Abstract
We report infrared microspectral features of nuclei in a completely inactive and contracted (pyknotic) state, and of nuclei of actively dividing cells. For pyknotic nuclei, the very high local concentration of DNA leads to opaqueness of the chromatin and, consequently, the absence of DNA signals in the IR spectra of very small nuclei. However, these nuclei can be detected by their scattering properties, which can be described by the Mie theory of scattering from dielectric spheres. This scattering depends on the size of the nucleus; consequently, quite different scattering cross-sections are calculated and observed for pyknotic and mitotic nuclei.
INTRODUCTION
Infrared microspectroscopy (IR-MSP) is poised to become a powerful method for objective diagnosis in medicine, since it monitors small changes in molecular composition between tissue types, and between normal and diseased tissue. Data published as early as 1998 have demonstrated that IR-MSP, coupled to sophisticated methods of data analysis, can reliably identify cancerous areas in biopsies (Lasch and Naumann, 1998). Furthermore, these studies have demonstrated that patient-to-patient spectral variations are smaller than those observed between different tissue types, and stages of disease.
Similar sensitivity of IR-MSP has been demonstrated in the identification and classification of bacteria, where it is possible to reliably identify bacterial microcultures to the strain level (Maquelin et al., 2002). This method has being used, for example, in hospitals in France to monitor outbreaks of bacteria-caused diseases (Sockalingum et al., 2004), and to identify benign and pathogenic bacterial strains in the dairy product industry in Germany (Kümmerle et al., 1998).
The use of IR-MSP to distinguish individual benign from malignant exfoliated cells has not yet been successful, although it was first attempted as early as the mid-1990s. These early attempts were aimed at creating a spectroscopic Papanicolaou (Pap) test for exfoliated cells from the human cervix (Wong et al., 1991) to increase the low predictive value of present cytological methods. Early attempts used macroscopic measurements of sample pellets containing thousands of cervical cells. However, due to uncontrollable heterogeneity of the sample, and a poor understanding of the underlying cell spectroscopic principles, the results to date have been spurious, and the correlation against an unreliable gold-standard has been fortuitous (Wong et al., 1991; Sindhuphak et al., 2003).
We have initiated the ground work for the understanding of the spectroscopic properties of single dried or living human cells by studying cultured human and animal cells as a function of typical parameters which are known to influence cell development (Boydston-White et al., 1999; Pacifico et al., 2003; Diem et al., 2002, 2004a; Lasch et al., 2002b; Miljković et al., 2004). Early in this work, we realized that human cells exhibit spectral behavior that appears to contradict the Beer-Lambert law (Diem et al., 1999). In particular, we found that the DNA signals in pyknotic nuclei cannot be observed. Pyknosis is the shrinkage of inactive nuclei due to increased condensation of DNA.
Furthermore, very atypical spectra were observed for certain cells, apparently related to the cell division (Boydston-White et al., 1999; Holman et al., 2000). The most unusual of these spectra, reported by Holman and co-workers and confirmed by us (Diem et al., 2004a), have not been explained hitherto. In this article, we present the framework for the spectral properties that do not follow Beer-Lambert's law, and those that result from light scattering effects.
EXPERIMENTAL METHODS
Cell culture as well as spectroscopic experimental details have been reported in the publications in which the unusual spectral properties of cells were first reported, and will not be iterated here. May it suffice to state that the unusual spectral properties were observed using three different microspectrometers at Hunter College of the City University of New York, and at synchrotron-based facilities. The instruments at Hunter College include a Bruker (Billerica, MA) IRScope II, coupled to a Bruker Vector 22 optical bench, a SensIR Technologies (Danbury, CT) IlluminatIR/Olympus BX40 system, and a Perkin Elmer (Shelton, CT) Spectrum One/Spotlight 300 microspectrometer. The synchrotron-based instrument was a Nicolet Continuum Microspectrometer at beamline U10B at the National Synchrotron Light Source at Brookhaven National Laboratories (Upton, NY).
RESULTS AND DISCUSSION
Non-Lambert-Beer absorption behavior of pyknotic nuclei
Several studies have reported the infrared spectra of nuclei of human cells, which show, as expected, spectral features due to DNA and proteins (Diem et al., 2002; Jamin et al., 1998; Lasch et al., 2002a). In the nuclei, the spectra of proteins dominate those of all other biochemical constituents; furthermore, the protein signals in the nucleus are nearly 10 times stronger than those collected outside the nucleus in a typical cell. This is expected, since the nucleus in a dried cell offers a longer pathlength to the probing IR radiation; furthermore, the nucleus contains an extremely high concentration of proteins. Typical protein signals (reported for the amide I band at ∼1650 cm−1) from a nucleus are between 0.2 and 0.4 absorbance units.
DNA signals in the nucleus are manifested by bands at ∼1235 and ∼1085 cm−1 due to the antisymmetric and symmetric stretching modes of the  moieties of DNA, respectively. An unambiguous assignment of these peaks to DNA in human tissue (Chiriboga et al., 2000) and cells (Lasch et al., 2002a) was established by selectively removing the DNA from the sample by digestion via the enzyme DNase, and correlating the disappearance of these peaks to the digestion. In active cells, the
moieties of DNA, respectively. An unambiguous assignment of these peaks to DNA in human tissue (Chiriboga et al., 2000) and cells (Lasch et al., 2002a) was established by selectively removing the DNA from the sample by digestion via the enzyme DNase, and correlating the disappearance of these peaks to the digestion. In active cells, the  stretching peaks of DNA may exhibit an amplitude of 0.05 absorbance units; the equally strong DNA peaks in the double-bond stretching regions cannot be observed directly since they overlap the amide I protein band.
stretching peaks of DNA may exhibit an amplitude of 0.05 absorbance units; the equally strong DNA peaks in the double-bond stretching regions cannot be observed directly since they overlap the amide I protein band.
However, we found that cells that are terminally differentiated and metabolically inactive often show no DNA signals in the nucleus at all. This was first observed for superficial cervical cells (Diem et al., 2002) and verified for oral mucosa cells (Lasch et al., 2002a). These cell types are similar in that they are classified as squamous epithelial cells. In squamous tissue, a basal layer of actively dividing cells, ∼0.1–0.2 mm below the surface, produce daughter cells that migrate to the surface. In this maturation process, the cells enlarge and become stratified (flattened). When these cells reach the surface of the tissue, they eventually die and are lost from the surface.
Although they are at the surface, their main task is to protect the underlying tissue; thus, they become inactive. These inactive cells can no longer divide, and their protein synthesis is at a minimal level. Therefore, the nuclei shrink to ∼5–8 μm in diameter. This state is referred to as a pyknotic, or completely inactive, nucleus. Pyknotic nuclei can be identified microscopically by their small size, and characteristic morphology. However, although these nuclei are no longer transcribing or duplicating their DNA, they still have the entire genome in a very condensed state.
A micrograph of an oral mucosa cell with a pyknotic nucleus is shown in Fig. 1 A, and corresponding IR spectra in Fig. 1 B. The similarity between cytoplasmic and nuclear spectra shown in Fig. 1 B, traces a and b, demonstrates that, indeed, the DNA is virtually invisible, whereas the DNA from the nucleus of an active cell, shown in trace c, is clearly observable.
FIGURE 1.
(A) Photomicrograph of dried oral mucosa cell. (B) IR spectrum of cytoplasm of oral mucosa cell (trace a); IR spectrum of nucleus of oral mucosa cell (trace b); and IR spectrum of nucleus of actively dividing cell (trace c).
We have interpreted the absence of DNA signals from pyknotic nuclei in terms of the following model calculation. The concentration of carbonyl groups from the DNA bases within a nucleus of 5 μm diameter is actually quite high, and can be estimated by assuming a nuclear volume,
 |
(1) |
and the lengths of the human DNA, 5 × 109 basepairs (based on an average of diploid and quadruploid cells). Thus, the concentration C of DNA carbonyl groups
 |
(2) |
Using the molar extinction coefficient ɛ for a carbonyl group (Xiang et al., 1993),
 |
(3) |
at 1650 cm−1 in the carbonyl stretching vibration, and Beer-Lambert's law
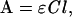 |
(4) |
the absorbance A of the DNA carbonyl groups in the nucleus is obtained as
 |
(5) |
Here, l is the path the light beam travels through the sample, and may be approximated by the diameter of the nucleus. This estimate assumes that the DNA is uniformly distributed in the nucleus, which is clearly not the case (see below).
Model IR spectroscopic studies on B-form DNA in vitro have demonstrated that the molar extinction coefficient for the symmetric and antisymmetric phosphodiester vibrations, at 1080 and 1235 cm−1, respectively, are similar to that of the carbonyl stretching vibrations (Wang and Keiderling, 1993). Thus, signal intensities similar to that in the carbonyl stretching region should be observed in the phosphodiester stretching vibrations.
However, the DNA is not distributed uniformly throughout the nucleus; rather, it is tightly wrapped around the histones to form chromatin. In chromatin, the DNA is extremely compact: depending on the degree of association (which depends on a cell's biological activity), the resulting structures may have a diameter between 30 and 800 nm. Assuming, for simplicity, that the DNA is packed into one spherical chromatin particle with a 500-nm diameter (with the same number of DNA basepairs), the concentration of carbonyl groups would increase 1000-fold, and the optical density 100-fold, since the pathlength decreases 10-fold:
 |
(6) |
One would expect that virtually no photons are transmitted within the strong absorption bands of DNA and proteins (which make up the chromatin) by particles of such high optical density. Furthermore, given the number of transitions and the width of the DNA and protein bands, the entire highly condensed chromatin would be virtually opaque. Thus, very little or no information on the condensed chromatin is contained in the infrared spectrum of a compact (pyknotic) nucleus. We have referred to the absence of DNA signals in pyknotic nuclei as dark DNA.
However, such highly condensed particles will exhibit large light scattering cross-sections. We have shown recently for mitotic cells that the strongest Raman signals are, indeed, observed for the most highly condensed chromatin (for example, in the metaphase and anaphase of mitosis; Diem et al., 2004b). Raman signals of the DNA of metabolically active, but nondividing cells, are quite weak.
Mie scattering from the entire nucleus, to be discussed in Light Scattering of Cells and Cellular Components, below, is expected to occur when the size of the nucleus is about the same as the wavelength of light, or in the infrared region of the spectrum. Elastic scattering of highly condensed, and much smaller chromatin particles, occurs at much lower wavelengths, and will not be discussed here.
Further evidence for dark DNA has been reported by a number of researchers. We found that avian erythrocytes (which contain inactive nuclei) and human erythrocytes (which are anucleated) exhibit virtually identical IR spectra (Fig. 2). Furthermore, Jamin et al. (2003) reported a large increase in the DNA signals of apoptotic cells. In apoptosis, DNA is degraded into oligonucleotides with a few hundreds of basepairs that subsequently diffuse out of the nucleus. The accumulation of these oligonucleotides was observed in high spatial resolution, high quality synchrotron FTIR microspectroscopic data. This observation suggests that the DNA, although highly condensed in the nucleus, is unobservable, yet its degradation products are readily detected by IR microspectroscopy.
FIGURE 2.
IR spectra of avian erythrocytes (with nuclei, shaded trace) and anucleated human erythrocytes (solid trace).
In Fig. 3, we present results that confirm that the nuclear DNA is not observed in absorption, yet the light scattering due to the nucleus is observable. The photomicrograph in Fig. 3 a shows a cell with the nucleus clearly visible. The aperture of the microspectrometer (PE Spectrum One/Spotlight 300) in single-point mode was set to straddle the nucleus, at 10 μm on edge. Individual spectra were subsequently acquired at positions indicated by the crosses along the black line. These spectra are shown in Fig. 3 b. This plot shows that the intensity of the spectral amplitude increases from the cytoplasm to the nucleus, and demonstrates again that in the strongest spectrum due to the nucleus, no DNA absorption signals are observed. However, we observe a broad spectral feature, centered at ∼2400 cm−1 between the amide I and the C-H stretching region, which occurs only in the nuclear spectrum. We attribute this feature to Mie-type scattering of the nucleus, to be discussed next.
FIGURE 3.
(a) Photomicrograph of unstained human buccal (oral mucosa) cell, with positions indicating the locations of spectral data acquisition. (b) Three-dimensional plot of IR spectra obtained at the positions indicated in a. Notice the absence of DNA features for the spectrum of the nucleus, and its broad scattering background, centered at ∼2400 cm−1.
Light scattering of cells and cellular components
Dielectric spheres are known to scatter electromagnetic radiation if the wavelength of the light is comparable to the size of the dielectric sphere. This scattering process was first described theoretically by Mie (1908).
The Mie scattering formalism can be approximated (Walstra, 1964) by the equation
 |
(7) |
where Q is the scattering cross-section, and
 |
(8) |
where a is the radius of the sphere, n the ratio of refractive indices inside and outside of the sphere, and λ the wavelength of the light. This set of equations, first derived by Walstra (1964), was selected since they are easily implemented, and approximate the scattering cross-section, predicted by the Mie theory, to within 1%. Typical scattering curves for spherical particles with a refractive index of 1.3 and computed according to Eq. 7 , are shown in Fig. 4. These curves show a first scattering maximum at a wavelength (expressed here in wavenumber) corresponding to the approximate size of the particle, followed by further scattering maxima of decreasing amplitudes toward higher wavenumber. We believe that scattering curves, such as those shown in Fig. 4, are responsible for the broad, undulating features frequently observed superimposed on the spectral absorption features of individual cells. A spectrum of a cell that includes such a scattering background, collected from 650 to 7000 cm−1, is shown in Fig. 5 (bottom trace). The absorption spectrum appears superimposed on a scattering contribution with a broad peak under the C-H stretching manifold. The onset of a second broadband appears at ∼6000 cm−1. The pure scattering spectrum, calculated for a nuclear radius of 4.2 μm, is shown in Fig. 5 (top trace). Based on an approximate size of the nucleus (r ≈ 4.5 ± 0.5 μm, judged by the selected aperture), we may estimate the refractive index of the nucleus to be on the order of 1.3 in the mid-to-near IR spectral region.
FIGURE 4.
Light scattering coefficients calculated according to Eq. 8, for dielectric spheres of radii indicated.
FIGURE 5.
IR spectrum of oral mucosa cell (bottom), and scattering spectrum for a dielectric sphere with a = 4.2 μm (top).
The aforementioned atypical absorption spectra of near-spherical, mitotic cells in the 900–1800 cm−1 region, first reported by Holman et al. (2000), can be explained similarly by scattering of a much larger spherical object. A spectrum of such a cell in the mid-IR region is shown in Fig. 6 (trace A). This spectrum is, obviously, superimposed on a broad background, which we believe is also due to scattering. A scattering spectrum, based on an object with a radius of 8.5 μm, is shown in Fig. 6 (trace B). Upon subtraction of this background, a spectrum is obtained with a reasonably straight baseline (Fig. 6, trace C). This spectrum shows extremely strong signals due to nucleic acids, indicating a very different level of activity in the cell's nucleus, possibly due to the onset of mitosis. (In the prophase of mitosis, the nucleus actually disappears, and all nuclear RNA spreads throughout the cell.) However, it is not clear at this point whether the scattering background is strictly due to the nucleus or the entire cell, or a combination of both. Spectra collected for other cells show the undulation of the background with more maxima and minima than expected from a single scattering object. Some of the spectra also show a small dispersion artifact, which was described by us recently (Romeo and Diem, 2005).
FIGURE 6.
(Trace A) Atypical IR spectrum of cultured cervical cancer cell, presumably at or close to mitosis. (Trace B) Scattering spectrum calculated for a = 8.5 μm. (Trace C) Difference spectrum, showing much improved background/baseline, and very strong nucleic acid features.
CONCLUSIONS
In this article, we demonstrate that spectra of cells may show effects that are usually absent or ignored in infrared spectroscopy, such as scattering and absorption behavior that does not obey Beer-Lambert's law. The scattering effects produce a broad, undulating background onto which the absorption features are superimposed. Depending on the positions of the scattering maxima, the observed spectral intensities may be distorted, and the baseline for absorption spectra may be difficult to define.
The nonlinear absorption observed for very compact nuclei suggests that nucleic acid features may be used as an indicator for a cell's level of activity: we have shown for many normal and cancerous cells, and cells at different stages of the cell cycle, that the level of nucleic acid signals can be related to the level of DNA condensation and, in turn, to the proliferation and activity level of a cell (Lasch et al., 2002; Miljković et al., 2004; Boydston-White et al., 2005). Similarly, the appearance of DNA signals from the cytoplasm of cells in late apoptosis (Jamin et al., 2003) indicates that after breakdown of nuclear DNA, and diffusion of DNA fragments into the cytoplasm, their signals become detectable in IR microspectroscopy.
These unusual spectra effects observed for individual cells, and the resulting heterogeneity of cellular spectra, need to be firmly understood before IR microspectroscopy can be utilized as a diagnostic indicator of disease.
Acknowledgments
Support of this research through grants from the American Cancer Society (ROG No. 99-119-01), from the National Institutes of Health (No. CA 81675, No. CA 090346, and No. GM 60654, all to M.D.), and from a “Research Centers in Minority Institutions” grant (National Center for Research Resources of the National Institutes of Health, award No. RR-03037) are gratefully acknowledged.
References
- Boydston-White, S., T. Gopen, S. Houser, J. Bargonetti, and M. Diem. 1999. Infrared spectroscopy of human tissue. V. Infrared spectroscopic studies of myeloid leukemia (ML-1) cells at different phases of the cell cycle. Biospectroscopy. 5:219–227. [DOI] [PubMed] [Google Scholar]
- Boydston-White, S., T. Chernenko, A. Regina, M. Miljković, C. Matthäus, and M. Diem. 2005. Microspectroscopy of single proliferating HeLa cells. Vibra. Spectrosc. In press.
- Chiriboga, L., H. Yee, and M. Diem. 2000. Infrared spectroscopy of human cells and tissue. VII FT-IR microspectroscopy of DNAse- and RNAse-treated normal, cirrhotic and neoplastic liver tissue. Appl. Spectrosc. 54:480–485. [Google Scholar]
- Diem, M., S. Boydston-White, and L. Chiriboga. 1999. Infrared spectroscopy of cells and tissues. Shining light onto an unsettled subject. Appl. Spectrosc. 53:148A–161A. [Google Scholar]
- Diem, M., P. Lasch, L. Chiriboga, and A. Pacifico. 2002. Infrared spectra and infrared spectral maps of individual normal and cancerous cells. Biopolym. Biospectrosc. 67:349–353. [DOI] [PubMed] [Google Scholar]
- Diem, M., M. Romeo, L. Miller, and P. Lasch. 2004a. Comparison of Fourier transform infrared (FTIR) spectra of individual cells acquired using synchrotron and conventional sources. IR Phys. Technol. 45:31–118. [Google Scholar]
- Diem, M., M. Romeo, S. Boydston-White, M. Miljović, and C. Matthäus. 2004b. A decade of vibrational microspectroscopy of human cells and tissue. Analyst. 129:880–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman, H. N., M. C. Martin, E. A. Blakely, K. Bjornstad, and W. R. McKinney. 2000. IR spectroscopic characteristics of cell cycle and cell death probed by synchrotron-based FTIR microspectroscopy. Biopolym. Biospectrosc. 57:329–335. [DOI] [PubMed] [Google Scholar]
- Jamin, N., P. Dumas, J. Moncuit, W. H. Fridman, J. L. Teillaud, G. L. Carr, and G. P. Williams. 1998. Chemical imaging of nucleic acids, proteins and lipids of a single living cell. Application of synchrotron infrared microspectrometry in cell biology. Cell. Mol. Biol. 44:9–14. [PubMed] [Google Scholar]
- Jamin, N., L. Miller, J. Moncuit, W. H. Fridman, P. Dumas, and J. L. Teillaud. 2003. Chemical heterogeneity in cell death: combined synchrotron IR and fluorescence microscopy studies of single apoptotic and necrotic cells. Biopolym. Biospectrosc. 72:366–373. [DOI] [PubMed] [Google Scholar]
- Kümmerle, M., S. Scherer, and H. Seiler. 1998. Rapid and reliable identification of food-borne yeasts by Fourier transform infrared (FT-IR) spectroscopy. Appl. Environ. Microbiol. 64:2207–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasch, P., and D. Naumann. 1998. FT-IR microspectroscopic imaging of human carcinoma in thin sections based on pattern recognition techniques. Cell. Mol. Biol. 44:189–202. [PubMed] [Google Scholar]
- Lasch, P., M. Boese, A. Pacifico, and M. Diem. 2002a. FT-IR spectroscopic investigations of single cells on the subcellular level. Vibra. Spectrosc. 28:147–157. [Google Scholar]
- Lasch, P., A. Pacifico, and M. Diem. 2002b. Spatially resolved IR microspectroscopy of single cells. Biopolym. Biospectrosc. 67:335–338. [DOI] [PubMed] [Google Scholar]
- Maquelin, K., L.-P. Choo-Smith, C. Kirschner, N. A. Ngo-Thi, D. Naumann, and G. J. Puppels. 2002. Vibrational spectroscopic studies of microorganisms. In Handbook of Vibrational Spectroscopy. J. M. Chalmers and P. R. Griffiths, editors. John Wiley and Sons Ltd., Chichester, UK. 1–27.
- Mie, G. 1908. Contribution to the optical properties of turbid media, in particular of colloidal suspensions of metals. Ann. Phys. (Leipzig). 25:377–452. [Google Scholar]
- Miljković, M., M. Romeo, C. Matthäus, and M. Diem. 2004. Infrared microspectroscopy of individual human cervical cancer (HeLa) cells suspended in growth medium. Biopolymers. 74:172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacifico, A., L. Chiriboga, P. Lasch, and M. Diem. 2003. Infrared spectroscopy of cultured cells. II. Spectra of exponentially growing, serum-deprived and confluent cells. Vibra. Spectrosc. 32:107–115. [Google Scholar]
- Romeo, M., and M. Diem. 2005. Correction of dispersive line shape artifacts observed in diffuse reflection Infrared spectroscopy and absorption/reflection infrared microspectroscopy. Vibra. Spectrosc. In press.
- Sindhuphak, R., S. Issaravanich, V. Udomprasertgul, P. Srisookho, S. Warakamin, S. Sindhuphak, R. Boonbundarlchai, and N. Dusitsin. 2003. A new approach for the detection of cervical cancer in Thai women. Gynecol. Oncol. 90:10–14. [DOI] [PubMed] [Google Scholar]
- Sockalingum, G. D., C. Sandt, V. Stoeckel, P. Imbs, P. Lemaire, H. Lepan, J. M. Pinon, M. Manfait, and D. Toubas. 2004. Combination of FTIR and rapid techniques for typing Candida albicans: followup on patients in ICU and Maternity Units. International Bunsen Discussion Meeting: Raman and IR Spectroscopy in Biology and Medicine, Jena, Germany.
- Walstra, P. 1964. Approximation formulae for the light scattering coefficient of dielectric spheres. Brit. J. Appl. Phys. 15:1545–1552. [Google Scholar]
- Wang, L., and T. Keiderling. 1993. Helical nature of poly(dI-dC) • poly(dI-dC). VCD results. Nucleic Acid Res. 21:4127–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, P. T. T., R. K. Wong, T. A. Caputo, T. Godwin, and B. Rigas. 1991. IR spectroscopy of exfoliated human cervical cells; evidence of extensive structural changes during carcinogenesis. Proc. Natl. Acad. Sci. USA. 88:10988–10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, T., D. J. Goss, and M. Diem. 1993. Strategies for the computation of VCD and infrared absorption of biological molecules: ribonucleic acids. Biophys. J. 65:1255–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]








