Abstract
Chlorocatechol 1,2-dioxygenase from Pseudomonas putida (Pp 1,2-CCD) is a dioxygenase responsible for ring cleavage during the degradation of recalcitrant aromatic compounds. We determined the zero-field splitting of the Fe(III) cofactor (|D| = 1.3 ± 0.2 cm−1) by electron paramagnetic resonance (EPR) experiments that along with other structural data allowed us to infer the Fe(III) coordination environment. The EPR spectrum of the ion shows a significantly decrease of the g = 4.3 resonance upon substrate binding. This result is rationalized in terms of a mechanism previously proposed, where catechol substrate is activated by Fe(III), yielding an exchange-coupled Fe(II)-semiquinone (pair). The Pp 1,2-CCD capacity of binding amphipatic molecules and the effects of such binding on protein activity are also investigated. EPR spectra of spin labels show a protein-bound component, which was characterized by means of spectral simulations. Our results indicate that Pp 1,2-CCD is able to bind amphipatic molecules in a channel with the headgroup pointing outwards into the solvent, whereas the carbon chain is held inside the tunnel. Protein assays show that the enzyme activity is significantly lowered in the presence of stearic-acid molecules. The role of the binding of those molecules as an enzyme activity modulator is discussed.
INTRODUCTION
Dioxygenases are bacterial nonheme iron enzymes responsible for the aerobic catabolism of several intermediates produced by the decomposition of aromatic compounds that are industrially released in the environment. Their paramount role in the degradation funnel of such toxic substances (Ornston and Stanier, 1966; Schlömann, 1994) is the main interest in studying their structural arrangement as well as their mechanism of action. Dioxygenases catalyze ring cleavage with the incorporation of both atoms of an oxygen molecule into the substrate. The ring can be cleaved at different points on the aromatic structure thus characterizing the enzyme as an extradiol or an intradiol dioxygenase (Nozaki et al., 1970). In both cases, a nonheme iron center is crucial for protein activity with extradiol enzymes containing a ferrous ion whereas intradiol proteins present a ferric ion (for recent reviews see Bugg, 2001; and Solomon et al., 2000).
Intradiol enzymes can be further divided into two structurally different families: protocatechuate 3,4-dioxygenases (3,4-PCDs) and catechol 1,2-dioxygenases (1,2-CTDs). The former comprises proteins composed by two homologous subunits (αβFe(III)), which in turn are arranged in large oligomeric structures. 3,4-PCDs catalyze hydroxybenzoates. Crystal structure data (Ohlendorf et al., 1988, 1994; Vetting et al., 2000) along with several spectroscopic studies (Whittaker and Lipscomb, 1984; True et al., 1990; Orville and Lipscomb, 1989) on 3,4-PCD enzymes have shown that the ferric ion is in a trigonal-bipyramidal geometry ligated by two histidine residues, two tyrosine residues, and a hydroxyl ion (2His,2Tyr,1OH). On the other hand, 1,2-CTDs are homodimers (αFe(III))2 (except for 1,2-CTD from Pseudomonas arvilla) and catalyze a diverse set of substrates such as catechol and its halogenated derivatives. Only recently the crystal structures of two members of the catechol dioxygenases family became available: 1,2-CTD from Acinetobacter sp. ADP1 (Ac 1,2-CTD) (Vetting and Ohlendorf, 2000) and chlorocatechol 1,2-dioxygenase from Rhodococcus opacus 1CP (Rho 1,2-CCD) (Ferraroni et al., 2004). These proteins are homodimers with a nonheme Fe(III) active site/monomer. Like the 3,4-PCDs, the iron center shows a (2Tyr,2His,1OH) coordination. A remarkable difference in substrate specificity has been noticed between 1,2-CTD and 1,2-CCD enzymes (Broderick and O'Halloran, 1991; Vetting and Ohlendorf, 2000; Ferraroni et al., 2004). The intradiol dioxygenases that catalyze chlorocatechol have a much broader substrate tolerance than the catechol 1,2-dioxygenases (Broderick and O'Halloran, 1991). The structural features yielding such substrate diversity are still not clear.
Both structures of catechol dioxygenases (Ac 1,2-CTD and Rho 1,2-CCD) reveal a novel hydrophobic helical zipper as a subunit linker, where it was found two bound phospholipid molecules. These molecules have their headgroup regions pointing outward into the solvent and the carbon chain directed inward toward each other (Vetting and Ohlendorf, 2000; Ferraroni et al., 2004). The correct identification of the phospholipid was not possible from the crystal data since no electron density was observed for their headgroups. The capacity of binding amphipathic molecules brought up a whole new set of issues concerning the possible binding of catechol dioxygenases to membranes (Vetting and Ohlendorf, 2000) and, of course, the role of that binding during ring cleavage.
In this article, we used electron paramagnetic resonance (EPR) to investigate the Fe(III) center and the existence of a hydrophobic tunnel in the structure of chlorocatechol 1,2-dioxygenase from Pseudomonas putida (Pp 1,2-CCD). We characterized the Fe(III) environment with the evaluation of its zero-field splitting, which analyzed together with the EPR and structural data for other catechol dioxygenases allowed us to confirm the Fe(III) coordination environment in Pp 1,2-CCD. We also measured the iron EPR spectrum in the presence of excess substrate, where we observed a decrease in the line intensity upon catechol binding. Furthermore, we made use of spin-labeled molecules to probe the existence of the hydrophobic tunnel in the structure of Pp 1,2-CCD. These spin-label EPR experiments were followed by measurements of the enzyme activity that consistently showed a significant decrease of catalytic activity in the presence of stearic acid molecules. Our data allowed us to speculate on the possible roles of the hydrophobic tunnel.
MATERIALS AND METHODS
Overexpression and protein purification
The recombinant protein expression was produced in Escherichia coli BL21(DE3). The protocol for protein expression and purification was described in Araújo et al. (2000). Briefly, the host cells harboring pTYBCLCA grew in LB medium, to which had been added 0.1 g/L of FeSO4 and 100 μg/ml ampicilin. The cells were suspended in Tris-NaCl buffer (20 mM Tris, 500 mM NaCl, 20 μM PMSF, pH 8.0), lysed by sonication and the cell debris were separated by centrifugation at 20,000 × g. The supernatant was loaded onto a chitin column (NBL) preequilibrated with the same buffer used for the resuspension of the cells. The column was washed with the resuspension buffer and subsequently with 20 mM Tris-HCl, pH 8.0 buffer containing 50 mM NaCl, 30 mM DTT, 20 μM PMSF. The CLCA protein was eluted with the same buffer after 15 h of incubation at 8°C. The active fractions were combined, concentrated, and loaded onto a Superdex 75 column (Pharmacia) (1.6 cm × 60 cm) in 20 mM Tris-NaCl buffer, pH 8.4 with NaCl 50 mM, and 1 mM PMSF. The major peak eluted was used for all measurements.
Enzyme assay
Catalytic activity was assayed by the method described in Nakazawa (Nakazawa and Nakazawa, 1970), where changes in absorbance at 260 nm due to cis-cis muconic acid production are monitored. All experiments were carried out at room temperature and catechol was used as the substrate for the dioxygenase reaction. One unit of enzymatic activity is defined as the amount of enzyme that catalyzes 1 μM of product per minute at 24°C. The specific activity is in turn defined as the number of enzymatic activity units per milligram of protein (Nakai et al., 1988).
EPR spectroscopy of Fe(III) center
X-band (9.5 GHz) electron paramagnetic resonance spectra of the Fe(III) ion were recorded on a Bruker ELEXSYS E580 (Bruker BioSpin, Rheinstetten, Germany) at liquid helium temperatures (4 K). The temperature was controlled by an Oxford ITC503 cryogenic system. EPR samples containing a convenient amount of the protein were frozen by immersion in liquid nitrogen and then placed in the spectrometer cavity. The frequency was measured with an HP5350B Microwave Frequency Counter. All EPR data were corrected by subtracting a baseline corresponding to the EPR signal of the buffer. Other acquisition conditions: modulation amplitude, 1.0 G; modulation frequency, 100 kHz. The g-values of the Fe(III) signal were determined by means of spectral simulation using the Bruker WinEPR-SimFonia program.
EPR spectroscopy of spin probes
The headgroup spin label 4-(N,N-dimethyl-N-hexadecyl) ammonium-2,2,6,6-tetramethylpiperidine-1-oxyl iodide (CAT16) and the chain labels n-doxyl stearic acid (n = 5, 12, 16-SASL) were purchased from Sigma-Aldrich (St. Louis, MO). The phospholipid labels 1-palmitoyl-2-(n-doxyl stearoyl) phosphatidylcholine (n = 5, 7, 10, 12, 16-PC) were purchased from Avanti Polar Lipids (Alabaster, AL). All labels were used without further purification. The chloroform present in the stock solutions of spin labels was removed by N2 flow followed by 1 h in a speedvac system to ensure complete removal of the solvent. A measured amount of the buffered CCD solution was added to the sample tube, and incubated for few minutes. A final volume of 100 μL of the samples containing mixtures of spin labels and enzyme solution in the molar ratio SASL/CCD 1:4 or PCSL/CCD 1:2 was drawn into a quartz flat cell, which was in turn placed in the EPR resonant cavity. Final enzyme concentration was 0.22 mM in all experiments. X-band EPR spectra of those samples were recorded on a Varian E109 spectrometer at room temperature. Acquisition conditions: modulation amplitude, 1.0 G; modulation frequency, 100 kHz; microwave power, 10 mW; field range, 100 G.
NLLS simulations
The EPR spectra of the spin probes in mixtures of probe/enzyme were simulated by means of a nonlinear least-squares program developed by Freed and co-workers (Meirovitch et al., 1984; Schneider and Freed, 1989; Budil et al., 1996). The parameters involved in the fitting procedure are as follows: hyperfine tensor components (Axx,Ayy,Azz), rotational diffusion rates (R⊥ and R‖), and a lorentzian (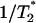 ) inhomogeneous broadening. The dynamics of the spin probe is characterized by R⊥ and R‖, which represent the rotational diffusion rates of the nitroxide radical around the axes perpendicular and parallel to the mean symmetry axis for the rotation. This symmetry axis is also the direction of preferential orientation of the spin label moiety (Schneider and Freed, 1989). For chain labels (n-SASL and n-PC) R⊥ accounts for the wagging motion of the long axis of the carbon chain. As for the label CAT-16, it represents the wagging motion of the headgroup region. The g-tensor components used in the simulations were obtained from Ge et al. (1990).
) inhomogeneous broadening. The dynamics of the spin probe is characterized by R⊥ and R‖, which represent the rotational diffusion rates of the nitroxide radical around the axes perpendicular and parallel to the mean symmetry axis for the rotation. This symmetry axis is also the direction of preferential orientation of the spin label moiety (Schneider and Freed, 1989). For chain labels (n-SASL and n-PC) R⊥ accounts for the wagging motion of the long axis of the carbon chain. As for the label CAT-16, it represents the wagging motion of the headgroup region. The g-tensor components used in the simulations were obtained from Ge et al. (1990).
RESULTS AND DISCUSSIONS
EPR characterization of the Fe(III) center
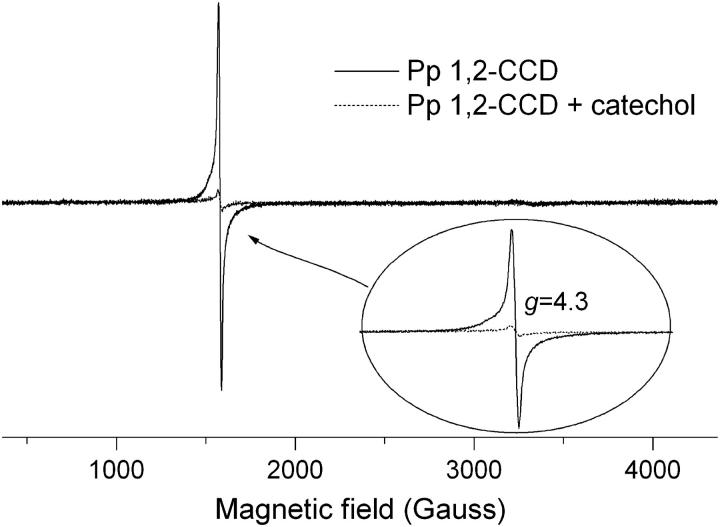
The EPR spectrum of the fully active protein is presented in Fig. 1. It shows the usual g = 4.3 line of Fe(III) in a rhombic environment as previously observed by Broderick and O'Halloran (1991). This signal is assigned to the middle Kramers doublet of the Fe(III) S = 5/2 spin system (Castner et al., 1960; Wickman et al., 1965). The iron-loaded protein was further characterized by evaluating its zero-field parameter D as described elsewhere (Peisach et al., 1971; Pinkowitz and Aisen, 1972). This method has been used to determine the ligands of Fe(III) ion in nonheme proteins (Peisach et al., 1971; Pinkowitz and Aisen, 1972; Blumberg and Peisach, 1973; Deligiannakis et al., 1997). The population of any Kramers doublet at a given temperature is related to the magnitude of D, which is in turn determined by the ligand atoms of the metal center. Under nonsaturating conditions, the changes in the Boltzmann populations of specific energy levels, in our case the middle Kramers doublet, can be measured by monitoring the changes in the respective EPR signal. The intensity of the EPR line for a S = 5/2 spin system is described by
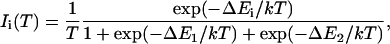 |
(1) |
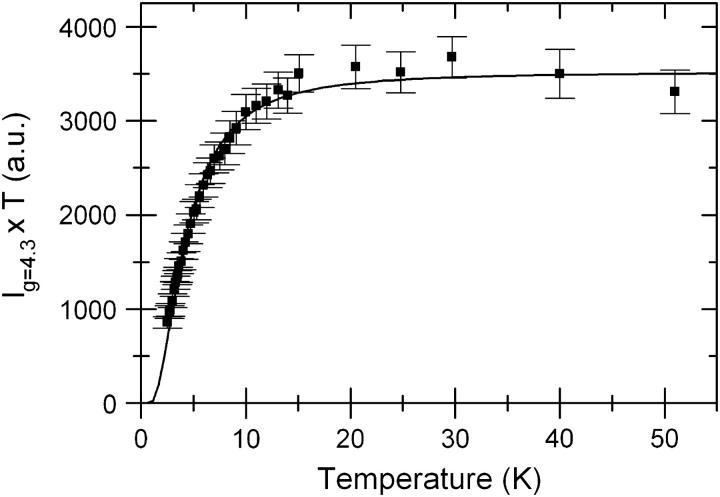
where i = 1 and 2 for the middle and higher Kramers doublet, respectively. In Fig. 2, we plot the product of the intensity of the g = 4.3 line and the temperature as a function of the temperature. The simulation of the Fe(III) spectrum (Fig. 1) yielded the set of g-values: gx = gy = gz = 4.294, which indicates a complete rhombic symmetry with zero-field parameters E/D = 1/3. In this case, the spin Hamiltonian for S = 5/2 spin systems predicts that the Kramers doublets are equally spaced in energy with splittings given by ΔE1 = ΔE2 = 4(7)1/2D/3. A least-squares fit to the data in Fig. 2 was then performed using a Boltzmann distribution (Eq. 1) over the three Kramers doublets. The energy splittings were determined as ΔE1 = ΔE2 = 4.7 cm−1, and the zero-field parameter |D| was thus calculated as (1.3 ± 0.2) cm−1. The good agreement between this value of D and those obtained for other intradiol dioxygenases such as protocatechuate 3,4-dioxygenase (3,4-PCD) from Pseudomonas aeruginosa (|D| = 1.5 cm−1) (Que et al., 1976) and for 3,4-PCD from Brevibacterium fuscum (|D| = 1.2 cm−1) (Whittaker et al., 1984) indicates that the Fe(III) coordination environment in Pp 1,2-CCD is also formed by two tyrosines, two histidines, and a water molecule. A sequence alignment of Pp 1,2-CCD, Ac 1,2-CTD and Rho 1,2-CCD shows that all residues involved in Fe(III) coordination are conserved in the structure of Pp 1,2-CCD that, together with the |D| evaluated above, confirms the coordination of the Fe(III) in the active site.
FIGURE 1.
X-band EPR spectra of the Fe(III) center from chlorocatechol 1,2-dioxygenanse. Upon catechol binding to CCD, the intensity of the sharp resonance at g = 4.3 is drastically diminished (dashed line). Experimental conditions are described in the text.
FIGURE 2.
Plot of the product of g = 4.3 signal intensity and temperature as a function of temperature for chlorocatechol 1,2-dioxygenase. The solid line is the least-squares fit of the experimental points to Eq. 1.
Reduction of the Fe(III) to Fe(II) during catalysis
To monitor changes in the EPR spectrum of the enzyme upon substrate binding, we mixed catechol (final concentration 3 mM) to the enzyme solution previously measured. The final mixture is then frozen by immersion in liquid nitrogen for subsequent EPR measurements. The EPR spectrum of the Pp 1,2-CCD in the presence of a catechol excess is shown in Fig. 1 (dashed line). The intensity of the g = 4.3 line decreases upon catechol binding to the enzyme without appearance of any other spectral feature or a line broadening. This indicates that Fe(III) is converted to Fe(II), which could be in a low-spin (S = 0) or a high-spin configuration (S = 2). A catalytic mechanism for intradiol catechol dioxygenases has been proposed by Jang et al. (1991), where the authors have used the functional model [FeIII(TPA)DBC]BPh4, (where TPA is tris(2-pyridylmethyl)amine and DBC is 3,5-di-tert-butylcatecholate dianion) to show that Fe(III) activates catechol yielding a Fe(II) semiquinone. The latter reacts with the oxygen molecule to give a hydroperoxide intermediate that undergoes Criegee rearrangement via acyl migration to give muconic anhydride. The formation of a semiquinone radical would be readily detected by EPR unless this radical is in close proximity with another paramagnetic species such as S = 2 Fe(II) ion. The results of Jang et al. (1991) give an estimate for the distance between the oxygen atom on the catechol ring and the Fe ion as ∼2 Å. In this case a strong exchange interaction takes place between the two paramagnetic species leading to massive line broadening (Bencini and Gatteschi, 1990) and a subsequent decrease in the EPR line intensity. Our data thus suggest that, upon catechol binding, Fe(III) is reduced to Fe(II) keeping the iron ion in its high-spin state, i.e., the crystal field strength is not significantly disturbed by the presence of the substrate. We believe that, after freezing, the mixture of enzyme and substrate contains two different populations of enzyme with the iron center in either redox states. To our knowledge, this is the first experimental evidence that shows the formation of Fe(II) during catechol catalysis by an intradiol catechol dioxygenase.
Hydrophobic channel
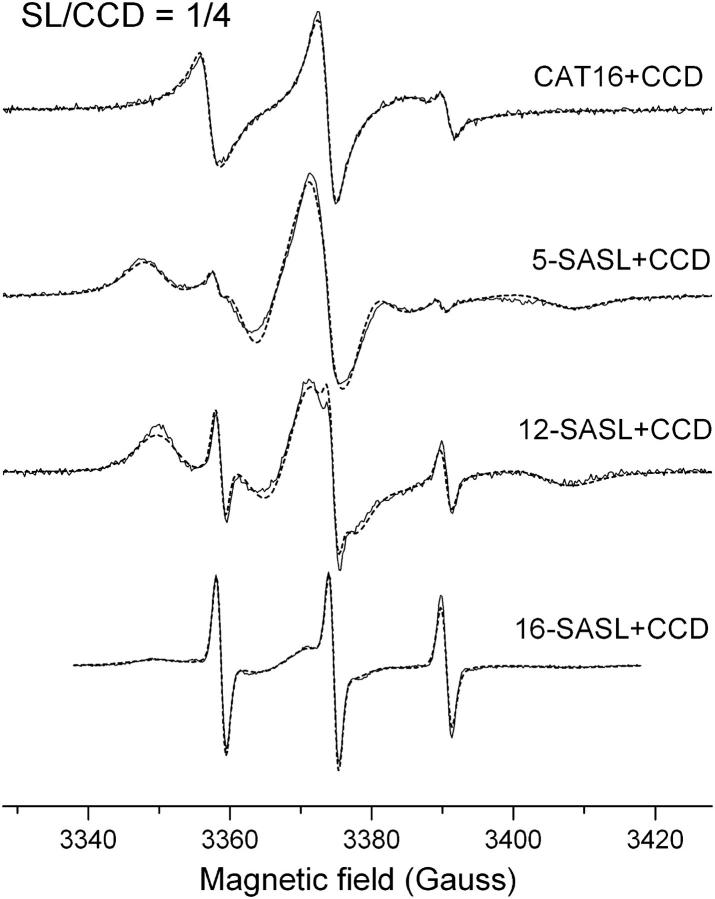
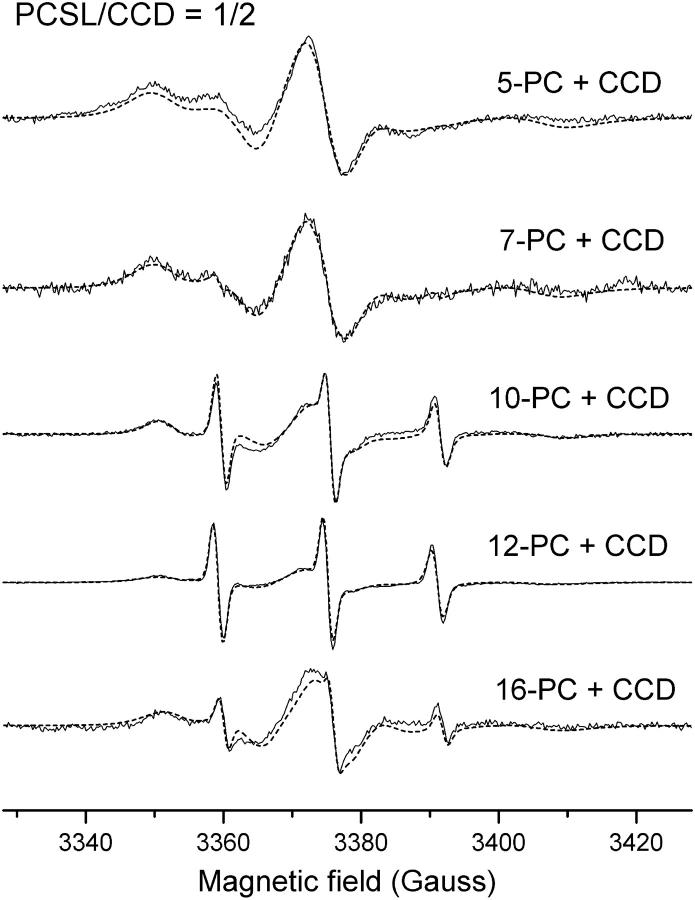
The binding of phospholipid molecules in the structure of two recent solved structures of intradiol dioxygenases (Vetting and Ohlendorf, 2000; Ferraroni et al., 2004) seems to be a common feature among this class of enzymes. To probe the existence of such hydrophobic channel in the structure of the Pp 1,2-CCD we measured the EPR spectra of labeled fatty acid and phospholipids in the presence of the protein. Fig. 3 shows the spectra of the probes CAT16, 5-SASL, 12-SASL, and 16-SASL from mixtures of label/CCD 1:4 in 20 mM Tris-HCl, 50 mM NaCl, pH 8.4. In all cases a two-component spectrum containing a fast and an immobilized component is observed. The fast component is associated with free-labeled molecules in solution, whereas the more immobilized one is assigned to probes bound to the enzyme. The addition of 8 mM urea to one of the samples led to a spectrum containing only the fast component (data not shown), thus indicating the dissociation of the enzyme/spin labeled fatty acid complex upon denaturation of the protein. Furthermore, the capacity of binding phospholipid molecules was also investigated by using phospholipid derivatives as spin probes. The spectra of the labels 5-, 7-, 10-, 12-, and 16-PC in mixtures of probes and Pp 1,2-CCD (label/CCD 1:2) are shown in Fig. 4 and the overall features are very similar to those observed for the single-chain labels. The size of the channel in the structure of Ac 1,2-CTD (8 × 35 Å) and the modeling of a phospholipid containing a chain with 14–15 atoms in the structure of Rho 1,2-CCD are compatible with the sizes of the spin probes used in our experiments (16 carbon atoms and length ∼30 Å). Thus, the spectra in Figs. 3 and 4 along with the structural information from two other catechol dioxygenases indicate that a hydrophobic channel also exists in the structure of Pp 1,2-CCD.
FIGURE 3.
Experimental (solid line) and simulated (dashed line) EPR spectra from CAT16, 5-, 12,- and 16-ASL bound to CCD in the molar ratio of spin label to CCD of 1:4 at pH 8.4 and 25°C.
FIGURE 4.
Experimental (solid line) and simulated (dashed line) EPR spectra from 5-, 7-, 10-, 12-, and 16-PCSL bound to CCD in the molar ratio of spin label to CCD of 1:2 at pH 8.4 and 25°C.
The NLLS simulation of the experimental spectra allowed us to obtain details about the structure of the hydrophobic channel. Table 1 contains the best-fit parameters for the pure single-chain probes and mixtures of either single-chain probes or phospholipid labels with Pp 1,2-CCD. A two-component theoretical spectrum was used in all cases where the enzyme was present. The fast-component parameters were kept fixed at those values shown in Table 1 A, whereas we varied the bound-label parameters. To avoid local minima during the minimization process, we started the fits from several sets of seed values for the bound-label parameters. This procedure was also useful in obtaining an error estimate for the parameters varied during the simulations. The experimental (solid line) and calculated (dashed line) spectra are in good agreement as observed in Figs. 3 and 4. An additional fitting parameter included a Gaussian inhomogeneous broadening (ΔG) that resulted in unexpectedly high values (Table 1). This kind of behavior has been previously reported in lipid/gramicidin A mixtures (Costa-Filho et al., 2003), where the authors attribute those unusual high values of ΔG to the greater heterogeneity in the local environment around the boundary lipid. We believe that is also the case observed in this work and the ΔG values should be analyzed with extra care.
TABLE 1.
Best-fit parameters from NLLS simulations of the EPR spectra obtained from mixtures of the enzyme Pp 1,2-CCD and several spin labels
| Label | Axx† | Ayy | Azz | A0‡ | R⊥ (109 s−1) | ΔG (Gauss) | |
|---|---|---|---|---|---|---|---|
| (A) Pure label | CAT16 | 6.5 | 5.5 | 38.8 | 16.9 | 2.50 | 1.4 |
| 5-SASL | 6.5 | 5.5 | 35.4 | 15.8 | 0.88 | 1.3 | |
| 12-SASL | 6.5 | 5.5 | 35.7 | 15.9 | 0.77 | 1.3 | |
| 16-SASL | 6.5 | 5.5 | 35.6 | 15.9 | 0.91 | 1.3 | |
| (B) SL/CCD = 1:4 | CAT16 | 6.5 | 5.6 | 38.3 | 16.8 | 0.077 | 0* |
| 5-SASL | 6.3 | 5.9 | 34.6 | 15.6 | 0.019 | 2.1 | |
| 12-SASL | 6.1 | 5.0 | 32.1 | 14.4 | 0.012 | 3.7 | |
| 16-SASL | 6.0 | 5.5 | 33.6 | 15.0 | 0.020 | 3.5 | |
| (C)PCSL/CCD = 1:2 | 5-PC | 6.3 | 5.5 | 34.8 | 15.5 | 0.020 | 3.3 |
| 7-PC | 6.3 | 6.3 | 33.5 | 15.4 | 0.017 | 3.7 | |
| 10-PC | 6.5 | 6.5 | 31.5 | 14.8 | 0.006 | 3.2 | |
| 12-PC | 6.6 | 6.6 | 31.9 | 15.0 | 0.012 | 4.7 | |
| 16-PC | 6.6 | 6.5 | 33.2 | 15.4 | 0.018 | 2.4 |
The magnetic g-tensor components used in the fits were gxx = 2.0086, gyy = 2.0063, gzz = 2.0025 (Ge et al., 1990).
Very small values, so kept fixed at 0.
A-tensor components are in Gauss.
A0 = (Axx + Ayy + Azz)/3.
Estimated errors: R⊥ (5%); Axx and Ayy (10%); Azz (5%); and ΔG (10%).
The average hyperfine parameter (A0) is a measure of the relative polarity around the nitroxide moiety. The greater A0 value, the more hydrophilic is the environment around the probe. From Table 1 B we can see that the headgroup label CAT16 is in a much more hydrophilic environment than the other probes with A0 value (16.8 G) comparable to the one obtained for the CAT16 label free in solution (16.9 G). This indicates that this region is pointing outwards into the solvent. The polarity profile inside the tunnel does not change significantly as one goes down the label chain, except for position n = 12 of the stearic acid label 12-SASL. There the polarity-related A0 value drops to 14.4 G, which can be due to the presence of hydrophobic side chains at that depth of the channel. An inspection of the hydrophobic tunnel in the crystal structure of Ac 1,2-CTD shows that there are at least two hydrophobic residues (Leu-62 and Leu-65) around position n = 12 of the carbon chain. In the Pp 1,2-CCD structure, one of those leucine residues is exchanged by a valine residue, and this pair of apolar residues could account for the decrease in A0 value at that depth of the channel. The A0 values obtained for the phospholipid labels show a similar polarity profile as a function of the depth in the tunnel, with a less abrupt decrease of the polarity around n = 12. The position n = 5 of the carbon chain is already inside the channel with A0 values for both labels lower than the value obtained for a solvent-exposed site. It is worth noting that, although presenting significant changes, some A0 values in Table 1 B–C are in the hydrophilic range thus indicating that the channel is not as hydrophobic as previously suggested by the crystal structure data for other catechol dioxygenases (Vetting and Ohlendorf, 2000; Ferraroni et al., 2004).
The dynamics of the amphipatic molecules inside the tunnel can be discussed in terms of the R⊥ and R‖ parameters. As also observed in other works where the NLLS program was used (Kar et al., 1985; Shin and Freed, 1989, Ge and Freed, 1999), we found that our results are insensitive to R‖. This value was then fixed at R‖ = 10 R⊥ during all simulations. The CAT16 label presents the highest R⊥ value due to its freedom of motion outside the channel. The positions n = 5 and 12 of the stearic acid probes show decreasing rotational diffusion rates, whereas at position n = 16 the label is somewhat less restricted than those other positions. The mobility profile for the phospholipid probes also present decreasing values of R⊥ until the n = 10 position is reached. From there on the rotational diffusion rates increase to a final value at n = 16 which is three times larger than the R⊥ at n = 10. The mobility of the 16-SASL is almost twice the one observed for the 16-PC label, which indicates that the end-chain region of the tunnel can easily accommodate the single-chain stearic acid, while offering a tighter fit for the larger phospholipid derivative. The mobility profile suggest that the tunnel is somehow narrower around n = 10 and then gets broader at the end of the carbon chain. This is in agreement with observations from the crystal structures of other catechol dioxygenases (Vetting and Ohlendorf, 2000; Ferraroni et al., 2004). The overall trends observed for A0 and R⊥ parameters from both types of probes (single- and double-chain) indicate that both occupy the same site in the protein structure.
To test the possible roles of the hydrophobic channel, we also measured the catalytic activity of the Pp 1,2-CCD in the presence of stearic acid molecules. A chloroform solution (50 μL) of 20 μM stearic acid was dried under N2 flow and pumped for 1 h in a high-vacuum system. The dried film was then resuspended with 50 μL of a 4 μM solution of Pp 1,2-CCD. The final solution was kept for 60 min at room temperature. The activity of the enzyme with bound stearic acid was measured as described in the Material and Methods section and then compared to the activity of the enzyme measured under the exact same condition except for the absence of the stearic acid molecules. These experiments resulted in consistent lower activities for the complex enzyme-stearic acid (4.0 U/mg) when compared to the sole enzyme (47.8 U/mg), which indicates that the fatty acid molecule acts as an inhibitor of the enzyme activity. In the structure of the hydrophobic tunnel (linker domain) there are two helices that also participate in the structure of the active site (Vetting and Ohlendorf, 2000; Ferraroni et al., 2004). The binding of the amphipatic molecule might induce conformational changes of those helices, which would affect the active site structure as well, thus diminishing the accessibility of that site to substrate. The EPR spectrum of the Fe(III) ion in the presence of bound stearic acid spin labels (data not shown) did not present any significant differences that could characterize such a change. If a conformational change of those helices upon stearic acid binding really takes place, it does not disturb the local geometry around the Fe(III) site.
We could speculate a little further and suggest that the binding of the Pp 1,2-CCD to a membrane would be the mechanism used by the bacteria to switch off the enzyme when not needed for catechol cleavage. High levels of substrate would make the protein dissociate from the membrane, thus becoming fully active. Furthermore, accumulations of the product cis,cis-muconate could be harmful and/or play a regulatory role in the cell as observed by Gaines et al. (1996) in Acinetobacter calcoaceticus. The regulation of the bound/unbound state of the enzyme could control the concentration of product into the cell. This fact can only be verified with further investigations of the Pp 1,2-CTD binding to model membranes, which are in progress. The hypothesis concerning the binding of catechol dioxygenaes to membranes has been proposed by Vetting and Ohlendorf (2000) and we believe our results shed some light on the possible implications of the binding of amphipatic molecules to catechol dioxygenases.
CONCLUSIONS
In this article, we studied the binding sites of the cofactor Fe(III) and of amphipatic molecules in the structure of Pseudomonas putida chlorocatechol 1,2-dioxygenase by means of electron paramagnetic resonance. The Fe(III) ion is in a complete rhombic symmetry as determined from the g = 4.3 EPR line. The evaluation of the zero-field splitting (|D| = 1.3 ± 0.2 cm−1) is in agreement with values previously determined for other dioxygenases indicating that the iron coordination in the Pp 1,2-CCD follows the usual arrangement observed in intradiol dioxygenases. Moreover, the EPR spectrum of the Fe(III) in the presence of substrate excess has its intensity severely reduced, suggesting that reduction of the Fe(III) to Fe(II) takes place during catalysis. This result can be rationalized on the basis of the mechanism proposed by Jang et al. (1991), where a semiquinone radical is formed as an intermediate of the catalytic pathway. The EPR signal of such a radical would be detectable unless the semiquinone is exchange coupled to the S = 2 Fe(II) ion, which is very likely to occur due to the proximity of both species inside the active site.
The binding of amphipatic molecules to Pp 1,2-CCD was also investigated with the use of spin labeled stearic acid and phospholipid. A hydrophobic channel also exists in the structure of the Pp 1,2-CCD as recently observed in the crystal structures of two other dioxygenases. Furthermore, the enzymatic activity of the protein in the presence of bound stearic acid molecules is dramatically decreased, indicating that the binding of phospholipids could be a mechanism to regulate the protein catalytic activity. The precise role of phospholipid binding in catechol dioxygenases still needs further investigations, but we believe our results support the hypothesis that the binding of phospholipids by intradiol dioxygenases is a common feature among the members of this class of enzyme and that such binding is probably used to control the catalysis inside the cell either keeping the levels of product, which might be toxic, low or delaying the activation of the protein while it is transported to a specific site into the cell.
Acknowledgments
The authors thank the Brazilian agencies FAPESP, CNPq, and CAPES for financially supporting this work. This work is part of a joint program PRONEX/FAPESP/CNPq (grant No. 03/09859-2).
References
- Araújo, A. P. U., G. Oliva, F. Henrique-Silva, R. C. Garratt, O. Cáceres, and L. M. Beltramini. 2000. Influence of the histidine tail on the structure and activity of recombinant chlorocatechol 1,2-dioxygenase. Biochem. Biophys. Res. Commun. 272:480–484. [DOI] [PubMed] [Google Scholar]
- Bencini, A., and D. Gatteschi. 1990. EPR of exchange coupled systems. Springer-Verlag, Berlin.
- Blumberg, W. E., and J. Peisach. 1973. The measurement of zero field splitting and the determination of ligand composition in mononuclear nonheme iron proteins. Ann. N. Y. Acad. Sci. 222:539–560. [DOI] [PubMed] [Google Scholar]
- Broderick, J. B., and T. V. O'Halloran. 1991. Overproduction, purification, and characterization of chlorocatechol dioxygenase, anon-heme iron dioxygenase with broad substrate tolerance. Biochemistry. 30:7349–7358. [DOI] [PubMed] [Google Scholar]
- Budil, D. E., S. Lee, S. Saxena, and J. H. Freed. 1996. Nonlinear-least squares analysis of slow-motion EPR spectra in one and two dimensions using a modified Levenberg-Marquardt algorithm. J. Magn. Reson. A120:155–189. [Google Scholar]
- Bugg, T. D. 2001. Oxygenases: mechanism and structural motifs for O2 activation. Curr. Opin. Chem. Biol. 5:550–555. [DOI] [PubMed] [Google Scholar]
- Castner, T., Jr., G. S. Newell, W. C. Holton, and C. P. Slichter. 1960. Note on the paramagnetic resonance of iron in glass. J. Chem. Phys. 32:668–673. [Google Scholar]
- Costa-Filho, A. J., R. H. Crepeau, P. P. Borbat, M. Ge, and J. H. Freed. 2003. Lipid-Gramicidin interactions: dynamic structure of the boundary lipid by 2D-ELDOR. Biophys. J. 84:3364–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deligiannakis, Y., A. Boussac, H. Bottin, V. Perrier, O. Bârzu, and A. M. Gilles. 1997. A new nonheme iron environment in Paracoccus denitrificans adenylate kinase studied by electron paramagnetic resonance and electron spin echo envelope modulation spectroscopy. Biochemistry. 36:9446–9452. [DOI] [PubMed] [Google Scholar]
- Ferraroni, M., I. P. Solyanikova, M. P. Kolomytseva, A. Scozzafava, L. Golovleva, and F. Briganti. 2004. Crystal structure of 4-chlorocatechol 1,2-dioxygenase from the chlorophenol-utilizing Gram-positive Rhodococcus opacus 1CP. J. Biol. Chem. 279:27646–27655. [DOI] [PubMed] [Google Scholar]
- Gaines 3rd, G. L., L. Smith, and E. L. Neidle. 1996. Novel nuclear magnetic resonance spectroscopy methods demonstrate preferential carbon source utilization by Acinetobacter calcoaceticus. J. Bacteriol. 178:6833–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, M., and J. H. Freed. 1999. Electron-spin resonance study of aggregation of Gramicidin in dipalmitoylphosphatidylcholine bilayers and hydrophobic mismatch. Biophys. J. 76:264–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, M., S. B. Rananavare, and J. H. Freed. 1990. ESR studies of stearic acid binding to bovine serum albumin. Biochim. Biophys. Acta. 1036:228–236. [DOI] [PubMed] [Google Scholar]
- Jang, H. G., D. D. Cox, and L. Que, Jr. 1991. A highly reactive functional model for the catechol dioxygenases. Structure and properties of [Fe(TPA)DBC]BPh4. J. Am. Chem. Soc. 113:9200–9204. [Google Scholar]
- Kar, L., E. Ney-Igner, and J. H. Freed. 1985. Electron spin resonance and electron-spin-echo study of oriented multilayers of Lα-dipalmitoylphosphatidylcholine water system. Biophys. J. 48:569–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirovitch, E., A. Nayeem, and J. H. Freed. 1984. An analysis of protein-lipid interactions based on model simulations of ESR spectra. J. Phys. Chem. 88:3454–3465. [Google Scholar]
- Nakai, C., T. Nakazawa, and M. Nozaki. 1988. Purification and properties of catechol 1,2-dioxygenase (pyrocatechase) from Pseudomonas putida MT-2 in comparison with that from Pseudomonas arvilla C-1. Arch. Biochem. Biophys. 267:701–713. [DOI] [PubMed] [Google Scholar]
- Nakazawa, T., and A. Nakazawa. 1970. Pyrocatechase (Pseudomonas). In Methods in Enzymology, Vol. 17A. S. P. Colowick and N. O. Kaplan, editors. Academic Press, New York. 518–522.
- Nozaki, M., S. Kotani, K. Ono, and S. Seno. 1970. Metapyrocatechase 3. Substrate specificity and mode of ring fission. Biochim. Biophys. Acta. 220:213–223. [DOI] [PubMed] [Google Scholar]
- Ohlendorf, D. H., J. D. Lipscomb, and P. C. Weber. 1988. Structure and assembly of protocatechuate 3,4-dioxygenase. Nature. 336:403–405. [DOI] [PubMed] [Google Scholar]
- Ohlendorf, D. H., A. M. Orville, and J. D. Lipscomb. 1994. Structure of protocatechuate 3,4-dioxygenase from Pseudomonas aeruginosa at 2.15 Å resolution. J. Mol. Biol. 244:586–608. [DOI] [PubMed] [Google Scholar]
- Ornston, L. N., and R. Y. Stanier. 1966. Conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. J. Biol. Chem. 241:3776–3786. [PubMed] [Google Scholar]
- Orville, A. M., and J. D. Lipscomb. 1989. Binding of isotopically labeled substrates, inhibitors, and cyanide by protocatechuate 3,4-dioxygenase. J. Biol. Chem. 264:8791–8801. [PubMed] [Google Scholar]
- Peisach, J., W. E. Blumberg, E. T. Lode, and M. J. Coon. 1971. An analysis of the electron paramagnetic resonance spectrum of Pseudomonas oleovorans Rubredoxin: a method for determination of the ligands of ferric iron in completely rhombic sites. J. Biol. Chem. 246:5877–5881. [PubMed] [Google Scholar]
- Pinkowitz, R. A., and P. Aisen. 1972. Zero-field splittings of iron complexes of Transferrins. J. Biol. Chem. 247:7830–7834. [PubMed] [Google Scholar]
- Que, L., Jr., J. D. Lipscomb, R. Zimmermann, E. Münck, N. R. Orme-Johnson, and W. H. Orme-Johnson. 1976. Mössbauer and EPR spectroscopy on protocatechuate 3,4-dioxygenase from Pseudomonas aeruginosa. Biochim. Biophys. Acta. 452:320–334. [DOI] [PubMed] [Google Scholar]
- Schlömann, M. 1994. Evolution of chlorocatechol catabolic pathways. Biodegradation. 5:301–321. [DOI] [PubMed] [Google Scholar]
- Schneider, D. J., and J. H. Freed. 1989. Calculating slow motional magnetic resonance spectra: a user's guide. In Biological Magnetic Resonance. Vol. 8. L. J. Berliner and J. Reuben, editors. Plenum Publishing, New York. 1–76.
- Shin, Y. K., and J. H. Freed. 1989. Thermodynamics of phosphatidylcholine-cholesterol mixed model membranes in the liquid crystalline state studied by the orientational order parameter. Biophys. J. 56:1093–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon, E. I., T. C. Brunold, M. I. Davis, J. N. Kemsley, S.-K. Lee, N. Lehnert, F. Neese, A. J. Skulan, Y. S. Yang, and J. Zhou. 2000. Geometric and electronic structure/function correlations in nonheme iron enzymes. Chem. Rev. 100:235–350. [DOI] [PubMed] [Google Scholar]
- True, A. E., A. M. Orville, L. L. Pearce, J. D. Lipscomb, and L. Que, Jr. 1990. An EXAFS study of the interaction of substrate with the ferric active site of protocatechuate 3,4-dioxygenase. Biochemistry. 29:10847–10854. [DOI] [PubMed] [Google Scholar]
- Vetting, M. W., D. A. D'Argenio, L. N. Ornston, and D. H. Ohlendorf. 2000. Structure of Acinetobacter strain ADP1 protocatechuate 3,4-dioxygenase at 2.2 angstrom resolution: Implications for the mechanism of an intradiol dioxygenase. Biochemistry. 39:7943–7955. [DOI] [PubMed] [Google Scholar]
- Vetting, M. W., and D. H. Ohlendorf. 2000. The 1.8 Å crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker. Structure. 8:429–440. [DOI] [PubMed] [Google Scholar]
- Whittaker, J. W., and J. D. Lipscomb. 1984. Transition-state analogs for protocatechuate 3,4-dioxygenase – Spectroscopic and kinetic-studies of the binding reactions of ketonized substrate-analogs. J. Biol. Chem. 259:4476–4486. [PubMed] [Google Scholar]
- Whittaker, J. W., J. D. Lipscomb, T. A. Kent, and E. Münck. 1984. Brevibacterium fuscum protocatechuate 3,4-dioxygenase: Purification, crystallization, and characterization. J. Biol. Chem. 259:4466–4475. [PubMed] [Google Scholar]
- Wickman, H. H., M. P. Klein, and D. A. Shirley. 1965. Paramagnetic resonance of Fe3+ in polycrystalline ferrichrome A. J. Chem. Phys. 42:2113–2117. [Google Scholar]