Abstract
The Drosophila GAGA factor (GAF) controls transcription and other chromosome functions by altering chromatin structure. We found that a second GAGA-binding protein of Drosophila, Pipsqueak (Psq), can directly bind to GAF and is associated with GAF in vivo. Genetic interaction studies provide evidence that Psq and GAF act together in the transcriptional activation and silencing of homeotic genes. A complete colocalization of Psq and GAF on polytene interphase chromosomes and mitotic chromosomes suggests that the two proteins cooperate as general partners not only at homeotic loci, but also at hundreds of other chromosomal sites.
Transcription and other chromosomal functions critically depend on alterations in chromatin structure collectively referred to as chromatin remodeling (for reviews see refs. 1 and 2). The Drosophila GAGA factor (GAF), which has initially been identified as a classical transcription factor (3–5), was soon suspected of acting through a chromatin remodeling mechanism (reviewed in ref. 6). In addition to its role in transcriptional activation, GAF was later implicated with such diverse functions as transcriptional silencing (7–9), enhancer blocking (10), and chromosome condensation and segregation during mitosis (11).
GAGA DNA-binding sites are essential for the transcriptional activation of many genes, including heat shock (hsp) genes (12) and developmental genes like Krüppel (Kr) and the homeotic gene Ultrabithorax (Ubx) (3, 13, 14). Binding of GAF counteracts the chromatin-mediated transcriptional repression of the Kr and hsp70 promoters (14, 15). At the hsp70 promoter, GAF directs the formation of an open chromatin structure, acting in concert with the nucleosome-remodeling factor NURF (16). A chromatin-opening function of GAF was further supported by the discovery that GAF is encoded by the Trithorax-like (Trl) gene, which is required for maintenance of the expression of homeotic genes (17). GAF thus shares an important function with the trithorax group (trxG) proteins, which are believed to act by counteracting chromatin-mediated gene silencing (18). Surprisingly, recent studies suggest that GAF is also required for the transcriptional silencing of homeotic genes by Polycomb response elements (PREs) (7–9, 19, 20), which is thought to be achieved by formation of a closed, heterochromatin-like structure (21). The repressive function of PREs is mediated by different multiprotein complexes that are assembled by at least 13 different Polycomb group (PcG) proteins (18). Because only one of these proteins, Pleiohomeotic, has a sequence-specific DNA-binding activity (22), a function of GAF may be to tether certain PcG complexes to their DNA target sites.
It has recently been observed that GAGA DNA elements can be bound by a second Drosophila protein, encoded by the pipsqueak (psq) gene (23). The Psq protein contains a novel DNA-binding domain, consisting of four tandem repeats of a 50-amino acid motif, which defines a growing family of eukaryotic helix-turn-helix proteins (24). In addition, Psq contains a conserved BTB (Broad-Complex, Tramtrack, Bric à brac) (25) or POZ (poxvirus and zinc finger) (26) domain, which is also present in GAF. The BTB domains of GAF and other proteins have been shown to mediate homologous and heterologous protein–protein interactions (26–29). Like the zinc finger DNA-binding domain of GAF, the Psq DNA-binding domain binds to DNA elements that contain repeats of the GAGAG consensus sequence. Functions of these elements could therefore be mediated by Psq and do not necessarily infer an involvement of GAF, unless a role of GAF is demonstrated by more direct evidence. However, the observation that Psq binding in vitro is restricted to GAGA elements that are longer than elements sufficient for GAF binding, suggested that Psq might only act through a certain subset of GAGA binding sites in vivo (23).
Here we present data arguing against this model. The binding patterns of Psq and GAF on polytene and mitotic chromosomes show a complete overlap. Psq and GAF can be coimmunoprecipitated from nuclear extracts, and they directly bind to one another in vitro through their BTB domains. The two proteins are, therefore, partners in a protein complex in vivo and are thus directed to common chromosomal target sites. Like Trl mutations, mutations in psq enhance the extra sex combs phenotype of the Polycomb allele Pc3. Moreover, psq strongly enhances the genetic interaction between Trl and Ubx, and it acts, like Trl, as an enhancer of position effect variegation (PEV). Taken together, these data are consistent with a model in which Psq and GAF cooperate in the transcriptional activation and silencing of homeotic genes. Because the chromosomal binding patterns of Psq and GAF are identical, this cooperation is likely to include the control of many other genes as well as functions of GAF and Psq during mitosis.
Materials and Methods
Antibodies.
The anti-GAF antibody used in this study was affinity-purified rat anti-GAGABTB-ZF-C-ter519 (30). Rabbit antisera AS1 and AS2, described in ref. 31, were used for the detection of Psq, but only results obtained with AS2 are shown. Polytene chromosome staining patterns with AS1 and AS2 were identical. Antibodies used for the detection of dAP-4 and BR-C protein are described in refs. 32 and 33.
Chromosome Staining.
Cy5-conjugated AffiniPure goat anti-rat IgG (Jackson ImmunoResearch) and Alexa fluor 488 goat anti-rabbit IgG conjugate (Molecular Probes) were used as secondary antibodies in chromosome staining that was analyzed by using a Leica CLSM 4D confocal microscope. The fluorophores Cy5 and Alexa 488 do not overlap in their emission spectra, allowing reliable discrimination between Psq- and GAF-derived signals. For localization of immunofluorescent signals performed with a Zeiss axiophot photomicroscope, polytene and mitotic chromosomes were also stained with the Psq antibody using Cy3-conjugated AffiniPure goat anti-rabbit IgG (Jackson ImmunoResearch) as secondary antibody and counterstained with the DNA dye Hoechst 33258 (Fig. 4A; and data not shown). Polytene chromosomes were stained as described in ref. 34, and mitotic chromosomes from larval brains were stained according to Protocol 1.9, Method 2, described in ref. 35, except that brains were hypotonically swollen for 6 min and fixed in methanol/acetic acid/H2O at a ratio of 4:4:2. Antibodies were used at a dilution of 1:200.
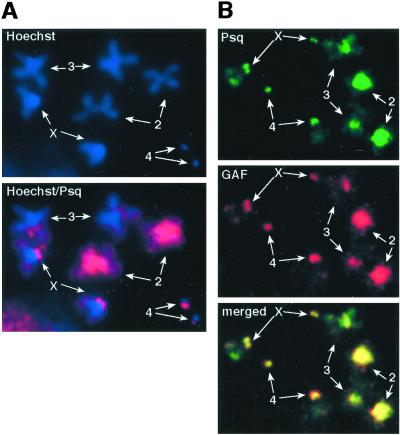
Figure 4.
Psq and GAF colocalize on mitotic chromosomes. Metaphase chromosomes from larval brain neuroblasts were immunostained for Psq and GAF. (A) The localization of signals derived with the Psq antibody on chromosomes counterstained with the DNA dye Hoechst 33258. (B) A double staining with Psq and GAF antibodies. Psq is bound to the same regions of centromeric heterochromatin that are bound by GAF (39, 40). The four pairs of homologous chromosomes are marked.
Immunoprecipitation and Direct Protein Interaction Assays.
Coimmunoprecipitation reactions (Fig. 2) containing 100 μl of salivary gland nuclear extract and 50 μl of anti-Psq antiserum were essentially performed as described in ref. 26, and SDS/PAGE and immunoblotting were performed by using standard procedures. To analyze the ability of full-length and truncated Psq to bind GAF519 (Fig. 3B), his-tagged derivatives were expressed in Escherichia coli and bound to a Ni2-NTA matrix. Columns with Psq-bound or, as a control, pure matrix, were then loaded with purified recombinant GAF519. After washing the columns with 60 vol of binding buffer (20 mM Tris⋅HCl, pH 7.9/100 mM imidazole/125 mM NaCl/5 mM MgCl2/10% glycerol), bound proteins were eluted with 1 M imidazole in the same buffer, fractionated by SDS/PAGE, and the presence of GAF519 was assayed by immunoblotting. GST-pulldown assays (Fig. 3C) were performed according to standard protocols. Further details can be obtained upon request.
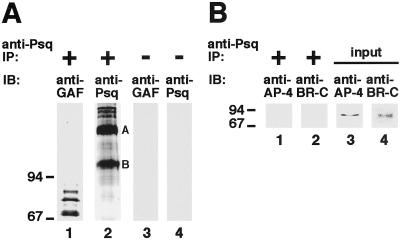
Figure 2.
Psq and GAF are specifically associated with one another in a protein complex in vivo. (A) Nuclear extract from larval salivary glands was immunoprecipitated (IP) by using Psq antibody (lanes 1 and 2), or mock precipitated under omission of the antibody (lanes 3 and 4). Recovered protein was fractionated by SDS/PAGE (7.5% gel), and the presence of GAF and Psq protein analyzed by immunoblotting (IB). The anti-GAF antibody detects the same pattern of GAF isoforms in the salivary glands that has been described (30). The anti-Psq antibody detects two strong bands, marked A and B, which correspond to the Psq isoforms A and B described in ref. 31. In addition, the antibody recognizes slower migrating protein species, which also seem to be present in ovarian extracts (31). (B) Same experiment as in A, except that a 15% gel was used for SDS/PAGE and anti-dAP-4 and anti-BR-C antibodies were used in immunoblotting (lanes 1 and 2). Lanes 3 and 4 show immunoblots of the nuclear extract protein used as an input in this experiment, demonstrating the presence of both dAP-4 and BR-C in the extract.
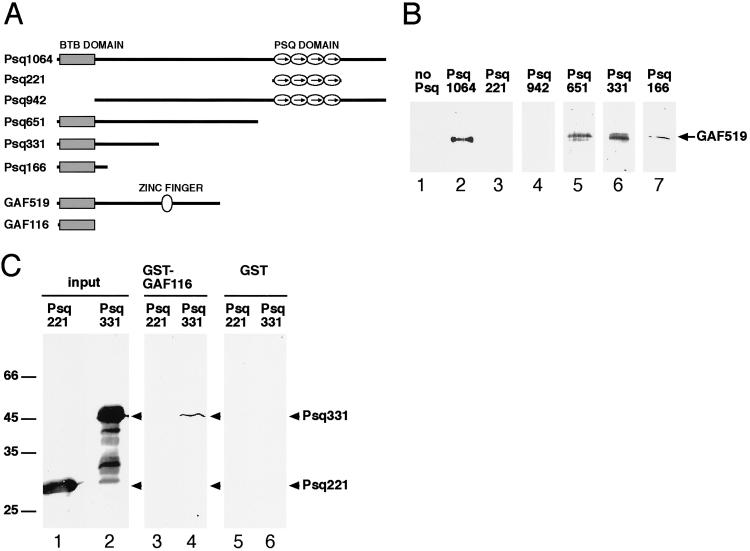
Figure 3.
Psq and GAF directly bind to one another in vitro. Full-length and truncated Psq and GAF proteins depicted in A were expressed in bacteria and assayed for their ability to interact with each other in B and C. (B) The indicated Psq proteins were expressed as his-tagged fusion proteins and analyzed for their ability to retain nontagged GAF519 on a Ni2-NTA column. After washing out unbound protein, bound protein was eluted and a Western blot of this protein was probed with the anti-GAF antibody. (C) A GST-GAF116 fusion protein, or GST alone, were immobilized on glutathione agarose beads and analyzed for their ability to pull-down Psq331 and Psq221. Protein recovered from the washed beads was fractionated by SDS/PAGE, blotted, and probed with the anti-Psq antibody (lanes 3–6). This antibody recognizes both the his-tagged purified Psq221 and Psq331 proteins, which were used as an input in this assay (lanes 1 and 2). Taken together, B and C show that the BTB domains of both Psq and GAF are sufficient, and that the BTB domain of Psq is required, to mediate a direct interaction between the two proteins.
Fly Strains and Genetic Interaction Studies.
Strains carrying w; Df(2R)psq-lolaΔ18/CyO[act-GFP] w+, w; psq0115/CyO[act-GFP] w+, and w; psqRF13/CyO[act-GFP] w+ were obtained from C. A. Berg (Univ. of Washington, Seattle); Pc3/TM3, Ser [act-GFP] w+, TrlR85/TM3, Ser [act-GFP] w+, and Ubx130/kg(v) red(1) Sb (Sbd-1) were obtained from S. Sakonju (Eccles Institute of Human Genetics, Univ. of Utah, Salt Lake City); psqF112; ry/SM5a:TM6B, Tb was obtained from U. Weber (Mount Sinai School of Medicine, New York); and In(1)wm4h was obtained from G. Korge (Institut für Biologie, Freie Universität Berlin, Berlin). To examine the interaction between psq and Pc, we carried out crosses between flies of the genotypes Pc3/TM3, Ser [act-GFP] w+ and Df(2R)psq-lolaΔ18/CyO[act-GFP] w+, w; psq0115/CyO[act-GFP] w+, or w; psqRF13/CyO[act-GFP] w+. To identify a possible dominant genetic interaction between psq and Ubx, we crossed flies of the genotypes Df(2R)psq-lolaΔ18/CyO[act-GFP] w+ and Ubx130/kg(v) red(1) Sb (Sbd-1). To analyze the effect of psq on the dominant interaction between Trl and Ubx (17), we first established strains of the genotypes Df(2R)psq-lolaΔ18/SM5; TrlR85/TM6B, Tb and psqF112/CyO[act-GFP]; TrlR85/TM3, Sb. Virgin females of these strains were then crossed with Ubx130/TM3, Sb, Ubx130/TM6B (Tb, Ubi-GFP), or Ubx130/kg(v) red(1) Sb (Sbd-1) males. Progeny of the genotypes given in Table 1 were examined for ectopic sex combs on the second and third legs, and for haltere and notal transformations. To study the effect of the psq alleles Df(2R)psq-lolaΔ18 and psq0115 on PEV, the CyO[act-GFP] w+ balancer chromosomes of the strains carrying these alleles were first replaced by CyO chromosomes lacking the w+ allele, and males were then crossed to In(1)wm4h/In(1)wm4h females. Male progeny of the genotype In(1)wm4h/y; +/CyO was backcrossed with females of the respective psq strain. Female progeny carrying the CyO balancer chromosome was then compared with female progeny from the previous cross, carrying the same chromosome. About half of the animals had the uniform dark red eye color of In(1)wm4h/w; +/CyO animals, the other half [In(1)wm4h/w; psq/CyO] showed a conspicuous mottled eye phenotype.
Table 1.
Genetic interaction of psq with Pc and Trl/Ubx
| Genotype | Penetrance, % (n)*
|
|
|---|---|---|
| Pc × psq | psq × Pc | |
| CyO/+;Pc3/+ | 2.3 (33) | |
| psq0115/+;Pc3/+ | 27.8 (45) | |
| psq0115/+;TM3,Ser/+ | 0 (44) | |
| CyO/+;Pc3/+ | 7.6 (36) | 8 (126) |
| psq-lolaΔ18/+;Pc3/+ | 29.8 (31) | 60 (138) |
| psq-lolaΔ18/+;TM3,Ser/+ | 0 (41) | 0 (195) |
| CyO/+;Pc3/+ | 4.8 (57) | |
| psqRF13/+;Pc3/+ | 22.7 (55) | |
| psqRF13/+;TM3,Ser/+ | 0 (55) | |
| SM5/+;Ubx130/TM6B | 0 (267) | |
| SM5/+;Ubx130/TrlR85 | 7.6 (329) | |
| psq-lolaΔ18/+;Ubx130/TM6B | 0 (319) | |
| psq-lolaΔ18/+;Ubx130/TrlR85 | 27.9 (366) | |
| CyO/+;Ubx130/+ | 0 (1034) | |
| psq-lolaΔ18/+;Ubx130/+ | 0.5 (779) | |
| CyO/+;Ubx130/TrlR85 | 8.7 (747) | |
| psqF112/+;Ubx130/TrlR85 | 41.1 (863) | |
Upper three groups of genotypes: percentage of legs of the second and third thoracic segment that carried an extra sex comb. These legs do not carry sex combs in the wild type. Probably because of the reduced maternal contribution of Psq protein, the penetrance of the extra sex combs phenotype is twice as high when crosses are set up with females instead of males carrying the psq-lolaΔ18 allele. Lower three groups: percentage of animals of the indicated genotype that showed haltere or notal transformations.
Number of animals of that particular genotype examined.
Results and Discussion
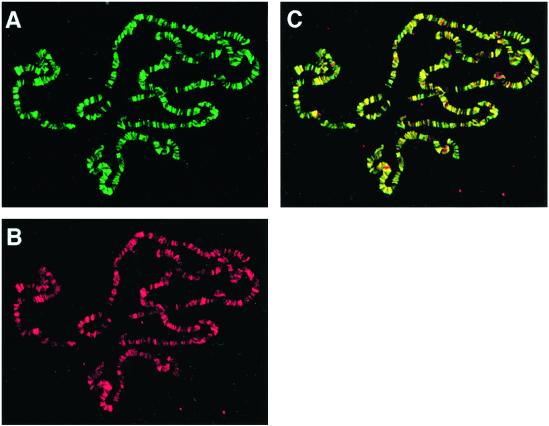
To identify chromosomal loci that are targets of both GAF and Psq, we doubly immunostained salivary gland polytene chromosomes with anti-Psq and anti-GAF antibodies. Surprisingly, the staining patterns obtained with the two antibodies appeared to be identical (Fig. 1). Like GAF, Psq binds to hundreds of loci on the polytene chromosomes. Analysis of the binding patterns by confocal microscopy confirmed that virtually every signal derived with GAF antibody coincided with a signal derived with Psq antibody and vice versa. Occasionally, single sites appeared to be stained by only one of the two antibodies. However, these sites varied between different chromosome preparations, and we therefore believe that they do not represent binding sites truly specific for only one of the two proteins. Western analyses showed that the overlapping staining patterns were not caused by cross-reaction of Psq antibody with GAF protein or GAF antibody with Psq protein (Fig. 2A; and data not shown).
Figure 1.
Psq and GAF bind to hundreds of identical loci on the larval polytene chromosomes. Chromosomes from salivary glands of late-third instar larvae were immunostained with anti-Psq antibody (green) (A) and anti-GAF antibody (red) (B). The two staining patterns are merged in C. Sites where both proteins colocalize appear yellow. Sites where the intensity of the two signals differs appear shaded in green or red. However, careful comparison of the patterns shown in A and B, and of other chromosome preparations not shown here, suggests that the staining patterns are indeed identical. This finding was confirmed by confocal microscopy (see Materials and Methods).
One possibility to explain the colocalization of GAF and Psq is that GAGA-binding sites are recognized in vivo by a protein complex that contains both proteins. We therefore prepared protein extracts from salivary gland nuclei and performed immunoprecipitation assays with the Psq antibody (Fig. 2A). This antibody efficiently coimmunoprecipitated Psq and GAF from these extracts (Fig. 2A, lanes 1 and 2). To test whether this effect was specific, we also analyzed whether two other transcription factors, dAP-4 and BR-C, were precipitated by the antibody. Both proteins are known to be expressed in salivary glands, from which the nuclear extract was derived (32, 33, 36). In addition, like Psq and GAF, all isoforms encoded by the BR-C contain a BTB protein interaction domain (37). However, neither dAP-4 nor BR-C was found to be precipitated by the Psq antibody (Fig. 2B, lanes 1 and 2). We conclude that the interaction between Psq and GAF is specific.
We next asked whether the association of Psq and GAF in vivo might be due to direct binding of the two proteins to one another. To address this question, we expressed histidine (his)-tagged full-length and truncated Psq proteins in bacteria (Fig. 3A), and tested the ability of these proteins to retain the GAF519 isoform on a Ni2-NTA column (Fig. 3B). Full-length Psq (Psq1064), but not polypeptides including only the Psq domain (Psq221) or lacking the BTB domain (Psq942), were able to efficiently bind GAF519 (Fig. 3B, lanes 2–4). When Psq1064 was successively truncated from the C terminus, all resulting polypeptides were able to bind GAF519, including Psq166, which essentially consists of the BTB domain (Fig. 3B, lanes 5–7). Because previous studies have shown that the BTB domain forms a protein–protein interaction interface (26, 27), this result suggests that binding is mediated by the BTB domains of both proteins. We therefore expressed a GST fusion of the GAF BTB domain in bacteria (GST-GAF116; see Fig. 3A) and tested whether this polypeptide can be coimmunoprecipitated with Psq331, a polypeptide that consists of the N-terminal third of Psq, which includes the BTB domain. As a control, we tested whether GAF116 can be coimmunoprecipitated with Psq221, which was shown before to be unable to interact with full-length GAF519 (Fig. 3B, lane 3). Fig. 3C shows that Psq331, but not Psq221, is bound by GAF116. Taken together, these results suggest that Psq and GAF are associated with one another in vivo through direct binding mediated by their BTB domains. The BTB domain of GAF has been shown to also mediate self-oligomerization of GAF that leads to formation of large protein complexes in vitro (28, 29). Our results suggest that Psq is a partner of GAF in similar complexes formed in vivo. Because both GAF and Psq have been shown to bind GA-rich sequences (4, 23), it is likely that both proteins contribute to the cooperative binding of multiple GAGA elements through these complexes. The complete colocalization of GAF and Psq at hundreds of chromosomal loci predicts that, in general, Psq and GAF act as partners. Consistent with this notion, the bxd PRE of Ubx (38) and the MCP silencer of Abdominal B (Abd-B) (S. Sakonju, personal communication) emerge as first examples of specific loci where Psq and GAF appear to interact.
To further test our model that Psq and GAF are partners, we asked whether Psq is also bound to the GA-rich satellite DNA of the centromeric heterochromatin, which is occupied by GAF during mitosis (39, 40). When mitotic chromosomes from larval brains are stained with GAF antibodies, strong fluorescent signals are observed in the pericentric regions (Fig. 4B and ref. 39). A similar staining of the pericentric regions was observed when chromosomes were stained with the Psq antibody (Fig. 4A). Double staining experiments showed that this staining cannot be distinguished from the staining pattern obtained with the GAF antibody (Fig. 4B). As on the polytene chromosomes, there thus seems to be a complete overlap of the GAF and Psq binding sites. Taken together, the results of the polytene and mitotic chromosome staining strongly suggest that Psq and GAF are general partners that not only share common functions in the control of target genes at euchromatic sites, but also in heterochromatin organization and mitosis.
To learn more about the functional relationship between GAF and Psq, we analyzed a possible genetic interaction between Trl and psq, and compared their roles in the control of homeotic gene expression. Animals doubly heterozygous for the null allele TrlR85 and the alleles psqRF13, psq0115, or Df(2R)psq-lolaΔ18 showed no readily apparent defects or reduced viability (data not shown). Thus, a single copy of each gene, together with a substantial maternal contribution of Trl and psq gene products (11, 31), seems to be sufficient for normal development. It has recently been shown that Trl has properties of a PcG gene. Thus, GAF is a component of some PcG complexes (8) and GAGA-binding sites are required to maintain the silencing activity of the bxd, iab-7, and MCP PREs (8, 9, 19). Consistent with a role of GAF in PRE function, Trl alleles enhance the extra sex combs phenotype of Pc mutants (20). To test whether psq shows a similar genetic interaction, we examined animals doubly heterozygous for the allele Pc3 and alleles psqRF13, psq0115, or Df(2R)psq-lolaΔ18. In all three combinations of psq alleles with Pc3, the frequency of animals with ectopic sex combs (Fig. 5A) was significantly higher than in the presence of the Pc3 allele alone (Table 1). The same interaction, as well as an interaction between polyhomeotic and psq, has been observed by others (ref. 38, and S. Sakonju, personal communication). On the basis of its interaction with Ubx and Abd-B, Trl has originally been identified as a member of the trxG of genes, which are required for the maintenance of homeotic gene expression (17). Animals doubly heterozygous for the alleles TrlR85 and Ubx130 show haltere-to-wing and notal transformations with a penetrance of about 8% (Table 1). In contrast, the same transformations are observed at a much lower frequency in animals doubly heterozygous for the psq allele Df(2R)psq-lolaΔ18 and Ubx130 (Fig. 5B; Table 1). We therefore asked whether replacement of one wild-type copy of psq by a mutant allele would have an influence on the dominant genetic interaction between TrlR85 and Ubx130. Both the psq alleles Df(2R)psq-lolaΔ18 and psqF112 clearly enhanced this interaction, leading to a 4- to 5-fold increase in the frequency of haltere and notal transformations (Table 1). The weak dominant genetic interaction observed between psq and Ubx thus seems to be indeed indicative of a requirement of psq wild-type function for Ubx expression. We conclude that psq has similar functions as Trl, not only in silencing homeotic genes, but also in their activation. This conclusion is further supported by the finding that both psq and Trl alleles are dominant enhancers [E(var)s] of PEV. The white (w) gene is suppressed in a clonally inherited manner when juxtaposed to centromeric heterochromatin by a chromosomal rearrangement, In(1)wm4h, leading to a variegated eye color. The ability to enhance (or suppress) PEV has been exploited to genetically identify factors that counteract (or promote) chromatin-mediated gene silencing (41). Like the Trl13C allele (17), the psq alleles Df(2R)psq-lolaΔ18 and psq0115 clearly enhance the variegated eye phenotype of In(1)wm4h (Fig. 5C). The similar behavior of psq and Trl in this assay is consistent with a common mechanistic basis for the actions of the products encoded by the two genes.
Figure 5.
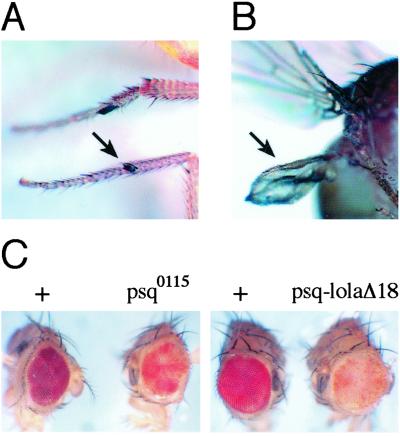
psq shows similar genetic interactions as Trl. (A) psq alleles enhance the extra sex combs phenotype of Pc. Ectopic sex comb on the second leg (arrow) of a psq0115/+; Pc3/+ fly. (B) psq alleles weakly interact with Ubx, and strongly enhance the genetic interaction between Trl and Ubx. Transformation of a haltere into wing (arrow) of a Df(2R)psq-lolaΔ18/+; Ubx130/+ fly. (C) psq alleles are dominant enhancers of PEV. Eyes of flies doubly heterozygous for In(1)wm4h and the indicated psq alleles (Right) show enhanced mottling of the eye color when compared with the eyes of flies of the same genetic background but lacking the psq alleles (Left).
The genetic, biochemical, and cytogenetic data presented here strongly suggest that Psq and GAF act together as partners in the control of homeotic and many other genes. As both GAF and Psq are encoded by essential genes (11, 31, 42), the functions of these proteins are not redundant. Psq may even be an obligatory partner of GAF. Future studies on GAF function will therefore have to include this partner, and may thus provide novel insights into the mechanism of action of this important chromatin factor. The recent finding that Psq is a member of a larger family of DNA-binding proteins, which includes many Drosophila BTB proteins with previously unknown DNA-binding activity (24), may help to further elucidate the composition and function of Psq/GAF-containing protein complexes.
Acknowledgments
We thank C. Benyajati and C. A. Berg for providing anti-GAF and anti-Psq antibodies; C. A. Berg, S. Sakonju, and U. Weber for fly stocks; R. Menzel and T. Siegmund for help with the confocal microscope; K. Dünnbier for technical assistance; G. Korge, S. Sakonju, and C. Thummel for helpful discussions and comments on the manuscript; and S. Sakonju for communicating results before publication. This work was supported by Deutsche Forschungsgemeinschaft Grant LE870/5-2 (to M.L.).
Abbreviations
- BTB
Broad-Complex, Tramtrack, Bric à brac
- GAF
GAGA factor
- PEV
position effect variegation
- PRE
Polycomb response element
- Psq
Pipsqueak
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Aalfs J D, Kingston R E. Trends Biochem Sci. 2000;25:548–555. doi: 10.1016/s0968-0004(00)01689-3. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg R D, Lorch Y. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 3.Biggin M D, Tjian R. Cell. 1988;53:699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- 4.Soeller W C, Oh C E, Kornberg T B. Mol Cell Biol. 1993;13:7961–7970. doi: 10.1128/mcb.13.12.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soeller W C, Poole S J, Kornberg T. Genes Dev. 1988;2:68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- 6.Granok H, Leibovitch B A, Shaffer C D, Elgin S C. Curr Biol. 1995;5:238–241. doi: 10.1016/s0960-9822(95)00048-0. [DOI] [PubMed] [Google Scholar]
- 7.Hagstrom K, Muller M, Schedl P. Genetics. 1997;146:1365–1380. doi: 10.1093/genetics/146.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horard B, Tatout C, Poux S, Pirrotta V. Mol Cell Biol. 2000;20:3187–3197. doi: 10.1128/mcb.20.9.3187-3197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mishra R K, Mihaly J, Barges S, Spierer A, Karch F, Hagstrom K, Schweinsberg S E, Schedl P. Mol Cell Biol. 2001;21:1311–1318. doi: 10.1128/MCB.21.4.1311-1318.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohtsuki S, Levine M. Genes Dev. 1998;12:3325–3330. doi: 10.1101/gad.12.21.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhat K M, Farkas G, Karch F, Gyurkovics H, Gausz J, Schedl P. Development (Cambridge, UK) 1996;122:1113–1124. doi: 10.1242/dev.122.4.1113. [DOI] [PubMed] [Google Scholar]
- 12.Wilkins R C, Lis J T. Nucleic Acids Res. 1997;25:3963–3968. doi: 10.1093/nar/25.20.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerrigan L A, Croston G E, Lira L M, Kadonaga J T. J Biol Chem. 1991;266:574–582. [PubMed] [Google Scholar]
- 14.Croston G E, Kerrigan L A, Lira L M, Marshak D R, Kadonaga J T. Science. 1991;251:643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- 15.Tsukiyama T, Becker P B, Wu C. Nature (London) 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- 16.Tsukiyama T, Wu C. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 17.Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. Nature (London) 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 18.Mahmoudi T, Verrijzer C P. Oncogene. 2001;20:3055–3066. doi: 10.1038/sj.onc.1204330. [DOI] [PubMed] [Google Scholar]
- 19.Busturia A, Lloyd A, Bejarano F, Zavortink M, Xin H, Sakonju S. Development (Cambridge, UK) 2001;128:2163–2173. doi: 10.1242/dev.128.11.2163. [DOI] [PubMed] [Google Scholar]
- 20.Strutt H, Cavalli G, Paro R. EMBO J. 1997;16:3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orlando V, Paro R. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- 22.Brown J L, Mucci D, Whiteley M, Dirksen M L, Kassis J A. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 23.Lehmann M, Siegmund T, Lintermann K G, Korge G. J Biol Chem. 1998;273:28504–28509. doi: 10.1074/jbc.273.43.28504. [DOI] [PubMed] [Google Scholar]
- 24.Siegmund T, Lehmann M. Dev Genes Evol. 2002;212:152–157. doi: 10.1007/s00427-002-0219-2. [DOI] [PubMed] [Google Scholar]
- 25.Godt D, Couderc J L, Cramton S E, Laski F A. Development (Cambridge, UK) 1993;119:799–812. doi: 10.1242/dev.119.3.799. [DOI] [PubMed] [Google Scholar]
- 26.Bardwell V J, Treisman R. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- 27.Ahmad K F, Engel C K, Prive G G. Proc Natl Acad Sci USA. 1998;95:12123–12128. doi: 10.1073/pnas.95.21.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espinas M L, Jimenez-Garcia E, Vaquero A, Canudas S, Bernues J, Azorin F. J Biol Chem. 1999;274:16461–16469. doi: 10.1074/jbc.274.23.16461. [DOI] [PubMed] [Google Scholar]
- 29.Katsani K R, Hajibagheri M A, Verrijzer C P. EMBO J. 1999;18:698–708. doi: 10.1093/emboj/18.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benyajati C, Mueller L, Xu N, Pappano M, Gao J, Mosammaparast M, Conklin D, Granok H, Craig C, Elgin S. Nucleic Acids Res. 1997;25:3345–3353. doi: 10.1093/nar/25.16.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horowitz H, Berg C A. Development (Cambridge, UK) 1996;122:1859–1871. doi: 10.1242/dev.122.6.1859. [DOI] [PubMed] [Google Scholar]
- 32.King-Jones K, Korge G, Lehmann M. J Mol Biol. 1999;291:71–82. doi: 10.1006/jmbi.1999.2963. [DOI] [PubMed] [Google Scholar]
- 33.Renault N, King-Jones K, Lehmann M. Development (Cambridge, UK) 2001;128:3729–3737. doi: 10.1242/dev.128.19.3729. [DOI] [PubMed] [Google Scholar]
- 34.Lehmann M, Korge G. EMBO J. 1996;15:4825–4834. [PMC free article] [PubMed] [Google Scholar]
- 35.Pimpinelli S, Bonaccorsi S, Fanti L, Gatti M. In: Drosophila Protocols. Sullivan W, Ashburner M, Hawley R S, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 2000. pp. 3–23. [Google Scholar]
- 36.Emery I F, Bedian V, Guild G M. Development (Cambridge, UK) 1994;120:3275–3287. doi: 10.1242/dev.120.11.3275. [DOI] [PubMed] [Google Scholar]
- 37.DiBello P R, Withers D A, Bayer C A, Fristrom J W, Guild G M. Genetics. 1991;129:385–397. doi: 10.1093/genetics/129.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hodgson J W, Argiropoulos B, Brock H W. Mol Cell Biol. 2001;21:4528–4543. doi: 10.1128/MCB.21.14.4528-4543.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Platero J S, Csink A K, Quintanilla A, Henikoff S. J Cell Biol. 1998;140:1297–1306. doi: 10.1083/jcb.140.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raff J W, Kellum R, Alberts B. EMBO J. 1994;13:5977–5983. doi: 10.1002/j.1460-2075.1994.tb06943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wakimoto B T. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- 42.Weber U, Siegel V, Mlodzik M. EMBO J. 1995;14:6247–6257. doi: 10.1002/j.1460-2075.1995.tb00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]