995) was used to treat the long-range electrostatic interactions. SHAKE (Ryckaert et al., 1977) was applied to constrain all bonds connecting hydrogen atoms to allow a time step of 2.0 fs. To reduce the computation, nonbonded forces were calculated using a two-stage reference system propogation algorithm approach (Barash et al., 2003) where the forces within a 10-Å radius were updated every step and those beyond 10 Å were updated every two steps. Temperature was controlled by Berendsen's thermostat (Berendsen et al., 1984) with a coupling constant of 2.0 ps. The trajectories were saved at 10.0-ps intervals and 23,000 snapshots were produced in each set of simulations for further analysis. The snapshots at the randomization/equilibration phase were also collected and used as references.
Order score of a pair of associated subunits
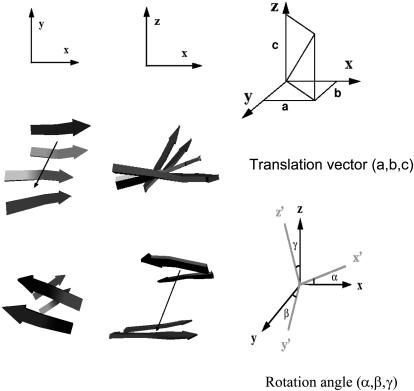
Because the dimers of the hexapeptide were restrained to be two-strand parallel or antiparallel β-sheets, their main chains were almost rigid. Hence, the translation and rotation were sufficient to characterize the geometry relationship between two subunits. The translation vector t between two dimer subunits was calculated based on their centers of mass. The relative rotation between two subunits was characterized by their reference frames, which were unitary matrices. The unitary matrices of dimer subunit one (U1) and two (U2) were calculated by the singular value decomposition of the correlation matrix (AT B) as follows:
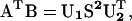 |
(1) |
where A(n × 3) and B(n × 3) were the coordinates of the main-chain atoms from the dimer subunit one and the dimer subunit two, respectively, S (3 × 3) is a diagonal matrix where the singular values of the matrix A or B are in diagonal with a decreasing order, U1 (U2) consists of the orthonormal column vectors h1, h2, h3 (k1, k2, k3).
In this case, h1 and k1, which were associated with the largest singular value (s1), were parallel to β-strand direction; h2 and k2 associated with the second largest singular value (s2) were almost parallel to the main-chain hydrogen-bonding direction; h3 and k3 associated with the smallest singular value (s3) were almost parallel to the norm of the β-sheet plane (hereafter defined as stacking direction) (Fig. 1).
FIGURE 1.
The translation vector (a, b, c) (x, β-strand direction; y, hydrogen-bond direction; z, β-sheet stacking direction) and rotation angles from the three directions (α, β, γ) were used to describe the association types and the association order score.
The cosine values (cosα, cosβ, and cosγ) of the angles between two corresponding vectors ((h1, k1) (h2, k2) (h3, k3)) were calculated to characterize the rotation matrix from dimer subunit one to dimer subunit two. The projections (a, b, c) of the translation vector t on each column vectors of the matrix U1 were calculated to characterize the translation along each direction from the dimer subunit one to the dimer subunit two. Based on these six parameters, the association relation for a pair of associated dimer subunits was further evaluated as follows (Fig. 1):
There are two ordered association modes for a pair of subunits. One mode is β-sheet extension (edge-to-edge), which produces a single-layer four-strand β-sheet. The other mode is β-sheet stacking (face-to-face), which produces a double-layer two-strand β-sheet. To classify a pair of subunits into either mode, the cutoffs on the six parameters (a, b, c, α, β, and γ) were set empirically (Fig. 1). In β-sheet extension association mode, the cutoffs are |α| < 35°, |β| < 60°, |a| < 18 Å (corresponding to the largest registry shift for two subunits to still be in contact), |b| < 10.5 Å (roughly two hydrogen-bond distance), and |c| < 6 Å (the largest deviation along the stacking direction allowed for the two subunits to still form a four-strand β-sheet). Similarly, the cutoffs for β-sheet stacking were defined as |α| < 45°, |β| < 60°, |a| < 18 Å, |b| < 6 Å, and |c| < 12.5 Å. The motivation to relax the angle along the strand direction from no more than 35 to 45° was that the stacking of β-sheets was less ordered than the β-sheet extension, which was mainly stabilized by cross-strand hydrogen bonds.
Furthermore, there were two possible configurations (parallel or antiparallel) for each type of ordered association (the β-sheet extension or the β-sheet stacking). The configuration type was defined by the directions of the two adjacent β-strands from the two associated subunits. In the case of the β-sheet extension, the configuration type was defined by the directions of the two central β-strands of a four-strand β-sheet. In the other case of the β-sheet stacking, the stacking configuration was defined by the two closest β-strands that were stacked.
A score function for the β-sheet extension (when satisfying the cutoffs for β-sheet extension mode) was given by Eq. 2:
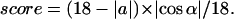 |
(2) |
The motivation for Eq. 2 is to consider the registry-shift defects of cross-strand hydrogen bonds, indicated by the relative shift along the β-strand direction and the relative rotation along the stacking direction. In this definition, the score is 1.0 when two subunits are aligned in-registry with perfect parallel or antiparallel orientation and becomes 0.0 when two subunits are either off-registry or orthogonal to one another. In a similar way, the β-sheet stacking score (when satisfying the cutoffs for β-sheet stacking mode) score was given by Eq. 3:
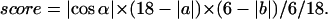 |
(3) |
The motivation for Eq. 3 was to consider the overlapping surface between the two subunits. The bigger the overlapping surface, the stronger the hydrophobic interaction between the two layers.
Order score for a snapshot
The order score for a snapshot was defined as the summation of the order score of all oligomers in the snapshot. The order score of an oligomer was defined as the summation of the order score of all unique pair of subunits in the oligomer. The average score over each set of simulations was calculated against the simulated time to monitor the formation of ordered oligomers.
Kinetics modeling
Following the work of Roberts (Roberts, 2003), the reaction scheme of an irreversible aggregation is shown in Eq. 4 where A(i)'s are soluble aggregates consisting of i subunits (i.e., “2i-mers”). The conformation transition from random coils to β-extended strands was not considered in this study. The third- and forth-order kinetics were also neglected due to their small rates. From Eq. 4, we consider two primary reaction pathways to form A(4): addition of one subunit at a step, or merge of two A(2).
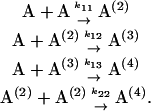 |
(4) |
Following Eq. 4, a set of differential equations with respect to time (t) are written in Eqs. 5.1–5.3. [A(i)] are the concentrations of 2i-mer. The reactions rates were estimated by a least-square fit of the concentration profiles of the 2i-mers to Eqs. 5.1–5.4.
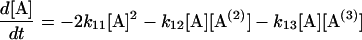 |
(5.1) |
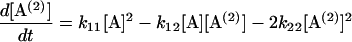 |
(5.2) |
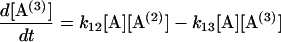 |
(5.3) |
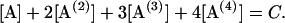 |
(5.4) |
RESULTS
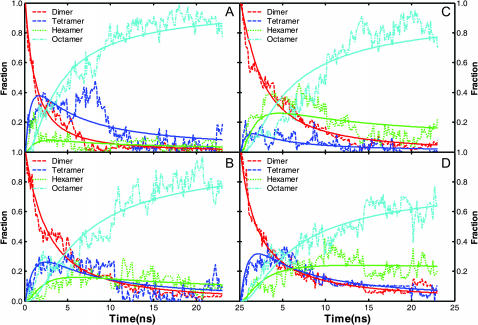
The association process is quantitatively characterized by the change of the oligomeric state of each subunit (two-strand β-sheet) in the solution. In this study, the total number of atom contacts between the peptide subunit is utilized to characterize the oligomeric state. The cutoff distance for atom-atom contact is set to the van der Waals distance plus 2.8 Å (the size of water molecule). Two subunits are considered associated when their atom contacts reach a predefined critical number that is set empirically to 400. Similarly, a subunit is classified as “associated” to an existing oligomer (n = 1, 2, 3 subunits) if the total number of atom contacts between the subunit and the oligomer reach the same critical number. The oligomeric state was defined as the number of subunits in the oligomer multiplied by 2 (i.e., two strands per subunit). The time evolution of the fraction of subunits in each oligomeric state (n = 2, 4, 6, 8) is shown in Fig. 2 for each type of subunit (NFGAIL parallel dimer (A), NFGAIL antiparallel dimer (B), NAGAIL parallel dimer (C), and NAGAIL antiparallel dimer (D)).
FIGURE 2.
The fraction of oligomer species. (A) NFGAIL parallel dimer; (B) NFGAIL antiparallel dimer; (C) NAGAIL parallel dimer; (D) NAGAIL antiparallel dimer. The fractions were averaged every nanosecond for clarity. Note that although the units here are fraction, the units in the text are in percentage.
As the octamer (four subunits) fraction increased monotonically, the dimer fraction (one subunit) decreased monotonically during the simulations, suggesting octamer formation dominated the process. Tetramers (two subunits with four strands) and hexamers (three subunits with six strands) started to form rather early. Their percentages continued to increase until ∼10.0 ns, then decreased to below 10%, which was caused by conversion into octamers as evidenced by the simultaneous increase of octamer. The final fraction of the octamers was above 60%. Neither the F23A mutation nor the subunit configuration (parallel/antiparallel dimer) changed the oligomerization process significantly when the formation of disordered oligomers dominated the process. However, the F23A mutation had a significant impact on the formation of well-ordered oligomers (discussed later).
Second-order kinetic rates
At the concentration represented in the simulations, the initial aggregations were almost irreversible association processes (Fig. 2). The reaction rates are estimated by the least-square fit of the concentration profiles to Eqs. 5.1–5.4 and are shown in Table 1. These rates provide a qualitative measure on the kinetics of the initial aggregation. 1), The initial aggregations were at least second-order processes. 2), Although the main pathway to the formation of octamers was through the addition of one subunit at a step, the other pathway was also possible in which two A(2) merged together. Both pathways were also observed in the study by Jang et al. (2004). 3), There was no significant difference in the initial aggregation kinetics between the peptide NFGAIL and the alanine mutant and the main products were disordered aggregates in both cases. The main difference between the wild type and the alanine mutant was the formation of the ordered aggregates, which will be discussed later. The configurations of the subunits did not affect the kinetics of the initial aggregation for the native peptide. However, for the alanine mutant, the parallel configuration of the subunit appeared to favor K12, whereas the antiparallel configuration appeared to favor K22.
TABLE 1.
Aggregation rates of the restrained subunits at 278 K
| Subumit | k11* | RMS error | k12 | RMS error | k13 | RMS error | k22 | RMS error |
|---|---|---|---|---|---|---|---|---|
| NFGAIL para dimer | 0.0065 | 35% | 0.0068 | 54% | 0.0452 | 80% | 0.0071 | 11% |
| NFGAIL antipara dimer | 0.0029 | 22% | 0.0068 | 53% | 0.0130 | 38% | 0.0077 | 18% |
| NAGAIL paral. dimer | 0.0022 | 23% | 0.0231 | 65% | 0.0111 | 42% | 0.0041 | 22% |
| NAGAIL anitpara dimer | 0.0039 | 17% | 0.0047 | 26% | 0.0035 | 28% | 0.0108 | 15% |
Rate unit (L mmol−1 ns−1).
Formation of well-ordered oligomers
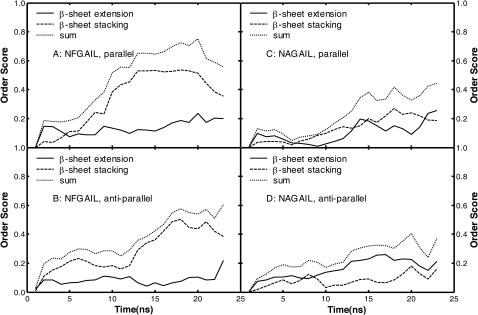
The order score for a pair of associated subunits is calculated based on the formula discussed in the Method section. The overall order score of an oligomer is the summation of the order scores of the distinct subunit pair in the oligomer. Formation of ordered oligomers by the four types of subunits, including NFGAIL parallel and antiparallel dimers (Fig. 3, A and B, respectively), and NAGAIL parallel and antiparallel dimers (Fig. 3, C and D, respectively), is characterized by the association order score. It is clear that the single F23A mutation had a significant impact on the ability of the mutant to pack β-sheets in an ordered way (Fig. 3, A–D, dashed lines). The wild-type NFGAIL demonstrated notably higher β-sheet stacking score than the mutant NAGAIL whereas their β-sheet extension scores were similar, indicating that the wild-type peptide had greater ability to produce ordered association by β-sheet stacking. Evidently, the Phe side chain played a critical role in directing the stacking of β-sheets. β-Sheet stacking contributed more to the order score in the formation of well-ordered oligomers of both sequences than that by β-sheet extension (Fig. 3, dashed lines versus solid lines), suggesting subunits were associated more likely by β-sheet stacking rather than by β-sheet extension.
FIGURE 3.
The association order scores. (A) NFGAIL parallel dimer; (B) NFGAIL antiparallel dimer; (C) NAGAIL parallel dimer; (D) NAGAIL antiparallel dimer.
There was no significant difference on the formation of the ordered oligomers by either parallel subunits or antiparallel subunits of both peptides (Fig. 3, A versus B, C versus D). Therefore, there was no clear preference on the orientation of the β-strands and both antiparallel and parallel β-sheets were equally likely to form. All four types of subunits (Fig. 3, A–D, solid lines) had similar patterns of β-sheet extension, implying that F23A mutation did not change the ability to form cross-strand main-chain hydrogen bonds, which is anticipated because the mutation has small effect on the hydrogen-bonding ability of the main chain. In summary, the F23A mutation strongly influenced the stacking interaction of the β-sheets and the wild type can form stacked β-sheet complex much more easily than the mutant, which is the precursor of the amyloid fibrils. This is consistent with the experimental studies (Azriel and Gazit, 2001) that the peptide NAGAIL formed amorphous aggregates rather than amyloid fibrils and the intersheet hydrophobic interactions are responsible for amyloid fibril formation and stability (Fraser et al., 1991a,b; Jang et al., 2004; Kowalewski and Holtzman, 1999).
Although 80% of the peptides were associated into octamers, only ∼8% of the NFGAIL octamers could be considered as well ordered based on their order scores (1.5 or higher, averaged over the two types of configurations). In comparison, only ∼2% of the octamers formed by the F23A mutant were well ordered. Thus the wild-type peptide increased the population of the ordered octamers by four times in comparison with its mutant. Interestingly, all well-ordered octamers were multilayer β-sheets and no single-layer octamers existed for either sequence. The fractions of the two, three, and four-layered structures formed by the NFGAIL were 4, 72, and 24%, respectively, whereas those formed by the F23A mutant were 78, 20, and 2%, respectively. Clearly, the wild-type peptide has notably higher tendency to form multilayered ordered structure than the F23A mutant. This, again, demonstrated that the Phe played a critical role to direct β-sheets to form orderly stacked multilayer oligomers and intersheet interaction played an important role in the formation of ordered β-sheet complex.
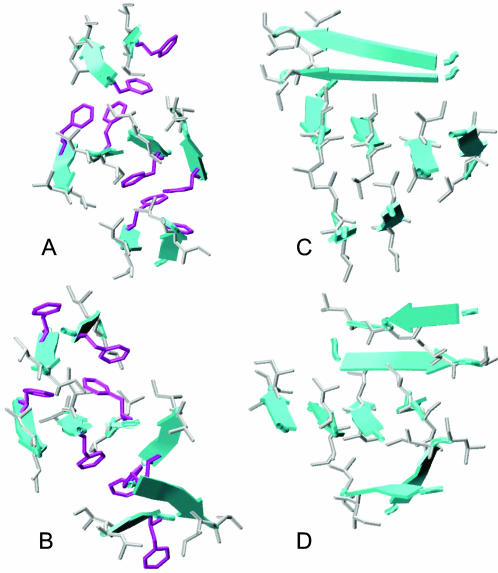
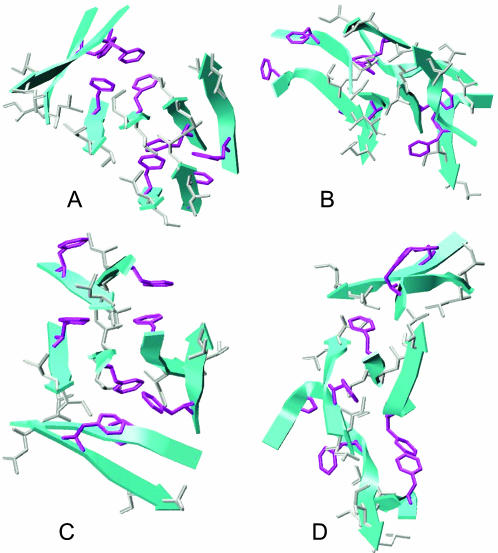
Fig. 4 shows the representative structures of well-ordered octamers. They were all three-layer compact structures where two-strand β-sheets were packed on both sides of the central four-strand β-sheet. The central layer was well formed by an edge-to-edge association of two dimers. The stacking of the dimers on the central layer was not well aligned in the F23A mutant. Consequently, it would be difficult for the misaligned three-layer β-sheet of the F23A mutant to grow into amyloid fibrils (Fig. 4, C and D).
FIGURE 4.
Representative structures of the ordered aggregates. (A) NFGAIL parallel dimer; (B) NFGAIL antiparallel dimer; (C) NAGAIL parallel dimer; (D) NAGAIL antiparallel dimer. Only the heavy atoms of the hydrophobic side chains are shown for clarity (Phe in purple; Leu and Ile in silver).
The structural characteristics of the ordered octamers were tabulated in Table 2. Because no restraints were imposed between the subunits, the spacing between the two central β-strands of a four-strand β-sheet reflected the true hydrogen-bonding spacing. The interstrand distance by the peptide NFGAIL was ∼4.5–4.7 Å, agreed well with the 4.60 and 4.83 Å obtained from x-ray fiber diffraction experiments (Sunde et al., 1997). The cross-layer intersheet distance of the peptide NFGAIL varied somewhat, depending upon the alignment of the peptides. It was 9.2 ± 1.1 Å when two parallel β-sheets stacked against each other in parallel, 8.8 ± 1.1 Å when two parallel β-sheets stacked against each other in antiparallel. This changed to 7.8 ± 0.6 Å when two antiparallel β-sheets stacked against each other in parallel and 9.8 ± 1.1 Å when two antiparallel β-sheets stacked against each other in antiparallel. These interlayer distances agreed well with the two distances, 8.60 and 11.4 Å (Sunde et al., 1997), observed in the fiber diffraction data of the decapeptide SNNFGAILSS. In contrast, the intersheet distances of the peptide NAGAIL were smaller: 6.7 ± 0.6 Å, 8.3 ± 1.0 Å, 7.0 ± 1.6 Å and 6.6 ± 1.2 Å, respectively. The smaller intersheet distances were the direct results of the F23A mutation, where the bulky side chain of Phe was replaced by a smaller side chain of Ala. The fractions of the well-ordered subunit pairs over all subunit pairs in the last 2 ns were also tabulated in Table 2.
TABLE 2.
The abundance and structure properties of the well-ordered associated subunit pairs (ASP)
| NFGAIL
| ||||||||
|---|---|---|---|---|---|---|---|---|
| NFGAIL parallel dimer
|
NFGAIL anti-parallel dimer
|
|||||||
| Subunit Associated mode
|
β-sheet extension
|
β-sheet stacking
|
β-sheet extension
|
β-sheet stacking
|
||||
| Parallel | Antiparallel | Parallel | Antiparallel | Parallel | Antiparallel | Parallel | Antiparallel | |
| No. of ASP* (full 23 ns) | 2520 | 3477 | 7081 | 1743 | 2902 | 118 | 2915 | 1477 |
| Interstrand (Å) | 4.5 ± 0.1 | 4.5 ± 0.1 | – | – | 4.6 ± 0.2 | 4.7 ± 0.3 | – | – |
| Intersheet (Å) | – | – | 9.2 ± 1.1 | 8.8 ± 1.1 | – | – | 7.8 ± 0.6 | 9.8 ± 1.1 |
| Fraction (last 2 ns)† | 0.06 | 0.05 | 0.09 | 0.03 | 0.05 | 0.02 | 0.07 | 0.06 |
| NAGAIL
| ||||||||
| NAGAIL parallel dimer
|
NAGAIL antiparallel dimer
|
|||||||
| SubunitAssociated mode
|
β-sheet extension
|
β-sheet stacking
|
β-sheet extension
|
β-sheet stacking
|
||||
| Parallel
|
Antiparallel
|
Parallel
|
Antiparallel
|
Parallel
|
Antiparallel
|
Parallel
|
Antiparallel
|
|
| No. of ASP (full 23 ns) | 3669 | 930 | 2538 | 1009 | 2698 | 3409 | 1025 | 560 |
| Interstrand (Å) | 4.6 ± 0.2 | 4.6 ± 0.2 | – | – | 4.5 ± 0.1 | 4.6 ± 0.1 | – | – |
| Intersheet (Å) | – | – | 6.7 ± 0.6 | 8.3 ± 1.0 | – | – | 7.0 ± 1.6 | 6.6 ± 1.2 |
| Fraction (last 2 ns) | 0.11 | 0.02 | 0.04 | 0.04 | 0.04 | 0.08 | 0.02 | 0.01 |
The number of observed specific type of associated subunit pairs (ASP) in the simulations.
The fraction of the well-ordered subunit pairs over all subunit pairs (summation of all possible subunit pairs provided by every oligomer) A dimer, a tetramer, a heptamer or an octamer provides 0, 1, 2, and 3 subunit pairs, respectively.
Because the central two strands of the observed four-strand β-sheets were formed without the assistance of restraining force, the relative orientation of the two strands can be indicative to the preference of the orientation. However, in our simulation, the central two strands showed no clear preference. They were in parallel in ∼6% of the time and in antiparallel in ∼4% of the time. Interestingly, the orientation of cross-layer stacking showed preference for the NFGAIL parallel subunits; ∼9% NFGAIL parallel subunits formed parallel double-layer β-sheets and only 3% formed antiparallel double-layer β-sheets. In comparison, ∼7% of the antiparallel subunits formed parallel double-layered β-sheets and 6% formed antiparallel double-layered β-sheets. All four configurations matched the general cross-β-structure, which is the hallmark of amyloid fibrils.
β-Sheet extensions
Formation of the β-sheets was facilitated by both the cross-strand main-chain hydrogen bonds and by side-chain interactions. The standard deviation of the interstrand distance along the hydrogen-bond direction for those unrestrained pairs was quite small (<0.3 Å in Table 2) suggesting strong hydrogen bonds. In addition, the side-chain packing favored the β-sheet extension by hydrophobic interaction. The hydrogen bonds were also stabilized by the side chains that protect the hydrogen bonds from solvent attack. The side-chain packing between the two central β-strands was characterized by atom contacts that are shown in Table 3. The mean atom contact between the two central β-strands of the peptide NFGAIL was 129 (when the two central strands formed a two-strand parallel β-sheet), and 103 (when the two central strands formed a two-strand antiparallel β-sheet). Intuitively, one may attempt to use this similarity to explain the comparable tendency of the peptide to form either parallel β-sheets (6% averaged over two types of subunits) or antiparallel β-sheets (4% averaged over two types of subunits). However, such explanation was not supported by the simulations on the mutant. Although the mean atom contacts between the two central β-strands of the F23A mutant was 106 and 40 for, respectively, the parallel and antiparallel β-sheets, the tendency to form either parallel or antiparallel β-sheets was similar (8% for parallel and 5% for antiparallel, averaged over both types of subunits). Nevertheless, both peptides had similar overall tendency to extend along the main-chain hydrogen-bond direction. The percentage of β-sheet extension associations for the wild type was 9%, whereas that of the mutant was 12% (Table 2). Therefore, the F23A mutation did not reduce the capability of the peptide to form cross-strand hydrogen bonds and β-sheet extension.
TABLE 3.
Atom contacts between the two central strands of a four-strand β-sheet
| Subunit Associated mode
|
NFGAIL dimer β-sheet extension
|
Subunit Associated mode
|
NAGAIL dimer β-sheet extension
|
||
|---|---|---|---|---|---|
| Parallel | Antiparallel | Parallel | Antiparallel | ||
| Phe-Phe | 7 | 7 | Ala-Ala† | 2 | 0 |
| With Phe | 49 | 70 | With Ala* | 10 | 18 |
| strand-strand | 129 | 103 | strand-strand | 106 | 40 |
The atom contacts with at least one sidechain atom from Phe/Ala.
Ala in F25A.
β-Sheet stacking
Side-chain stacking plays an important role in the formation of multilayer β-sheets. Interestingly, the standard deviation of the intersheet spacing was much larger (<1.1 Å in Table 2) compared with that of the interstrand spacing along the hydrogen-bond direction (<0.31 Å in Table 2). This is because the side-chain interactions were mainly hydrophobic interactions and the local energy minima are wider than that of the hydrogen bonds. The side-chain atom contacts between the stacked β-sheets are shown in Table 4. The mean side-chain atom contacts of the parallel NFGAIL dimer subunits were 295 and 188 for parallel and antiparallel stacking, respectively. Those of antiparallel NFGAIL dimer subunits were 301 when the dimers were stacked in parallel and 244 when they were stacked antiparallel. Obviously, these were at similar levels that further demonstrated the lack of preference over either parallel or antiparallel orientation. In the case of F23A mutant, these contacts were 232, 199, 251, and 139 for, respectively, parallel dimers stacked in parallel, parallel dimers stacked in antiparallel, antiparallel dimers stacked in parallel, and antiparallel dimers stacked in antiparallel. The reduction in the number of atom contacts, combined with the fact that the Phe side chain is considerably more hydrophobic than Ala side chain, made the mutant less likely to form stable stacked β-sheets. The mean atom contacts between Phe side chains were 43, about five times the contacts between Ala side chains; Ala side chains only had an average of 9 atom contacts. The mean atom contacts between Phe and other residues were 99 and that between Ala and other residues were 66. Overall, the F23A mutant had 77 fewer contacts than the wild type. Clearly, the interactions with the aromatic Phe side chains were the key factors to direct the ordered stacking between the β-sheets. This might be the key reason why a single F23A mutation prohibited the mutant NAGAIL to form amyloid fibrils.
TABLE 4.
Atom contacts in the two-layered β-sheets (two stacked subunits)
| NFGAIL
| ||||
|---|---|---|---|---|
| Subunit Associated mode
|
NFGAIL parallel dimer β-sheet stacking
|
NFGAIL antiparallel dimer β-sheet stacking
|
||
| Parallel | Antiparallel | Parallel | Antiparallel | |
| Phe-Phe | 39 | 42 | 86 | 4 |
| With Phe* | 73 | 97 | 70 | 155 |
| dimer-dimer | 295 | 188 | 301 | 244 |
| NAGAIL
| ||||
| Subunit Associated mode
|
NAGAIL parallel dimer β-sheet stacking
|
NAGAIL antiparallel dimer β-sheet stacking
|
||
| Parallel
|
Antiparallel
|
Parallel
|
Antiparallel
|
|
| Ala-Ala† | 0 | 10 | 25 | 0 |
| With Ala* | 79 | 33 | 34 | 115 |
| dimer-dimer | 232 | 199 | 251 | 139 |
Atom contact with only one sidechain atom from Phe/Ala.
Ala in F25A.
Stability of the well-ordered oligomer of the peptide NFGAIL
To test the stability of the well-ordered oligomers formed in the previous simulations (Fig. 4, A and B), two additional simulations for each oligomer were conducted for 22.0 ns at 320 K in which the main-chain torsion angles were restrained to the extended conformation (Φ ≈ −110°, Ψ ≈ 135°) in one simulation and no restraints were imposed in the other. The last snapshots at 22.0 ns of the four simulations were shown in Fig. 5. There were no significant structural changes in the two simulations with the restraints on backbone conformation. The cross-strand main-chain hydrogen bonds were well maintained, as were the β-sheets. The only notable changes were the minor translation and rotation of the two double-strand β-sheets (Fig. 5, A (top layer) and C (bottom layer)). In the unrestrained simulations, despite the loss of some cross-strand hydrogen bonds and partial unfolding of the β-sheets, most residues were still clustered together and maintained the extended conformations. The overall structures appeared to be reasonably stable with or without the restraints.
FIGURE 5.
Last snapshots at 22 ns of the dissociation simulations of the ordered aggregates of NFGAIL parallel dimer (A and B) and NFGAIL antiparallel dimer (C and D). Only the side chains of Phe, Ile, and Leu are shown for clarity. The main-chain torsion angles were restrained to the extended conformations in two simulations (A and C) and there were no restraints in the other two simulations (B and D).
DISCUSSION
A growing body of evidence (Canet et al., 2002; Dima and Thirumalai, 2002; Zurdo et al., 2001) indicates that the amyloid fibril precursors are more extended than the native-like partially folded states. Conformations derived from high temperature and acidic urea denaturation seem to be biased toward the extended main-chain conformations (Brant and Flory, 1965; Fandrich et al., 2003; Schwarzinger et al., 2002; Shi et al., 2002; Smith et al., 1996), which are more amyloidogenic. In this study, we enhanced the formation of the ordered oligomers by restraining the main-chain conformation so that the assembly process of a highly ordered β-sheet complex could be probed within the current computing power. Further enhancement was made by using two-strand β-sheets as subunits in this study. This allowed us to study the roles of intersheet stacking including hydrophobic interactions and β-sheet extension through hydrogen bonding in the formation of the highly ordered β-sheet complex. Previous studies (Fraser et al., 1991a,b; Jang et al., 2004; Kowalewski and Holtzman, 1999) showed that the intersheet hydrophobic interactions are responsible for amyloid fibril formation and stability. A delicate balance between the two types of interactions was crucial in the formation of the highly ordered β-sheet complex (Jang et al., 2004; Wu et al., 2004). Too strong of each type of interactions would lead to amorphous or globular aggregates. This is consistent with our results: among the well-ordered subunit pairs of the native peptide, about half was formed by β-sheet extension, whereas the other half was formed by β-sheet stacking (Table 2).
Based on experiments, Gazit recently suggested that the stacking of aromatic residues might play a key role in the formation of amyloid fibrils (Gazit, 2002). Our simulations showed that the role the Phe played in the formation of the well-ordered oligomers was to direct ordered stacking of β-sheets through both the specific interactions between the aromatic side chains and the nonspecific packing of the Phe with other hydrophobic residues. In contrast, the cross-strand main-chain hydrogen bonds were mainly responsible for β-sheet extension.
Nagarajaram and co-workers have analyzed the interstrand packing distance between two stacking β-sheets in globular proteins (Nagarajaram et al., 1999). They found that when Ala and Gly are at the packing interface of two β-strands belonging to two stacking β-sheets, they generally give rise to structural distortions of the β-strands. In comparison, when Phe, Leu, and Ile are at the packing interface, the β-strands are perfectly aligned and have a greater intersheet distance. Our simulation results agree very well with these observations. The F23A mutation caused the intersheet stacking of the mutant NAGAIL to be somewhat disordered and reduced the intersheet distance from an average of 8.9 Å for NFGAIL to an average of 7.2 Å for NAGAIL (Table 2). Again, the Phe played a critical role in the β-sheet stacking.
Although all amyloid fibrils share the same cross-β-structure, the detailed arrangement of β-strands can vary a great deal as shown in our simulations. Of particular interest was the lack of clear preference toward either parallel or antiparallel orientation in our simulations. This agreed with the experimental observation that short (15 or fewer residues) peptides can form both parallel and antiparallel β-sheets in amyloid fibrils (Petkova et al., 2004).
Furthermore, the association process was mainly driven by hydrophobic interactions, therefore its main products were disordered aggregates (Table 2) even if the strands had been restrained to a perfect β-extended conformation. Thus, this study further supported our proposal that dissociation of the disordered aggregates could be the limiting step of the formation of ordered oligomers (Wu et al., 2004).
A possible application of our studies might be to prevent the amyloid seed formation by inhibiting β-sheet stacking with an aromatic inhibitor. Porat and co-workers (Porat et al., 2004) have designed aromatic inhibitors to interact with Phe to prevent amyloid formation. They found that phenol red with three carbon rings could strongly inhibit human IAPP fibril formation and could rescue the β-cells from the cytotoxic effect of the external addition of the hIAPP. This suggests that Phe plays an essential role in the formation of ordered oligomers, which is consistent with our observation. Based on our simulation results, we propose that the role of phenol red is to inhibit β-sheet stacking rather than β-sheet extension.
CONCLUSION
We found that a critical role played by Phe on the formation of well-ordered oligomers was to direct the ordered stacking of β-sheets. This could be the key reason that a single F23A mutation prohibited the mutant NAGAIL from forming amyloid fibrils. We found that many molecular arrangements of the amyloidogenic peptide satisfied the “cross-β” structure, which is the hallmark of amyloid fibrils. The tendency for the peptide to form either parallel or antiparallel β-sheets was comparable, as was the tendency for β-sheets to stack either in parallel or antiparallel orientation. Overall, ∼85% of the native peptide formed octamers, where only 8% of the octamers were well ordered and the others were disordered. This further supported our previous proposal that the rate-limiting step of forming well-ordered oligomers is the dissociation of disordered oligomers (Wu et al., 2004). Among the well-ordered oligomers, triple-layer oligomers were predominant. We propose that these triple-layer oligomers could be candidates of the initial nucleus and may resemble soluble toxic oligomers.
Among the well-ordered subunit pairs, half of them were formed by β-sheet extension, whereas the others were formed by β-sheet stacking. The early disordered oligomers were mainly stabilized by cross-strand/cross-layer hydrophobic interactions, whereas well-ordered oligomers were further stabilized by the cross-strand hydrogen bonds and side-chain packing between the stacked β-sheets. Furthermore, the stability of the well-ordered octamers was validated by the simulations with less or no conformational restraints at a higher temperature.
Acknowledgments
We appreciate helpful comments from the reviewers.
This work has been supported by research grants from the National Institutes of Health (GM64458 and GM67168 to Y.D.).
References
- Azriel, R., and E. Gazit. 2001. Analysis of the minimal amyloid-forming fragment of the islet amyloid polypeptide: an experimental support for the key role of the phenylalanine residue in amyloid formation. J. Biol. Chem. 276:34156–34161. [DOI] [PubMed] [Google Scholar]
- Barash, D., L. J. Yang, X. L. Qian, and T. Schlick. 2003. Inherent speedup limitations in multiple time step/particle mesh Ewald algorithms. J. Comput. Chem. 24:77–88. [DOI] [PubMed] [Google Scholar]
- Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, A. DiNola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Comp. Phys. 81:3684–3690. [Google Scholar]
- Brant, D. A., and P. J. Flory. 1965. Configuration of random polypeptide chains. I. Experimental results. J. Am. Chem. Soc. 87:2788–2791. [Google Scholar]
- Canet, D., A. M. Last, P. Tito, M. Sunde, A. Spencer, D. B. Archer, C. Redfield, C. V. Robinson, and C. M. Dobson. 2002. Local cooperativity in the unfolding of an amyloidogenic variant of human lysozyme. Nat. Struct. Biol. 9:308–315. [DOI] [PubMed] [Google Scholar]
- Case, D. A., D. A. Pearlman, J. W. Caldwell, T. E. Cheatham III, W. S. Ross, C. L. Simmerling, T. A. Darden K. M. Merz, R. V. Stanton, A. L. Cheng, J. J. Vincent, M. Crowley, et al. 2002. AMBER 7. University of California, San Francisco, CA.
- Dima, R. I., and D. Thirumalai. 2002. Exploring protein aggregation and self-propagation using lattice models: phase diagram and kinetics. Protein Sci. 11:1036–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, F., N. V. Dokholyan, S. V. Buldyrev, H. E. Stanley, and E. I. Shakhnovich. 2002. Molecular dynamics simulation of the SH3 domain aggregation suggests a generic amyloidogenesis mechanism. J. Mol. Biol. 324:851–857. [DOI] [PubMed] [Google Scholar]
- Dobson, C. M. 1999. Protein misfolding, evolution and disease. Trends Biochem. Sci. 24:329–332. [DOI] [PubMed] [Google Scholar]
- Duan, Y., C. Wu, S. Chowdhury, M. C. Lee, G. Xiong, W. Zhang, R. Yang, P. Cieplak, R. Luo, T. Lee, J. Caldwell, J. Wang, and P. Kollman. 2003. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase QM calculations. J. Comput. Chem. 24:1999–2012. [DOI] [PubMed] [Google Scholar]
- Essmann, U., L. Perera, M. L. Berkowitz, T. A. Darden, H. Lee, and L. G. Pedersen. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103:8577–8593. [Google Scholar]
- Fandrich, M., V. Forge, K. Buder, M. Kittler, C. M. Dobson, and S. Diekmann. 2003. Myoglobin forms amyloid fibrils by association of unfolded polypeptide segments. Proc. Natl. Acad. Sci. USA. 100:15463–15468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, P. E., L. K. Duffy, M. B. Omalley, J. Nguyen, H. Inouye, and D. A. Kirschner. 1991a. Morphology and antibody recognition of synthetic beta-amyloid peptides. J. Neurosci. Res. 28:474–485. [DOI] [PubMed] [Google Scholar]
- Fraser, P. E., J. T. Nguyen, W. K. Surewicz, and D. A. Kirschner. 1991b. Ph-dependent structural transitions of Alzheimer amyloid peptides. Biophys. J. 60:1190–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazit, E. 2002. A possible role for pi-stacking in the self-assembly of amyloid fibrils. FASEB J. 16:77–83. [DOI] [PubMed] [Google Scholar]
- Gsponer, J., U. Haberthur, and A. Caflisch. 2003. The role of side-chain interactions in the early steps of aggregation: molecular dynamics simulations of an amyloid-forming peptide from the yeast prion Sup35. Proc. Natl. Acad. Sci. USA. 100:5154–5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppener, J. W. M., B. Ahren, and C. J. M. Lips. 2000. Islet amyloid and type 2 diabetes mellitus. N. Engl. J. Med. 343:411–419. [DOI] [PubMed] [Google Scholar]
- Jang, H. B., C. K. Hall, and Y. Q. Zhou. 2004. Assembly and kinetic folding pathways of a tetrameric beta-sheet complex: molecular dynamics simulations on simplified off-lattice protein models. Biophys. J. 86:31–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayed, R., E. Head, J. L. Thompson, T. M. McIntire, S. C. Milton, C. W. Cotman, and C. G. Glabe. 2003. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 300:486–489. [DOI] [PubMed] [Google Scholar]
- Kelly, J. W. 1998. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr. Opin. Struct. Biol. 8:101–106. [DOI] [PubMed] [Google Scholar]
- Klimov, D. K., and D. Thirumalai. 2003. Dissecting the assembly of A beta(16–22) amyloid peptides into antiparallel beta sheets. Structure. 11:295–307. [DOI] [PubMed] [Google Scholar]
- Kowalewski, T., and D. M. Holtzman. 1999. In situ atomic force microscopy study of Alzheimer's beta-amyloid peptide on different substrates: new insights into mechanism of beta-sheet formation. Proc. Natl. Acad. Sci. USA. 96:3688–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. P., T. A. Darden, L. Bartolotti, D. Kominos, and L. G. Pedersen. 1999. An atomic model for the pleated beta-sheet structure of A beta amyloid protofilaments. Biophys. J. 76:2871–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo, A., B. Razzaboni, G. C. Weir, and B. A. Yankner. 1994. Pancreatic-islet cell toxicity of amylin associated with type-2 diabetes mellitus. Nature. 368:756–760. [DOI] [PubMed] [Google Scholar]
- Mason, J. M., N. Kokkoni, K. Stott, and A. J. Doig. 2003. Design strategies for anti-amyloid agents. Curr. Opin. Struct. Biol. 13:526–532. [DOI] [PubMed] [Google Scholar]
- Nagarajaram, H. A., B. V. B. Reddy, and T. L. Blundell. 1999. Analysis and prediction of inter-strand packing distances between beta-sheets of globular proteins. Protein Eng. 12:1055–1062. [DOI] [PubMed] [Google Scholar]
- Nguyen, H. D., and C. K. Hall. 2004. Molecular dynamics simulations of spontaneous fibril formation by random-coil peptides. Proc. Natl. Acad. Sci. USA. 101:16180–16185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova, A. T., G. Buntkowsky, F. Dyda, R. D. Leapman, W. M. Yau, and R. Tycko. 2004. Solid state NMR reveals a pH-dependent antiparallel beta-sheet registry in fibrils formed by a beta-amyloid peptide. J. Mol. Biol. 335:247–260. [DOI] [PubMed] [Google Scholar]
- Porat, Y., S. Efrat, and E. Gazit. 2004. Aromatic inhibition of amyloid formation by the human islet amyloid polypeptide. Biophys. J. 86:339A–340A. [Google Scholar]
- Roberts, C. J. 2003. Kinetics of irreversible protein aggregation: analysis of extended Lumry-Eyring models and implications for predicting protein shelf life. J. Phys. Chem. B. 107:1194–1207. [Google Scholar]
- Rochet, J. C., and P. T. Lansbury. 2000. Amyloid fibrillogenesis: themes and variations. Curr. Opin. Struct. Biol. 10:60–68. [DOI] [PubMed] [Google Scholar]
- Ryckaert, J.-P., G. Ciccotti, and H. J. C. Berendsen. 1977. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comp. Phys. 23:327–341. [Google Scholar]
- Schwarzinger, S., P. E. Wright, and H. J. Dyson. 2002. Molecular hinges in protein folding: the urea-denatured state of apomyoglobin. Biochemistry. 41:12681–12686. [DOI] [PubMed] [Google Scholar]
- Shi, Z. S., C. A. Olson, G. D. Rose, R. L. Baldwin, and N. R. Kallenbach. 2002. Polyproline II structure in a sequence of seven alanine residues. Proc. Natl. Acad. Sci. USA. 99:9190–9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, L. J., K. A. Bolin, H. Schwalbe, M. W. MacArthur, J. M. Thornton, and C. M. Dobson. 1996. Analysis of main chain torsion angles in proteins: prediction of NMR coupling constants for native and random coil conformations. J. Mol. Biol. 255:494–506. [DOI] [PubMed] [Google Scholar]
- Sunde, M., L. C. Serpell, M. Bartlam, P. E. Fraser, M. B. Pepys, and C. C. F. Blake. 1997. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 273:729–739. [DOI] [PubMed] [Google Scholar]
- Tenidis, K., M. Waldner, J. Bernhagen, W. Fischle, M. Bergmann, M. Weber, M. L. Merkle, W. Voelter, H. Brunner, and A. Kapurniotu. 2000. Identification of a penta- and hexapeptide of islet amyloid polypeptide (IAPP) with amyloidogenic and cytotoxic properties. J. Mol. Biol. 295:1055–1071. [DOI] [PubMed] [Google Scholar]
- Thirumalai, D., D. K. Klimov, and R. I. Dima. 2003. Emerging ideas on the molecular basis of protein and peptide aggregation. Curr. Opin. Struct. Biol. 13:146–159. [DOI] [PubMed] [Google Scholar]
- Westermark, P., C. Wernstedt, T. D. Obrien, D. W. Hayden, and K. H. Johnson. 1987. Islet amyloid in type-2 human diabetes mellitus and adult diabetic cats contains a novel putative polypeptide hormone. Am. J. Pathol. 127:414–417. [PMC free article] [PubMed] [Google Scholar]
- Wu, C., H. Lei, and Y. Duan. 2004. Formation of partially-ordered oligomers of amyloidogenic hexapeptide (NFGAIL) in aqueous solution observed in molecular dynamics simulations. Biophys. J. 87:3000–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanuy, D., B. Y. Ma, and R. Nussinov. 2003. Short peptide amyloid organization: stabilities and conformations of the islet amyloid peptide NFGAIL. Biophys. J. 84:1884–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanuy, D., and R. Nussinov. 2003. The sequence dependence of fiber organization. A comparative molecular dynamics study of the islet amyloid polypeptide segments 22–27 and 22–29. J. Mol. Biol. 329:565–584. [DOI] [PubMed] [Google Scholar]
- Zurdo, J., J. I. Guijarro, J. L. Jimenez, H. R. Saibil, and C. M. Dobson. 2001. Dependence on solution conditions of aggregation and amyloid formation by an SH3 domain. J. Mol. Biol. 311:325–340. [DOI] [PubMed] [Google Scholar]