Abstract
The interaction of bovine serum albumin (BSA) with cationic surfactant cetylpyridinium bromide (CPB) in aqueous solution (pH 7.00) was studied quantitatively with ultraviolet (UV)-visible, far-UV, and near-UV circular dichroism, fluorescence, small angle x-ray scattering, and nuclear magnetic resonance measurement. It was found that CPB at low and high concentrations could induce the unfolding and refolding of BSA, respectively. We suggest that in the unfolding process, there existed BSA-CPB complex with the “necklace and bead” structure in which the unfolded BSA wrapped around CPB micelles, and that the hydrophobic interaction between the complexes led to the formation of large aggregates. The aromatic headgroup of CPB interacted with the tryptophan residues of BSA, resulting in the aromatic ring stacking between BSA and CPB. During the refolding process, the BSA molecule was penetrated into the rod micelle of CPB and the hydrophobic moiety of the BSA molecule was exposed outside while its hydrophilic part was hidden inside, thereby disrupting the aromatic ring stacking.
INTRODUCTION
Globular proteins are used as functional ingredients in health care and pharmaceutical products because of their ability to catalyze biochemical reactions, to be adsorbed on the surface of some substances, and to bind other molecules and form molecular aggregates (Dalgleish, 1996). The protein-surfactant interactions are important because they can shed light on the functional properties. Over the past decade, protein-surfactant complexes have been studied extensively in the field of chemistry, biochemistry, and food science. It is well known that surfactants bind strongly to protein, thereby inducing conformational changes in the protein. These conformational changes often result in changes of polarity and stability of the protein. Many methods have been used to investigate these changes, such as calorimetry for thermal stability (Kelley and McClements, 2003), turbidimetry for protein aggregation (Valstar et al., 2000), fluorometry for polarity (Ray and Chakrabarti, 2003), and NMR for structure of complex (McCoy and Wyss, 2002), etc. Different models to describe the saturated protein-SDS complex have been summarized by Guo (Guo and Chen, 1990) as follows: 1), the “rodlike particle model” (Reynolds and Tanford, 1970a,b); 2), the “flexible helix model” (Lundahl et al., 1986); 3), the “necklace model” (Shirahama et al., 1974); and 4), the “α-helix/random coil model” ((Mattice et al., 1976). Among them, the necklace model seems to be best supported by experimental results, obtained with techniques such as small-angle neutron scattering study (Ibel et al., 1990; Chen and Teixeira, 1986), viscometry (Shinagawa et al., 1993), and NMR (Turro and Lei, 1995), etc. It was reported that NMR could distinguish the two possible “necklace and bead” structures (Turro and Lei, 1995); in one the surfactant aggregate wraps around the hydrophobic parts of protein because of hydrophobic interactions, and in the other the protein wraps around the surfactant micelles. Recently, Gelamo and his co-workers (Gelamo et al., 2002) described the modeling of bovine serum albumin (BSA) with low concentration of anionic surfactant (SDS) and cationic surfactants (CTAC and HPS), which was monitored by fluorescence spectroscopy of the intrinsic tryptophan residues.
However, the surfactants used are typically anionic surfactants such as SDS because it has a stronger interaction with the protein compared to cationic surfactants (Nozaki et al., 1974; Ananthapadmanabhan, 1993). In addition, the surfactants are usually controlled to be at the low concentrations. It is valuable for us to know the changes of BSA at the high concentration of surfactants. Herein, we investigated in deeper details the interaction of BSA with aromatic cationic surfactants CPB from low to high concentrations, mainly at the concentrations of its first and second critical micelle concentration (CMC). The study also proposed the structural models of protein-CPB complex and evaluated the effect of the aromatic ring stacking between BSA and CPB.
MATERIALS AND METHODS
Materials
Unless otherwise noted, all reagents and solvents used in this study were analytical grade obtained from Sigma (St. Louis, MO). A Tris-HCl buffer was 0.05 mol/L and its pH was adjusted to 7.00 with HCl using a Delta 320-S acidity meter (Mettler Toledo, Shanghai, China). CPB was obtained from Chemical Company of China (Shanghai, China).
Methods
Ultraviolet-visible (UV-vis) spectroscopy was recorded on a Hitachi UV-4100 spectrophotometer at room temperature in a 1-cm quartz cuvette. Static fluorescence spectroscopy was recorded with a F4500 spectrofluorimeter (Hitachi, Tokyo, Japan) in a 1-cm quartz cuvette and the excitation and emission slits are both 5 nm with the scan speed of 1200 nm/min. Far-UV and near-UV circular dichroism (CD) spectroscopy was carried out on a JASCO J-725S spectropolarimeter (Easton, MD). The 1H-NMR spectroscopy was recorded with a Bruker (Billerica, MA) Avance-600 spectrometer (600 MHz) using D2O as solvent. Chemical shifts were reported in ppm (δ) relative to internal standard tetramethylsilane at 0.00 ppm. CMC data were measured on a processor tensiometer-K12 (with a precision of 0.01 mN × m−1; Krüss, Hamburg, Germany) by the Wilhelmy plate. Small angle x-ray scattering (SAXS) curves were obtained at the SAXS beam line of a Philips PW3830 x-ray generator (Eindhoven, The Netherlands) (50 K, 40 mA), at room temperature (22 ± 1°C), with radiation wavelength λ = 1.542 Å and sample-to-detector distance of 233 nm with an MBRAUN (Stratham, NH) position sensitive detector. The obtained curves (data collection of 16 min) were corrected and normalized by taking into account the decrease of the x-ray beam intensity during the experiment. Due to the samples' attenuation, the parasitic background (buffer solution) was subtracted.
RESULTS AND DISCUSSIONS
UV absorption and CD spectroscopy
The strong binding of surfactants to proteins usually leads to substantial changes in protein conformation. In this study, we found that there was a dramatic decrease in the absorbance of BSA at 200 nm that corresponds to the absorption of protein backbone after the addition of CPB (see Fig. 1 a) (Gaggelli et al., 2002; Caspi and Ben-Jacob, 2000; Tao, 1981). The decrease was considered to be the result of the conformational changes due to the increase of the α-helix content (Gaggelli et al., 2002; Tao, 1981). This surfactant-induced unfolding process could be illuminated by the curve of the absorbance variation ΔA caused by the addition of CPB versus the logarithm of the concentration of surfactant (Fig. 1 b). Five characteristic regions were observed, indicating the binding of surfactant as a ligand to BSA. The curve in the left four regions (from 1 to 4) was in good agreement with the binding isotherms reported previously (Jones, 1975). The main unfolding process of the protein was believed to occur in the cooperative binding region 3 and the normal micelle formation occurred as excess surfactant was added in the region 4. Thus, it is believed that at the inflexion point between the 3 and 4 regions, the maximum unfolding occurs when CPB concentration is 1.00 × 10−5 mol/L, which is near the first CMC of CPB (1.50 (±0.01) × 10−5 mol/L) in the absence of protein as determined by the Wilhelmy-plate method. It is deduced that the unfolding is completed when CPB reaches its first CMC.
FIGURE 1.
Absorption spectra of BSA Conditions are: BSA, 1.0 × 10−3 g/mL; Tris-HCl, 5.0 × 10−3 mol/L, pH = 7.00. (a) BSA with different concentration of CPB (from 1 to 18 × 10−5 mol/L); 0, 0.10, 0.20, 0.30, 0.40, 0.50, 0.60, 0.70, 0.80, 1.00, 1.50, 5.0, 8.0, 9.0, 9.1, 9.2, 9.3, and 9.4. (b) The curve of absorbance increment (ΔA) versus the concentration of CPB (ΔA = A0 − A, A0 and A are the absorbances without and with CPB, respectively).
When CPB concentration was 9.0 × 10−5 mol/L, the backbone absorption peak recovered and appeared at ∼225 nm. The absorbance and the peak position moved toward shorter wavelengths with the increase of CPB concentration. And these changes were presented in region 5 of Fig. 1 b. Folding reoccurs for BSA, possibly meaning a renaturing process. However, in this region BSA differs from native BSA because this peak does not come back to 200 nm, indicating the changes in polarity of the microenvironment. In addition, the second CMC of CPB in the absence of BSA was obtained to be 8.50 (±0.13) × 10−5 mol/L with pyrene as a fluorescence probe according to the reported method (Li et al., 1985). So this alteration of secondary structure of BSA occurred near the second CMC of CPB.
Far-UV CD spectroscopy gave quantitative information on the secondary structure mentioned above (Fig. 2 a). The spectra (normalized to residue molar ellipticity) were analyzed with the software DICROPROT by self-consistent method to get structural information from the spectral changes (Deleage, 1993). At the first CMC, the CD spectra gave a large value of molar ellipticity at 222 and 208 nm, indicating an augmentation of the α-helix content in BSA structure from 49 to 78%; at the same time, the β-sheet content decreased from 9.8 to 7.6%. When CPB concentration reached the second CMC, the molar ellipticity decreased, yielding a broad minimum at 217 nm, where α-helix was calculated to be 37% and β-sheet to be 10.5%.
FIGURE 2.
Far-UV CD (a) and near-UV CD (b) spectra of BSA and BSA in CPB. BSA (1); BSA in 1.0 × 10−5 mol/L CPB (2); BSA in 1.0 × 10−4 mol/L CPB (3).
To obtain more information about the structural alteration of BSA, the near-UV CD spectra, which can infer the tertiary structure of BSA (Dockal et al., 2000; Kelly and Price, 1997; Price, 2000), were recorded at different CPB concentrations, and the results were shown in Fig. 2 b. It was shown that there were two minima at 261 and 268 nm as well as two shoulders at 277 and 284 nm. An increase in the ellipticity at 261 and 268 nm and a decrease from 280 to 300 nm at the first CMC of CPB denote a change of tertiary structure, especially the loss in asymmetry around disulfide bridges and the changes in the environment of the tryptophan residue. Under this condition, BSA is less compact compared to the native state. When CPB concentration reached its second CMC, there was a slight loss in the CD signals at 261 and 268 nm compared with those of native BSA, reflecting the perturbations that existed around numerous disulfide bridges, which resulted in the revivification of tertiary structure. Apparently, this revivification is not the same as that of native BSA based on their different near-UV CD spectra. In addition, a small raise in ellipticity in the region between 280 and 300 nm was observed, which was ascribed to the changes in the environment of the tryptophan residues.
Based on the results mentioned above, we suggest that at low CPB concentration, the surfactant molecules bind on the surface of protein through electrostatic and hydrophobic interactions, causing the protein to expand somewhat. Large amounts of CPB molecules are adsorbed on the surface of the protein with the increasing concentrations of CPB, resulting in the formation of the aggregates (such as micelle-like clusters). The intrachain hydrophobic interaction is replaced by the repulsive force, leading to the expanding of polypeptide and the unfolding of BSA, and finally destroys the tertiary structure.
When CPB concentration reaches its second CMC in BSA-CPB system, the surfactant molecules aggregate in the big rod micelles with strong hydrophobic inner core, resulting in stronger hydrophobic interaction between CPB and BSA in comparison to their electrostatic force. In addition, the dimension of rod micelle is similar to that of BSA (May and Ben-Shaul, 2001). Far-UV and near-UV CD studies also indicate that the secondary and tertiary structures of BSA under this condition were compressed and refolded. On the basis of above reasons, we propose a structural model of CPB-BSA complex at the second CMC of CPB in which BSA molecule penetrates into the rod micelle of CPB. In this packing model, the rod micelle limits the expanding of BSA molecule. On the other hand, the stronger hydrophobic interaction makes the hydrophobic part of the BSA molecule expose outside while its hydrophilic part is totally buried inside. Thus, BSA is refolded again, but the refolding is different from that in the aqueous environment, and refolded structure is different from the native BSA according to their far-UV and near-UV CD spectra.
1H-NMR studies
Here, we chose the NMR method to obtain the information on the mobility of different parts of surfactant molecule, e.g., headgroup region versus tails, and in turn, provided the information on the location of the protein in relation to the surfactant headgroups and the tails in the complex. In this article NMR spectra of 1.0 × 10−5 and 9.0 × 10−5 mol/L CPB and their complexes with 1.0 × 10−4 g/mL BSA were shown in Fig. 3. Corresponding to the pyridine in the headgroup of CPB, there were three clusters signals (Sun et al., 2004); the peaks at the chemical shift of ∼9.169, 8.774, and 8.298 ppm in Fig. 3 (1) corresponded to the H-2, H-1, and H-3, respectively. When CPB was mixed with BSA near the first CMC, three signals were broadened and moved to upfield significantly. The broadening effect indicates a significant reduction in the headgroup mobility and the involvement of the headgroup in the protein-surfactant aggregate, which supports the model that the protein wraps around the micelles or premicelles.
FIGURE 3.
1H-NMR spectra. (1) 1.0 × 10−5 mol/L CPB; (2) 1.0 × 10−5 mol/L CPB in 1.0 × 10−5 g/mL BSA; (3) 9.0 × 10−5 mol/L CPB in 1.0 × 10−5 g/mL BSA.
The upfield shift of these three signals of CPB in the presence of BSA is considered to be the result of aromatic ring stacking (Liu et al., 1999) between the pyridine in CPB and the residues (such as tryptophan, tyrosine, or phenylalanine) in BSA. Governed by the position of the protons relative to the ring, the magnitude of the chemical shift Δδ may serve as a measure of the extent of stacking. The strength of aromatic ring stacking in solution may be evaluated by stacking efficiency  (Ladokhin et al., 1999), which can be calculated from Eq. 1:
(Ladokhin et al., 1999), which can be calculated from Eq. 1:
 |
(1) |
where 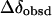 and
and 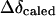 are observed upfield shift value and the shift expected for complete stacking, respectively. And
are observed upfield shift value and the shift expected for complete stacking, respectively. And 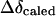 can be estimated according to a simple model of aromatic ring stacking:
can be estimated according to a simple model of aromatic ring stacking:
 |
(2) |
The association constant K is defined by:
 |
(3) |
 can be calculated by
can be calculated by  usingEq. 4:
usingEq. 4:
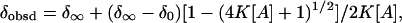 |
(4) |
where  and
and  represent the shift in monomeric A and in a complete stacking, respectively.
represent the shift in monomeric A and in a complete stacking, respectively.
In our system, K in Eq. 3 was calculated by fluorophotometry (Guo et al., 2003) according to Eq. 5:
 |
(5) |
where F and F0 are the fluorescence intensity of BSA in the presence and absence of CPB, respectively, and [Q] is the concentration of CPB. The obtained stacking efficiencies were listed in Table 1.
TABLE 1.
Upfield shifts (observed and calculated) for aromatic ring stacking and its efficiency
| H | 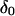 |
 |
 |
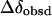 |
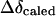 |
 |
|---|---|---|---|---|---|---|
| H-1 | 8.774 | 8.689 | 8.577 | 0.085 | 0.197 | 0.43 |
| H-2 | 9.169 | 8.990 | 8.934 | 0.179 | 0.235 | 0.76 |
| H-3 | 8.298 | 8.213 | 8.183 | 0.085 | 0.115 | 0.73 |
From this table, it can be seen that  for H-2 and H-3 are 0.179 and 0.085, and the corresponding efficiency are 0.76 and 0.73, respectively. This indicates that these two protons are the closest to the aromatic rings of BSA.
for H-2 and H-3 are 0.179 and 0.085, and the corresponding efficiency are 0.76 and 0.73, respectively. This indicates that these two protons are the closest to the aromatic rings of BSA.
When CPB reached its second CMC, these signals become as narrow as those of the pure CPB except a little upfield moving, indicating that the headgroups of CPB gain their mobility again due to the departure from the BSA surface. This result is considered as the form of rod micelles at CPB's second CMC. And the upfield moving in the spectra is considered to be the result of the weak aromatic rings stacking between the CPB molecules themselves.
Fluorescence spectroscopy
Fluorescence measurements showed that the addition of CPB could markedly influence the fluorescence of BSA. At the excitation wavelength of 290 nm, BSA was excited and the emission peak was at 350 nm. When a low concentration of CPB was added, the BSA emission was enhanced (Fig. 4 a). This enhancing effect reached a maximum at 1.25 × 10−5 mol/L CPB and remained constant until 7.50 × 10−5 mol/L, then the fluorescence of BSA was quenched suddenly, as shown in Fig. 4 b. The second-order derivative fluorescence spectra were recorded to analyze the details of this enhancement, which were shown in Fig. 4 c. The peak at 280 nm was attributed to tyrosine residue and the peak at 320 nm was attributed to tryptophan residue (Wang et al., 2000). In the presence of CPB, the peak of tryptophan residue increases significantly, indicating that it is the main contributor for the BSA fluorescence. The I− quenching test proved that the dosage of I− required to quench the fluorescence of unfolded BSA in the first CMC of CPB was about twice that required for native BSA. There are two tryptophan residues in one BSA molecule: Trp-134 in the eighth helix of D129-R144 in domain I, and Trp-213 in the second helix of E206-F221 in domain II (McLachlan and Walker, 1977). Only the helix of Trp-134 is active to interact with ligand in native BSA (similar to known crystallographic structure of human serum albumin (PDB1BJ5)) (shown in Fig. 5). As a ligand, CPB with low concentration primarily acts on Trp-134 through the aromatic ring stacking, which was proved by our NMR measurement above. With the increase of CPB concentration, Trp-213 gradually is exposed to the surface of BSA and interacts with CPB due to the unfolding of BSA. When CPB reaches its second CMC and above, BSA is refolded and tryptophan residues are hidden from CPB, and accordingly the aromatic ring stacking between CPB and the tryptophan residues disappears.
FIGURE 4.
The fluorescence spectra of BSA and BSA in CPB. (a) BSA (1); BSA in CPB (2) (vis CPB as blank). (b) BSA fluorescence enhancement factor at different CPB concentration. (c) The second derivative fluorescence, BSA (1); BSA-CPB (2). Conditions are: BSA, 1.0 × 10−4 g/mL; CPB, 1.0 × 10−3 mol/L; Tris-HCl, 5.0 × 10−3 mol/L, pH = 7.00.
FIGURE 5.
Structure of BSA and its unfolded structure in CPB, and the aromatic ring stacking between tryptophan residues and CPB, and rod micelle of CPB-BSA.
SAXS data analysis
The overall changes of the protein structure can also be revealed by means of the SAXS measurement. The SAXS intensity I(q) of an isotropic solution of a monodispersed particle of a small anisometry depends on both the form and structure factors (Guinier and Fournet, 1955):
 |
(6) |
where P(q) is a function of the particle size and shape (form factor) and S(q) is a function of the interparticle interactions (structure factor) ( the scattering vector; 2θ, the scattering angle). In this study, the concentrations of both of BSA and CPB were low enough that they could be considered as a widely separated system (not interaction system, S(q) ≈ 1). Hence, I(q) can be represented as (Improta et al., 1998):
the scattering vector; 2θ, the scattering angle). In this study, the concentrations of both of BSA and CPB were low enough that they could be considered as a widely separated system (not interaction system, S(q) ≈ 1). Hence, I(q) can be represented as (Improta et al., 1998):
 |
(7) |
The inverse Fourier transform of I(q) yields P(r), which is the frequency of vectors connecting small-volume elements within the entire volume of the scattering particles (Glatter and Krathy, 1982):
 |
(8) |
The behavior of P(r) provides the information about the shape of the scattering particles, thus it is extremely sensitive to the disposition of domains within a multidomain structure, and it goes to zero at the maximum linear dimension Dmax of the molecule. Moreover, the particle radius of gyration Rg is derived from P(r) and gives a simple measure of the distribution of volume elements within the molecule away from its center-of-mass, which is defined as the root-mean square of all elemental volumes from the center-of-mass of the particle, weighted by their scattering densities.  can be represented as (Glatter and Krathy, 1982):
can be represented as (Glatter and Krathy, 1982):
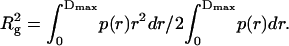 |
(9) |
The SAXS experimental results are presented in Fig. 6. It can be seen that there are obvious difference between the curves of BSA in the absence and presence of CPB at both the first CMC and above the second CMC. Compared to the native BSA, two distinct regions in the SAXS curve are marked when CPB is added. One has a greater slope than that of native BSA in the small q region, suggesting an increase in the scattering particle size, and another one has a different decay in the scattering curve, indicating changes of the protein shape. The data were analyzed through the distance distribution function p(r) using the GNOM program for Eqs. 8 and 9 (GNOM program is available free at ftp://ftp.embl-hamburg.de/szx.) When CPB is at its first CMC, calculated values of Rg and Dmax are 41.4 and 145 Å for BSA-CPB, and 30.2 and 95 Å for the native BSA, indicating a CPB-induced protein expanding. Whereas, when CPB concentration reaches its second CMC, Rg and Dmax decrease to 34.1 and 99 Å, which are a little bigger than that of native BSA but obviously smaller than those in BSA-CPB at its first CMC. Therefore, there is clear evidence that the expansion of BSA is limited and the size of the complex is decreased. In addition, it can also be seen from Fig. 6 that both Rg and the scattering intensity (I) have significant enhancement at the first CMC of CPB, which is considered to be the result of the aggregation between the clusters of BSA-CPB via the interaction between the hydrophobic patches exposed on the polypeptides chains in BSA. On the contrary, both Rg and I decrease at the second CMC of CPB, which indicates the decreasing of the aggregation mentioned above.
FIGURE 6.
Small angle x-ray scattering curves from 1 wt% of BSA, in the absence (1) and presence of 1.0 × 10−5 mol/L CPB (2) and 1.0 × 10−4 mol/L CPB (3).
Fig. 7 showed the scattering curves of aqueous solution containing CPB at both its first CMC and above second CMC with and without BSA. It can be seen that q value of CPB is smaller in comparison to that of the BSA-CPB complex at first CMC of CPB, indicating the CPB micelle in pure CPB solution is bigger than that in the presence of BSA. Whereas at the second CMC of CPB, q value of CPB is bigger than that of the complex, indicating the dimension of rod CPB micelle in the presence of BSA is bigger than that of rod micelle of pure CPB.
FIGURE 7.
Small angle x-ray scattering curves of 1.0 × 10−5 mol/L CPB (1) and BSA (2), and 1.0 × 10−4 mol/L CPB (3) and BSA (4).
SAXS measurement indicates that the aggregate formed among the BSA-CPB complexes at the first CMC of CPB is significantly larger than that at the second CMC of CPB. The above result provides an evidence for the structure models proposed. At the first CMC, the small micelles are bound on the surface of BSA to form a BSA-CPB complex, in which the expanded BSA wraps the micelles. The hydrophobic interaction among the hydrophobic patches exposed on the polypeptide chains of BSA causes BSA-CPB complexes to aggregate. At the second CMC of CPB, the BSA molecules are packed into the rod micelle of CPB that has the positively charged outer surface. The repulsive forces among rod micelles make them apart from each other. Therefore, the dimensions of the aggregates formed, respectively, at first and second CMC of CPB have obvious difference due to the different structure models.
CONCLUSIONS
In this work, the overall interaction between cationic aromatic surfactant CPB and BSA is quantitatively tracked, which is emphasized at the first CMC and the second CMC of CPB using multitechniques. The UV-vis absorption spectroscopy indicated the initial unfolding and later refolding processes of BSA induced by CPB, which also caused the changes in the fluorescence of BSA. By means of far-UV and near-UV CD spectroscopy, we investigated the changes of the secondary and tertiary structure of BSA at both the first and the second CMC of CPB. NMR measurement provided the evidence for both the structure models and aromatic ring stacking in protein-surfactant complex. SAXS measurement was used to investigate the shape, size, and formation of BSA-CPB aggregates. On the basis of the investigations mentioned above, we proposed two structural models of BSA-CPB complex and discussed the aromatic ring stacking between CPB and BSA in both unfolding and refolding. Therefore, this work is valuable for the understanding of the interaction mechanism between protein and surfactants, and potential for the applications in drug delivery, cosmetics, and detergency.
Acknowledgments
We thank Dr. Min Wang (Dept. of Chemistry, Duke University, Durham, NC), Dr. Fang Huang (MRC Laboratory of Molecular Biology, Cambridge, UK), and Dr. Rutao Liu (Albert Einstein College of Medicine, Yeshiva University, New York) for their kindly help and suggestions on the writing and revision of this article.
This work was supported by Natural Science Foundation of Shandong Province, and by Visiting Scholar Foundation of Key Laboratory at Shandong University.
References
- Ananthapadmanabhan, K. P. 1993. Protein-surfactant interactions. CRC Press, Boca Raton, FL. 319–366.
- Caspi, S., and E. Ben-Jacob. 2000. Conformation changes and folding of proteins mediated by Davydov's solution. Phys. Lett. A. 272:124–129. [Google Scholar]
- Chen, S. H., and J. Teixeira. 1986. Structure and fractal dimension of protein-detergent complexes. Phys. Rev. Lett. 57:2583–2586. [DOI] [PubMed] [Google Scholar]
- Clegg, R. M. 1995. Fluorescence resonance energy transfer. Curr. Opin. Biotechnol. 6:103–110. [DOI] [PubMed] [Google Scholar]
- Dalgleish, D. G. 1996. Food emulsions. In Emulsions and Emulsion Stability. J. Sjoblom, editor. Marcel Dekker, New York.
- Deleage, G. 1993. DICROPROT, version 2.5. Institute of Biological Chemistry of Proteins, Lyon, France. http://www.ibcp.fr. [Online].
- Dockal, M., D. C. Carter, and F. Ruker. 2000. Conformational transitions of the three recombinant domains of human serum albumin depending on pH. J. Biol. Chem. 275:3042–3050. [DOI] [PubMed] [Google Scholar]
- Gaggelli, E., F. Berti, N. D'Amelio, N. Gaggelli, G. Valensin, L. Bovalini, A. Paffetti, and L. Trabalzini. 2002. Metabolic pathways of carcinogenic chromium. Environ. Health Perspect. 110:733–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelamo, E. L., C. H. T. P. Silva, H. Imasato, and M. Tabak. 2002. Interaction of bovine (BSA) and human (HSA) serum albumins with ionic surfactants: spectroscopy and modeling. Biochim. Biophys. Acta. 1594:84–99. [DOI] [PubMed] [Google Scholar]
- Glatter, O., and O. Krathy. 1982. Small Angle X-Ray Scattering. O. Glatter and O. Krathy, editors. Academic Press, London, UK.
- Guinier, A., and G. Fournet. 1955. Small Angle Scattering of X-Ray. A. Guinier and G. Fournet, editors. Wiley, New York.
- Guo, X. H., and S. H. Chen. 1990. The structure and thermodynamics of protein-SDS complexes in solution and the mechanism of their transports in gel electrophoresis process. Chem. Phys. 149:129–139. [Google Scholar]
- Guo, C. C., H. P. Li, X. B. Zhang, and R. B. Tong. 2003. Synthesis of meso-5, 10, 15, 20-tetra [4-(N-pyrrolidinyl)phenyl] porphyrin and its interaction with bovine serum albumin. Chem. J. Chinese Universities. 24:282–287. [Google Scholar]
- Ibel, K., R. P. May, K. Kirschner, H. Szadkowski, E. Mascher, and P. Lundahl. 1990. Protein-decorated micelle structure of sodium-dodecyl-sulfate-protein complexes as determined by neutron scattering. Eur. J. Biochem. 190:311–318. [DOI] [PubMed] [Google Scholar]
- Improta, S., J. K. Krueger, M. Gaute, and R. A. Atkinson. 1998. The assembly of immunoglobulin-like modules in titin: implications for muscle elasticity. J. Mol. Biol. 284:761–777. [DOI] [PubMed] [Google Scholar]
- Jones, M. N. 1975. A theoretical approach to the binding of amphipathic molecules to globular proteins. Biochem. J. 151:109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, D., and D. J. McClements. 2003. Interactions of bovine serum albumin with ionic surfactants in aqueous solutions. Food Hydrocolloids. 17:73–85. [Google Scholar]
- Kelly, S. M., and N. C. Price. 1997. The application of circular dichroism to studies of protein folding and unfolding. Biochim. Biophys. Acta. 1338:161–185. [DOI] [PubMed] [Google Scholar]
- Ladokhin, A. S., M. E. Selsted, and S. H. White. 1999. CD spectra of indolicidin antimicrobial peptides suggest turns, not polyproline helix. Biochemistry. 38:12313–12319. [DOI] [PubMed] [Google Scholar]
- Li, G., Y. Lin, Y. Gu, J. Shen, and J. Yang. 1985. The determination of the second CMC of surfactants by fluorescence. Journal of Shandong University. 4:111–115 (Natural Science Edition). [Google Scholar]
- Liu, D. H., D. A. Williamson, M. L. Kennedy, T. D. Williams, M. M. Morton, and D. R. Benson. 1999. Aromatic side chain-porphyrin interactions in designed hemoproteins. J. Am. Chem. Soc. 121:11798–11812. [Google Scholar]
- Lundahl, P., E. Greijer, M. Sandberg, S. Cardell, and K. O. Eriksson. 1986. A model for ionic and hydrophobic interactions and hydrogen-bonding in sodium dodecyl sulfate-protein complexes. Biochim. Biophys. Acta. 873:20–26. [Google Scholar]
- Mattice, W. L., J. M. Riser, and D. S. Clark. 1996. Conformational properties of the complexes formed by proteins and sodium dodecyl sulfate. Biochemistry. 15:4264–4272. [DOI] [PubMed] [Google Scholar]
- May, S., and A. Ben-Shaul. 2001. Molecular theory of the sphere-to-rod transition and the second CMC in aqueous micellar solutions. J. Phys. Chem. B. 105:630–640. [Google Scholar]
- McCoy, M. A., and D. F. Wyss. 2002. Structures of protein-protein complexes are docked using only NMR restraints from residual dipolar coupling and chemical shift perturbations. J. Am. Chem. Soc. 124:2104–2105. [DOI] [PubMed] [Google Scholar]
- McLachlan, A. D., and J. E. Walker. 1977. Profile analysis: detection of distantly related proteins. J. Mol. Biol. 112:543–558.875033 [Google Scholar]
- Nozaki, Y., J. A. Reynolds, and C. Tanford. 1974. The interaction of a cationic detergent with bovine serum albumin and other proteins. J. Biol. Chem. 249:4452–4459. [PubMed] [Google Scholar]
- Price, N. C. 2000. Conformational issues in the characterization of proteins. Biotechnol. Appl. Biochem. 31:29–40. [DOI] [PubMed] [Google Scholar]
- Ray, S., and A. Chakrabarti. 2003. Erythroid spectrin in miceller detergents. Cell Motil. Cytoskeleton. 54:16–28. [DOI] [PubMed] [Google Scholar]
- Reynolds, J. A., and C. Tanford. 1970a. Binding of Dodecyl Sulfate to Proteins at High Binding Ratios. Possible Implications for the State of Proteins in Biological Membranes. Proc. Natl. Acad. Sci. USA. 66:1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, J. A., and C. Tanford. 1970b. The Gross Conformation of Protein-Sodium Dodecyl Sulfate Complexes. J. Biol. Chem. 245:5161–5165. [PubMed] [Google Scholar]
- Shinagawa, S., K. Kameyama, and T. Takagi. 1993. Effect of salt concentration of buffer on the binding of sodium dodecyl sulfate and on the viscosity behavior of the protein polypeptide derived from bovine serum albumin in the presence of the surfactant. Biochim. Biophys. Acta. 1161:79–84. [DOI] [PubMed] [Google Scholar]
- Shirahama, K., K. Tsujii, and T. Takagi. 1974. Free-boundary electrophoresis of sodium dodecyl sulfate-protein polypeptide complexes with special reference to SDS-polyacrylamide gel electrophoresis. J. Biochem. (Tokyo). 75:309–319. [DOI] [PubMed] [Google Scholar]
- Sun, C. X., J. H. Yang, X. Wu, B. Y. Su, and S. F. Liu. 2004. Fluorescent enhancement effect in terbium-gadolinium-protein-cetylpyridine bromide system and its application for the determination of protein at nanogram level. Chem. Phys. Lett. 398:343–350. [Google Scholar]
- Tao, W. S. 1981. Protein Macular Basic. The People's Education Press, China.
- Turro, N. J., and X. G. Lei. 1995. Spectroscopic probe analysis of protein-surfactant interactions: the BSA/SDS system. Langmuir. 11:2525–2533. [Google Scholar]
- Valstar, A., M. Almgren, W. Brown, and M. Vasilescu. 2000. The interaction of bovine serum albumin with surfactants studied by light scattering. Langmuir. 16:922–927. [Google Scholar]
- Wang, H. Y., Q. S. Hui, L. D. Liu, Y. Sun, and L. Ma. 2000. Simultaneous determination of tryptophan and tyrosine by second order derivative fluorimetry. Spectroscopy and Spectral Analysis. 6:427–430. [PubMed] [Google Scholar]









