Abstract
Overexpression of the transmembrane receptor tyrosine kinase ErbB2 is common in multiple malignancies, including breast and ovarian cancer. ErbB2 is resistant to degradation mediated by c-Cbl, the E3 ubiquitin ligase responsible for ligand-induced ubiquitination of ErbB1 (epidermal growth factor receptor). Because of its resistance to degradation, ErbB2 is the preferred dimerization partner for other members of the ErbB family, and its overexpression in vivo is associated with poor prognosis. We now show that the chaperone-binding ubiquitin ligase CHIP efficiently ubiquitinates and down-regulates ErbB2. CHIP expression shortens the half-life of both nascent and mature ErbB2 protein. In vitro ubiquitination assay shows that CHIP serves as a ubiquitin ligase for ErbB2, and both exogenously expressed and endogenous CHIP coprecipitate with the kinase. Furthermore, CHIP association with ErbB2 requires a chaperone intermediate and is increased by the chaperone-binding drug geldanamycin, a potent stimulator of ErbB2 ubiquitination and degradation. These data describe a previously unrecognized pathway, amenable to pharmacologic manipulation, that mediates ErbB2 stability.
ErbB2 is a transmembrane receptor tyrosine kinase that heterodimerizes with other members of the ErbB family and promotes the transduction of proliferative and survival signals (1). ErbB2 is overexpressed in a significant proportion of adenocarcinomas and clinical studies have demonstrated that elevated ErbB2 expression correlates with poor prognosis in multiple malignancies, including breast and ovarian cancer (2, 3). The kinase has therefore been identified as a valuable molecular target for the treatment of these cancers (4–6).
Epidermal growth factor binding to ErbB1 homodimers stimulates receptor down-regulation, and this binding depends on recruitment of the E3 ubiquitin ligase c-Cbl to the phosphorylated receptors, followed by Cbl-mediated ErbB1 ubiquitination and degradation (7–10). In contrast, although certain tumor-inhibitory ErbB2 antibodies, such as Herceptin, enhance recruitment of c-Cbl to ErbB2 and accelerate ErbB2 internalization and degradation (11), in the absence of such antibodies phosphorylated ErbB2 only weakly associates with c-Cbl and thus is resistant to c-Cbl-induced down-regulation (1). Indeed, ErbB2 heterodimerization with ErbB1 antagonizes ErbB1/c-Cbl association and promotes receptor longevity and recycling to the cell surface (12). For this reason, and because point mutations that constitutively activate ErbB2 kinase activity are rarely found in ErbB2-overexpressing tumors (13), inhibition of ErbB2 kinase activity per se might be expected to prove less beneficial than approaches that focus on down-regulating the receptor. Thus, identification of novel means to regulate ErbB2 stability should provide additional opportunities for successfully interdicting signaling through ErbB2-containing receptor complexes.
We recently reported that stability of mature ErbB2 requires association of the kinase with the molecular chaperone Hsp90 (14). The Hsp90-binding drug geldanamycin (GA) rapidly destabilizes ErbB2 secondary to disruption of Hsp90/ErbB2 association and concomitant with stimulation of Hsp/Hsc70 association with the kinase (14). GA-induced destabilization of ErbB2 is preceded by its stimulation of ErbB2 ubiquitination, and drug effects can be at least partially blocked by proteasome inhibition (15). Recently, Ballinger and coworkers (16, 17) described a chaperone-interacting protein (CHIP) that contains an amino-terminal tetratricopeptide (TPR) domain and a carboxyl-terminal U box domain. CHIP binds to the chaperones Hsp/Hsc70 and Hsp90 by means of its TPR motif, while also displaying E3 ubiquitin ligase activity mediated by its U box domain. Indeed, CHIP is a member of what is now recognized to be a family of E3 proteins, distinct from those ubiquitin ligases containing either HECT (homologous to E6-AP carboxyl terminus) or RING finger domains (18, 19).
CHIP has been shown to induce the ubiquitination and proteasome-mediated degradation of the glucocorticoid receptor and the cystic fibrosis transmembrane-conductance regulator, which, like ErbB2, are Hsp90 client proteins (17, 20). Both CHIP and GA also promote a similar remodeling of Hsp90-containing multichaperone complexes to release the cochaperone p23, whose association with Hsp90 favors stabilization of client proteins (17, 21, 22). For these reasons, we have investigated the possibility that CHIP may normally regulate ErbB2 stability and/or may be recruited to the ErbB2/chaperone complex by GA, thus explaining how this Hsp90 inhibitor promotes ubiquitination and degradation of ErbB2. Indeed, our results show that CHIP is associated with ErbB2 protein in cells. This association is most likely mediated by a chaperone intermediate, and more importantly, it is enhanced by GA treatment. Lastly, CHIP induces ErbB2 ubiquitination in vitro, and its over-expression efficiently down-regulates ErbB2 protein in vivo.
Materials and Methods
Cell Culture and Transfection.
COS7 cells (ATCC) were maintained at 37°C and 5% CO2 in DMEM containing 1 mM sodium pyruvate and 10% FBS. Cells were transfected with various expression plasmids by using the FuGene reagent (Roche Molecular Biochemicals), according to the manufacturer's instructions. CHIP−/− primary fibroblast cells were derived from CHIP knockout mice, and maintained in DMEM + 10% FBS. Transfection of CHIP−/− cells was accomplished by using an engineered adenovirus containing wild-type CHIP gene (Ad-CHIP). SKOV3 ovarian cancer cells were kindly provided by M. Birrer (National Cancer Institute), and they were maintained in RPMI medium 1640 containing 10% FBS. The proteasome inhibitor N-acetyl-l-leucyl-l-leucyl-norleucinal (ALLnL) was purchased from Sigma. GA was obtained from the Drug Synthesis and Chemistry Branch, National Cancer Institute.
Preparation of Expression Constructs.
ErbB2 constructs have been described (14). c-Cbl constructs are kind gifts from S. Lipkowitz (National Cancer Institute) (9). CHIP point mutants, K30A and H260Q, were created with the GeneEditor in vitro site-directed mutagenesis system (Promega). Histidine-tagged CHIP was made by inserting the whole coding region into the pcDNA3.1/myc-His vector (Invitrogen). The coding region was made by PCR with the upper primer GGCGAAGCTTCGAGGCGCGGAGCTTGGGAG and the lower primer GGGCTCGAGGTAGTCCTCCACCCAGCCATTCT.
Immunoprecipitation and Western Blotting.
Western blotting techniques and ErbB2 immunoprecipitation have been described (14). To precipitate His-tagged CHIP proteins, transfected COS7 cells were lysed with PBS containing 1% Nonidet P-40, and Talon beads (CLONTECH) were added to cleared cell lysate. The mixture was rotated at 4°C for 2 h, pelleted beads were washed four times with lysis buffer, and precipitated proteins were dissolved in SDS sample buffer. Proteins were resolved by SDS/PAGE, transferred to poly(vinylidene difluoride) membrane, and probed with respective antibodies. For ubiquitin blot, proteins were transferred to nitrocellulose membrane, autoclaved (121°C, 30 min), and then probed with rabbit anti-ubiquitin polyclonal antibody (Sigma). Anti-ErbB2 (NeoMarker Ab-3 for extracellular domain, Oncogene Ab-3 for intracellular domain), anti-Hsp90 (StressGen Biotechnologies, Victoria, Canada, SPA-835), anti-Hsc70 (Santa Cruz Biotechnology, K19), anti-Hsp70 (Santa Cruz Biotechnology, K20), and anti-β-tubulin (Oncogene, Ab-1) were used for immunoblotting.
Pulse–Chase Analysis.
COS7 cells in 10-cm dishes were transfected with 5 μg of pcDNA3-ErbB2 plasmid DNA. Four hours later, half of the dishes received Ad-CHIP at a multiplicity of infection of 100, and rocked at 6 rpm for another 4 h. Twenty-four hours after ErbB2 transfection, cells were labeled with [35S]methionine for 45 min, washed, and chased for 0, 1, 2, 4, and 6 h, as described (23). To precipitate ErbB2 protein, 0.5 mg of cell lysate was mixed with Ab-2 and Ab-3 antibodies (Oncogene) and incubated at 4°C for 2 h, followed by addition of protein G-agarose beads (Invitrogen) to pellet the immune complex. Immunoprecipitates were separated by SDS/PAGE, and dried gels were autoradiographed as described (23).
In Vitro Ubiquitination Assay.
The sequence encoding the intracellular domain of ErbB2 was amplified by PCR and inserted into pET-32a vector (Novagen). ErbB2 protein was translated in vitro by using the TNT T7 Quick Coupled Transcription/Translation System (Promega), and then immunoprecipitated with rabbit anti-ErbB2 antibody (C-18, Santa Cruz Biotechnology). In vitro ubiquitination assay was performed according to Jiang et al. (19), with UbcH5a (Affiniti, Exeter, U.K.) as the E2 ubiquitin conjugating enzyme. Protein samples were separated by SDS/PAGE, transferred to nitrocellulose, and blotted with mouse anti-ErbB2 antibody (Ab-3, Oncogene).
Results
Overexpression of CHIP Down-Regulates ErbB2 Proteins.
We first determined whether ectopic expression of CHIP would affect the stability of ErbB2. To this end, we monitored ErbB2 protein level by Western blotting in COS7 cells cotransfected with plasmids expressing CHIP and ErbB2 genes. As is shown in Fig. 1A, coexpression of CHIP decreased the expression of wild-type ErbB2.
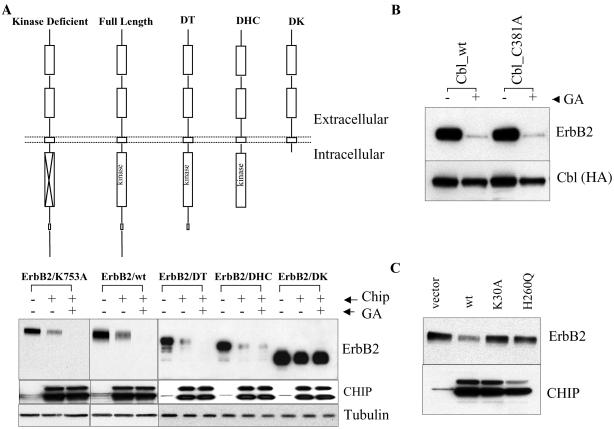
Figure 1.
The E3 ubiquitin ligase CHIP down-regulates ectopically expressed ErbB2 protein in COS7 cells. Cotransfected COS7 cells in six-well plates were lysed with TMNS buffer (50 mM Tris, pH 7.5/150 mM NaCl/20 mM Na2MoO4/0.09% Nonidet P-40). Cell lysate was separated by SDS/PAGE and probed with antibodies against ErbB2 (L87, Neomarker), hemagglutinin epitope (HA)-tagged c-Cbl (mouse anti-HA), tubulin (mouse monoclonal; Oncogene), or CHIP (rabbit polyclonal). (A) Cells were transfected with different ErbB2 constructs (Upper), with or without cotransfection of wild-type CHIP. One set of cells was treated with 1 μM GA for 6 h before lysis. (B) Cells were transfected with ErbB2 and c-Cbl constructs. GA treatment was as in A. (C) Cells were transfected with ErbB2, together with different constructs of CHIP or vector alone.
To determine the region in the cytoplasmic portion of ErbB2 that confers CHIP sensitivity, we examined the sensitivity of several carboxyl-terminal ErbB2 truncation mutants to coexpressed CHIP. Although removal of ErbB2's hydrophilic tail [necessary for Herceptin-induced c-Cbl interaction (11)] had no effect, deletion of ErbB2's kinase domain completely abrogated its sensitivity to CHIP (Fig. 1A). The sensitivity (or lack thereof) of the various ErbB2 deletion mutants to CHIP exactly parallels their described sensitivity to GA (14). Moreover, when added to CHIP transfectants in the current experiment, GA accentuated the loss of ErbB2 (Fig. 1A). In contrast to c-Cbl, which only down-regulates enzymatically active ErbB2, CHIP efficiently down-regulated ErbB2/K753A, a mutated receptor lacking kinase activity (14). Finally, neither wild-type c-Cbl nor the dominant-negative c-Cbl point mutant, Cbl(C381A), which lacks E3 activity, were able to affect positively or negatively the ability of GA to rapidly down-regulate ErbB2 (Fig. 1B). Taken together, these findings rule out a contributory role for c-Cbl E3 ligase activity in CHIP-mediated ErbB2 down-regulation.
An Intact CHIP Is Required for the Down-Regulation of ErbB2 Protein.
The CHIP protein contains an amino-terminal TPR domain, which binds to molecular chaperones, and a carboxyl-terminal U box domain, which is involved in its ubiquitination function. If CHIP's ability to down-regulate ErbB2 requires association with a chaperone intermediate, then disruption of CHIP's TPR domain should abrogate its effectiveness. Likewise, if CHIP were directly responsible for ErbB2 ubiquitination, then disruption of its U box domain should similarly abolish its activity toward ErbB2. To test these hypotheses, we made point mutations in both the TPR and U box domains of CHIP and examined the activity of these mutants toward ErbB2. CHIP(H260Q), a U box mutant, fails to bind its cognate E2, thus losing its E3 activity (18), whereas CHIP(K30A), a TPR domain mutant, does not bind to either Hsp90 or Hsp/Hsc70 (see below and unpublished data). Neither point mutant was able to down-regulate ErbB2 (Fig. 1C), demonstrating that both domains are indispensable for CHIP activity toward the kinase and thus implicating a chaperone requirement in this process.
CHIP Is Associated with ErbB2 Proteins in Cells.
We next examined whether CHIP associates in vivo with ErbB2. This association was first tested by coprecipitation experiments in which we immunoprecipitated ErbB2 from COS7 cells cotransfected with various ErbB2 constructs and with wild-type CHIP, and looked for the presence of CHIP in the immunopellets. We found that CHIP was indeed a component of the immunocomplex precipitated by ErbB2 antibody (Fig. 2A). Moreover, the CHIP-binding profile correlated perfectly with the pattern of ErbB2 sensitivity to CHIP (see Fig. 1A).
Figure 2.
CHIP interacts with ErbB2 in vivo by a chaperone intermediate. (A) CHIP specifically associates with ErbB2 proteins. COS7 cells in 150 × 25 mm plates were transfected with CHIP, plus full-length or truncated ErbB2. Twenty-four hours after transfection, cells were lysed with TMNS buffer. For immunoprecipitation (IP), 1 mg of cell lysate was incubated with 2 μg of anti-ErbB2 mAb (Ab2/Ab5, Oncogene), and the immunocomplexes were precipitated with protein G-agarose beads. Precipitated proteins were dissolved in SDS sample buffer and separated by SDS/PAGE. Blot was probed with ErbB2 antibody to check immunoprecipitation efficiency. IB, immunoblotting. Association of CHIP with ErbB2 was detected by rabbit polyclonal anti-CHIP antibody. (B) Association of CHIP with ErbB2 requires intact TPR domain. COS7 cells were transfected with wild-type ErbB2 plus different CHIP constructs. Immunoprecipitation and Western blotting were performed as in A. (C) GA increases binding of CHIP to ErbB2 protein. Transfected COS7 cells were treated with 1 μM GA and 100 μM ALLnL for 1 h before being lysed with TMNS buffer. Immunoprecipitation and Western blotting were as in A. (D) Endogenous CHIP and ErbB2 associate with each other in the presence of GA. SKOV3 cells were treated with ALLnL and GA, alone or in combination, and processed as in C. Immunoprecipitation and Western blotting were performed as in A. (E) Effects of GA on association of CHIP with chaperone and ErbB2 proteins. COS7 cells were cotransfected with ErbB2 and Myc-His-tagged CHIPs. Twenty hours after transfection, one set of transfected cells was treated with 1 μM GA for 30 min. Cells were lysed with PBS containing 1% Nonidet P-40, and CHIP proteins were precipitated from the supernatant by using Talon beads (CLONTECH). Precipitated proteins were separated on SDS/4–20% polyacrylamide gels, transferred to a poly(vinylidene difluoride) membrane, and probed with different antibodies. (F) CHIP alters composition of the chaperone complexes that bind ErbB2. COS7 cells were transfected with full-length ErbB2, with or without wild-type CHIP. Twenty hours after transfection, cells were treated with 1 μM GA and 100 μM ALLnL for 6 h. Immunoprecipitation was as in A. Association of CHIP, Hsc/Hsp70, and Hsp90 was detected with appropriate antibodies.
Association of CHIP with ErbB2 Proteins Is Mediated by a Chaperone Complex.
CHIP has been reported to bind to minimally ubiquitinated proteins by way of its U box domain (17). To investigate whether CHIP might associate with a minimally ubiquitinated ErbB2 directly, or instead interact indirectly by a chaperone intermediate, we determined the ability of the two CHIP TPR and U box domain mutants to interact with ErbB2. In accordance with the data shown in Fig. 1C, we found that ErbB2 immunoprecipitation failed to coprecipitate the TPR mutant, CHIP(K30A), whereas the U box mutant, CHIP(H260Q), reproducibly coprecipitated with ErbB2 to a greater extent than did wild-type CHIP (Fig. 2B). Because neither point mutant affected ErbB2 steady-state level (see Fig. 1C), the requirement of an intact TPR domain for CHIP's association with ErbB2 supports the hypothesis that this interaction is indeed indirect and is mediated by a chaperone intermediate. A 1-h preincubation of the cells with GA before lysis increased the amount of both wild-type CHIP and CHIP(H260Q) coprecipitating with ErbB2 (Fig. 2C), consistent with a model in which GA promotes CHIP recruitment to chaperone–client protein complexes.
Although transient transfection experiments are informative, they do not always accurately reflect the steady-state association of two proteins. Therefore, we next investigated whether endogenous CHIP and ErbB2 could be coprecipitated in the presence or absence of GA. For this purpose, we screened several cell lines for CHIP and ErbB2 expression and found that the human ovarian carcinoma cell line SKOV3 expressed moderate levels of both proteins. Immunoprecipitation of ErbB2 from untreated cells revealed minimal CHIP association, but, similar to our findings with exogenously expressed CHIP and ErbB2 (see Fig. 2C), brief exposure of the cells to GA dramatically enhanced the association of the two proteins (Fig. 2D, compare lane 4 with lane 2, and lane 3 with lane 1). GA did not affect the level of CHIP expression in cell lysate (data not shown). A proteasome inhibitor was included in this experiment to stabilize the association of CHIP and ErbB2 in the presence of GA.
To confirm the specificity of the CHIP/ErbB2 interaction, we placed a polyhistidine tag at the carboxyl terminus of CHIP, and, after transient cotransfection with ErbB2 in COS7 cells, we affinity-precipitated His-tagged CHIP with cobalt beads (Fig. 2E). The data clearly show that ErbB2 coprecipitates with His-tagged wild-type CHIP and CHIP(H260Q), but not with CHIP(K30A). As seen in the reciprocal experiments (Fig. 2 B and C), more ErbB2 was consistently found in association with CHIP(H260Q) than with wild-type CHIP, suggesting that ErbB2 association with enzymatically active CHIP is normally dynamic. Additionally, Hsp90 and Hsp/Hsc70 were found in all CHIP pull-downs that contained ErbB2 (Fig. 2E).
CHIP Remodels the Chaperone Complex Associating with ErbB2.
GA treatment disassociates Hsp90 while increasing the association of Hsp/Hsc70 with the ErbB2 protein (14). It has been postulated that this change in chaperone complex composition negatively affects ErbB2 stability. Because CHIP can bind to either Hsc70 or Hsp90, we wondered whether CHIP expression would modify the chaperone complex associated with ErbB2 in a fashion similar to GA. To examine this question, we immunoprecipitated ErbB2 from COS7 cells cotransfected with or without CHIP, and detected the presence of the chaperone proteins in the precipitated pellets. As shown in Fig. 2F, expression of wild-type CHIP decreased the Hsp90 level, at the same time as it increased the coprecipitation of Hsp/Hsc70 (Fig. 2F, compare lane 3 with lane 1). These results indicate that expression of CHIP in cells induces a prodegradation chaperone complex to associate with the ErbB2 protein, as does exposure to GA (Fig. 2F, lane 2).
CHIP Induces ErbB2 Ubiquitination.
In the presence of GA, ErbB2 protein is rapidly ubiquitinated before its degradation by the proteasome (15). To determine whether CHIP supports ErbB2 ubiquitination in vivo, we tested whether coexpression of CHIP alters the ubiquitination status of ErbB2. Because ubiquitinated proteins are highly unstable and are quickly degraded by the proteasome, the proteasome inhibitor ALLnL was added to the cells 6 h before lysis. As shown in Fig. 3A, coexpression of CHIP and ErbB2 in COS7 cells significantly increased the level of ErbB2 ubiquitination (compare lane 3 with lane 2), indicating that the down-regulation of ErbB2 caused by CHIP occurs by the ubiquitination pathway. Consistent with our previous data, wild-type CHIP, but not CHIP(H260Q), supported ErbB2 ubiquitination. Coexpression of CHIP(K30A) and ErbB2 caused a moderate increase of ErbB2 ubiquitination (Fig. 3A, lane 4), for reasons not completely understood. However, transfection of CHIP(H260Q) abrogated the ability of GA to promote ErbB2 ubiquitination (Fig. 3A, lane 9), thus acting in a dominant-negative fashion. In contrast, CHIP(K30A) did not affect GA-induced ErbB2 ubiquitination (Fig. 3A, lane 8), presumably because it failed to interact with the ErbB2/chaperone complex.
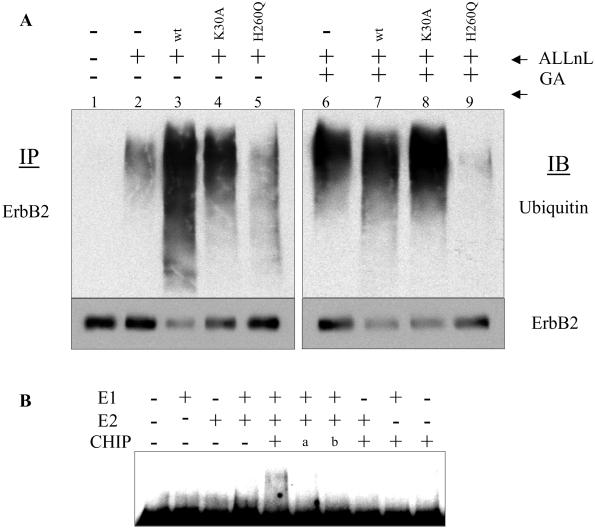
Figure 3.
CHIP induces ErbB2 ubiquitination. (A) CHIP enhances ubiquitination of ErbB2 protein in vivo. COS7 cells in 150 × 25 mm plates were transfected with 5 μg of ErbB2 and 15 μg of CHIP plasmid DNA. Twenty hours after transfection, cells were treated with 100 μM ALLnL, with or without 1 μM GA, for 6 h. Cells were then lysed with 2.5 ml of TMNS buffer. ErbB2 proteins were immunoprecipitated (IP) as described for Fig. 2A. Precipitated proteins were separated on an SDS/4–20% polyacrylamide gel, transferred to nitrocellulose membrane, and autoclaved to maximize the detection of polyubiquitinated proteins. The membrane was probed with rabbit anti-ubiquitin antibody, followed by mouse anti-ErbB2 mAb. IB, immunoblotting. (B) CHIP serves as an E3 ubiquitin ligase for ErbB2 in an in vitro ubiquitination assay. The entire intracellular domain of ErbB2 was translated in rabbit reticulocyte lysate, immunoprecipitated, and subjected to in vitro ubiquitination. The ubiquitination of ErbB2 (seen as slower migrating species) was detected by Western blotting with an ErbB2-specific antibody. a, Glutathione S-transferase (GST)-CHIP(1–197), lacking the U box domain; b, GST-CHIP(198–303), lacking the TPR domain.
To confirm that CHIP can directly ubiquitinate ErbB2, we performed an in vitro ubiquitination assay. Because the intracellular portion of the ErbB2 protein was required for CHIP-induced down-regulation (see Fig. 1), we tested whether this part of the molecule, in vitro translated in rabbit reticulocyte lysate, could be ubiquitinated by CHIP in the presence of E1 and E2 enzymes. As shown in Fig. 3B, inclusion of CHIP in the assay caused ubiquitination of ErbB2 (represented by slower migrating forms of the protein) that was E1- and E2-dependent, indicating that CHIP can serve as an E3 ubiquitin ligase for ErbB2. Neither U box domain-deleted nor TPR domain-deleted CHIP (labeled as “a” and “b,” respectively, in Fig. 3B) demonstrated E3 activity in this assay. The lack of ErbB2 ubiquitination in the absence of added CHIP rules out inadvertent coprecipitation of an E3 activity from rabbit reticulocyte lysate. On the other hand, the fact that both Hsp90 and Hsc70 were found in the ErbB2 immunoprecipitates (data not shown), together with the finding that TPR domain-deleted CHIP was ineffective in this assay, strengthens the chaperone requirement for CHIP activity.
Redundant Pathways Exist for the Degradation of ErbB2 Proteins.
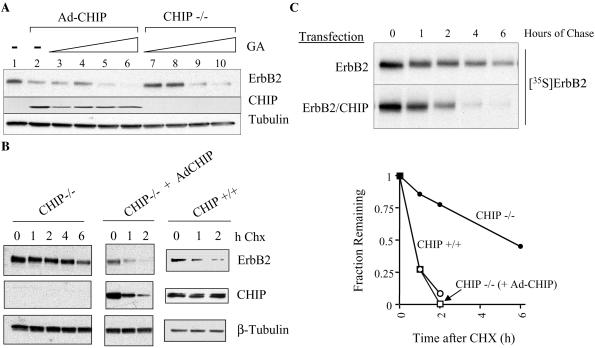
Because our data suggest that CHIP is involved in GA-induced ErbB2 degradation, we wished to determine whether CHIP is the only E3 protein able to mediate GA-induced ErbB2 down-regulation. For this purpose, we examined the GA sensitivity of endogenous ErbB2 in CHIP−/− fibroblasts. Unexpectedly, the kinase remained GA-sensitive in CHIP−/− cells (Fig. 4A, lanes 7–10). Nevertheless, reintroduction of CHIP by adenovirus infection into these cells markedly reduced the steady-state level of ErbB2 (Fig. 4A, compare lanes 1 and 2). The decrease of ErbB2 steady-state level reflects CHIP-dependent destabilization of both mature and nascent ErbB2 proteins. Thus, the half-life of mature ErbB2 protein (as determined by monitoring the decline in steady-state ErbB2 level after cycloheximide treatment to block protein synthesis) was significantly reduced upon reintroduction of CHIP into CHIP−/− cells (Fig. 4B). Indeed, the stability of mature ErbB2 protein was nearly identical in CHIP+/+ fibroblasts and in CHIP−/− cells whose CHIP protein was replaced by adenoviral infection (Fig. 4B, graph). Similarly, coexpression of CHIP with ErbB2 in COS7 cells decreased the half-life of newly synthesized ErbB2 protein by more than 50% (Fig. 4C). These data indicate that both newly synthesized and mature ErbB2 proteins are sensitive to CHIP, much as they are also both sensitive to the destabilizing effects of GA (24).
Figure 4.
CHIP is not the only E3 capable of mediating GA-induced down-regulation of ErbB2 protein. (A) CHIP−/− cells were infected with an adenovirus containing the CHIP gene (Ad-CHIP). Uninfected (control) or CHIP-infected cells were treated with increasing concentrations of GA (0.0625–0.5 μM) for 4 h before lysis. Endogenous ErbB2 protein in cell lysate was detected by Western blotting. (B) CHIP reduces the half-life of mature ErbB2 protein. CHIP−/− cells were infected as in A. Four hours later, they, and uninfected CHIP −/− and +/+ cells, were treated with 100 μg/ml cycloheximide (Chx), and lysed at various times. Levels of ErbB2, CHIP, and β-tubulin were detected by Western blotting of total cell lysate. (C) CHIP reduces the half-life of nascent ErbB2 protein. COS7 cells, transfected with ErbB2 alone (Upper) or together with CHIP (Lower), were pulse-labeled with [35S]methionine for 45 min, and then chased in nonradioactive complete medium for various times. ErbB2 protein was immunoprecipitated, and separated by SDS/PAGE. (Upper) The dried gel was exposed to Kodak X-Omat AR film. (Lower) Quantitation of the results.
Discussion
The ErbB2 receptor tyrosine kinase, unlike the closely related ErbB1, is resistant to ligand-induced down-regulation mediated by c-Cbl. Instead, it helps to recycle ErbB1 back to the cell surface and participates in additional rounds of signal transduction (12). However, we and others have shown that ErbB2 can be quickly down-regulated by the Hsp90-binding ansamycin antibiotic GA (24–26). Moreover, we showed recently that GA treatment decreases ErbB2 association with Hsp90 while increasing its association with Hsp/Hsc70, and that this effect occurs before the degradation of ErbB2 protein (14). The data described here show that ectopic expression of an Hsp90/Hsc70-interacting E3 ubiquitin ligase, CHIP, induces a similar phenotype, with respect to both ErbB2 instability and modification of the chaperone complexes associated with the ErbB2 protein. Although CHIP and ErbB2 can be found in association when CHIP is overexpressed in the absence of GA (although GA enhances this association), endogenous CHIP and ErbB2 detectably coprecipitate with each other only after GA exposure.
CHIP binding to Hsp90 or Hsp/Hsc70 is mutually exclusive (17), but because both Hsp90 and Hsp/Hsc70 appear in the CHIP pull-down experiments, it is not clear which chaperone mediates CHIP/ErbB2 association. Because CHIP has an equivalent affinity for both chaperones (U. Hartl and J. Young, personal communication), it is possible that they interact sequentially to mediate ErbB2 degradation. GA pretreatment decreased the amount of Hsp90 appearing in wild-type CHIP pull-downs, whereas the amount of both ErbB2 and Hsp/Hsc70 in these pull-downs remained the same (Fig. 2E, compare lane 6 with lane 2). In the reciprocal experiment, immunoprecipitation of ErbB2 in cells not transfected with CHIP coprecipitated much more Hsp90 than Hsp/Hsc70 (Fig. 2F, lane 1), confirming published observations (14). In contrast, when ErbB2 was immunoprecipitated from CHIP-transfected or GA-treated cells, Hsp/Hsc70 was the dominant chaperone coprecipitated, with a reduction in the amount of coprecipitated Hsp90 (Fig. 2F, compare lane 1 with lanes 2 and 3). Thus, both CHIP and GA increase the amount of Hsp/Hsc70 associated with ErbB2, but decrease the association of Hsp90. In contrast to wild-type CHIP, enzymatically inactive CHIP(H260Q) associated more strongly with Hsp90 without altered association of Hsp/Hsc70, and GA enhanced this phenomenon (Fig. 2E). Taken together, these data suggest that enzymatically active CHIP may first bind to Hsp90 (in the chaperone/client protein complex) and, subsequent to or during ErbB2 ubiquitination, shift its association to Hsp/Hsc70 while simultaneously promoting the release of Hsp90 from the complex. However, a definitive understanding of the role of each chaperone in the targeting of CHIP to ErbB2 awaits further experimentation. Even in the in vitro ubiquitination assay of CHIP activity, both Hsp90 and Hsc70 were present in the ErbB2 immunoprecipitates (and the TPR domain mutant of CHIP failed to ubiquitinate ErbB2), making it impossible to distinguish between the requirement for one or both chaperones in this process.
It is also not clear how the ubiquitinated ErbB2/chaperone complex associates with the proteasome. Although BAG-1, a cochaperone that binds to both Hsc70 and the proteasome, may participate in the delivery of the CHIP–ErbB2 complex to the proteasome for degradation (27), proteasome-dependent, CHIP-mediated degradation can be reconstituted in vitro without BAG-1 and without a significant change in the kinetics of degradation (C.P., unpublished observations).
In summary, we show here that CHIP, in the absence of GA and dependent on its chaperone-binding and ubiquitin ligase domains, can destabilize ErbB2 protein in vivo and promote its polyubiquitination in vitro. GA and CHIP seem to cooperate in this activity, because overexpression of the inactive E3 point mutant CHIP(H260Q) blocks GA-induced ubiquitination of ErbB2. However, because GA-induced down-regulation of ErbB2 still occurs in CHIP−/− cells, significant redundancy must exist in the machinery regulating chaperone-dependent Hsp90 client protein degradation. Indeed, although it has recently been proposed that CHIP is a general “quality-control” E3, responsible for ubiquitinating unfolded or misfolded proteins in a chaperone-dependent triage process (28), we recently reported that CHIP does not mediate the ubiquitination of another GA-sensitive Hsp90 client protein, HIF-1α (29). These findings suggest an additional, yet to be understood, level of specificity that determines CHIP interaction with specific Hsp90 client proteins.
Nonetheless, a role for CHIP in the regulation of ErbB2 stability is suggested by the finding that endogenous ErbB2 in CHIP+/+ fibroblasts displays a significantly shorter half-life than in CHIP−/− cells. Further, reintroduction of CHIP into these cells restores ErbB2 half-life to that seen in wild-type fibroblasts. Finally, as is true for ErbB2's GA sensitivity, both mature and newly synthesized ErbB2 proteins are sensitive to CHIP. Identification of CHIP as a component of a chaperone-dependent triage cycle that regulates ErbB2 stability suggests novel therapeutic approaches to the down-regulation of this kinase. Pharmacologic enhancement of CHIP–ErbB2 interaction should be a potent means for specifically reducing ErbB2 expression.
Abbreviations
- CHIP
C-terminal Hsc70-interacting protein
- TPR
tetratricopeptide
- GA
geldanamycin
- ALLnL
N-acetyl-l-leucyl-l-leucyl-norleucinal
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Muthuswamy S K, Gilman M, Brugge J S. Mol Cell Biol. 1999;19:6845–6857. doi: 10.1128/mcb.19.10.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klapper L N, Kirschbaum M H, Sela M, Yarden Y. Adv Cancer Res. 2000;77:25–79. [PubMed] [Google Scholar]
- 3.Ouyang X, Gulliford T, Zhang H, Smith G, Huang G, Epstein R J. Mol Cell Biochem. 2001;218:47–54. doi: 10.1023/a:1007249004222. [DOI] [PubMed] [Google Scholar]
- 4.Kuter I. Oncologist. 2001;6:338–346. doi: 10.1634/theoncologist.6-4-338. [DOI] [PubMed] [Google Scholar]
- 5.Mendelsohn J, Baselga J. Oncogene. 2000;19:6550–6565. doi: 10.1038/sj.onc.1204082. [DOI] [PubMed] [Google Scholar]
- 6.Baselga J, Tripathy D, Mendelsohn J, Baughman S, Benz C C, Dantis L, Sklarin N T, Seidman A D, Hudis C A, Moore J, et al. J Clin Oncol. 1996;14:737–744. doi: 10.1200/JCO.1996.14.3.737. [DOI] [PubMed] [Google Scholar]
- 7.Joazeiro C A, Wing S S, Huang H, Leverson J D, Hunter T, Liu Y C. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 8.de Melker A A, van Der Horst G, Calafat J, Jansen H, Borst J. J Cell Sci. 2001;114:2167–2178. doi: 10.1242/jcs.114.11.2167. [DOI] [PubMed] [Google Scholar]
- 9.Levkowitz G, Waterman H, Ettenberg S A, Katz M, Tsygankov A Y, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, et al. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 10.Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon W Y, Beguinot L, Geiger B, Yarden Y. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klapper L N, Waterman H, Sela M, Yarden Y. Cancer Res. 2000;60:3384–3388. [PubMed] [Google Scholar]
- 12.Lenferink A E, Pinkas-Kramarski R, van de Poll M L, van Vugt M J, Klapper L N, Tzahar E, Waterman H, Sela M, van Zoelen E J, Yarden Y. EMBO J. 1998;17:3385–3397. doi: 10.1093/emboj/17.12.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemoine N R, Staddon S, Dickson C, Barnes D M, Gullick W J. Oncogene. 1990;5:237–239. [PubMed] [Google Scholar]
- 14.Xu W, Mimnaugh E, Rosser M F, Nicchitta C, Marcu M, Yarden Y, Neckers L. J Biol Chem. 2001;276:3702–3708. doi: 10.1074/jbc.M006864200. [DOI] [PubMed] [Google Scholar]
- 15.Mimnaugh E G, Chavany C, Neckers L. J Biol Chem. 1996;271:22796–22801. doi: 10.1074/jbc.271.37.22796. [DOI] [PubMed] [Google Scholar]
- 16.Ballinger C A, Connell P, Wu Y, Hu Z, Thompson L J, Yin L Y, Patterson C. Mol Cell Biol. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connell P, Ballinger C A, Jiang J, Wu Y, Thompson L J, Hohfeld J, Patterson C. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 18.Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama K I. J Biol Chem. 2001;276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- 19.Jiang J, Ballinger C A, Wu Y, Dai Q, Cyr D M, Hohfeld J, Patterson C. J Biol Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 20.Meacham G C, Patterson C, Zhang W, Younger J M, Cyr D M. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 21.An W G, Schulte T W, Neckers L M. Cell Growth Differ. 2000;11:355–360. [PubMed] [Google Scholar]
- 22.Johnson J L, Toft D O. Mol Endocrinol. 1995;9:670–678. doi: 10.1210/mend.9.6.8592513. [DOI] [PubMed] [Google Scholar]
- 23.Xu W, Mimnaugh E G, Kim J S, Trepel J B, Neckers L M. Cell Stress Chaperones. 2002;7:91–96. doi: 10.1379/1466-1268(2002)007<0091:hngrti>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chavany C, Mimnaugh E, Miller P, Bitton R, Nguyen P, Trepel J, Whitesell L, Schnur R, Moyer J, Neckers L. J Biol Chem. 1996;271:4974–4977. doi: 10.1074/jbc.271.9.4974. [DOI] [PubMed] [Google Scholar]
- 25.Miller P, DiOrio C, Moyer M, Schnur R C, Bruskin A, Cullen W, Moyer J D. Cancer Res. 1994;54:2724–2730. [PubMed] [Google Scholar]
- 26.Kuduk S D, Zheng F F, Sepp-Lorenzino L, Rosen N, Danishefsky S J. Bioorg Med Chem Lett. 1999;9:1233–1238. doi: 10.1016/s0960-894x(99)00185-7. [DOI] [PubMed] [Google Scholar]
- 27.Demand J, Alberti S, Patterson C, Hohfeld J. Curr Biol. 2001;11:1569–1577. doi: 10.1016/s0960-9822(01)00487-0. [DOI] [PubMed] [Google Scholar]
- 28.Murata S, Minami Y, Minami M, Chiba T, Tanaka K. EMBO Rep. 2001;2:1133–1138. doi: 10.1093/embo-reports/kve246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isaacs J S, Jung Y J, Mimnaugh E G, Martinez A, Cuttitta F, Neckers L M. J Biol Chem. 2002;277:29936–29944. doi: 10.1074/jbc.M204733200. [DOI] [PubMed] [Google Scholar]