Abstract
A confocal laser scanning microscopy method has been developed for the quantitation of green fluorescent protein (GFP) as a reporter of gene activity in living three-dimensional structures such as sea urchin and starfish embryos. This method is between 2 and 50 times more accurate than conventional confocal microscopy procedures depending on the localization of GFP within an embryo. By using coinjected Texas red dextran as an internal fluorescent standard, the observed GFP intensity is corrected for variations in laser excitation and fluorescence collection efficiency. To relate the recorded image intensity to the number of GFP molecules, the embryos were lysed gently, and a fluorometric analysis of their contents was performed. Confocal laser scanning microscopy data collection from a single sea urchin blastula required less than 2 min, thereby allowing gene expression in dozens of embryos to be monitored in parallel with high spatial and temporal resolution.
Gene-transfer studies have provided a powerful vehicle for exploring the relationship between DNA sequence and gene function in a variety of living organisms. However, current analytical methods are destructive to living samples and therefore cannot quantify gene products in both space and time. In situ hybridization, for example, localizes mRNA expression only in fixed specimens, making it impossible to monitor gene-expression levels within the same embryo at different times. Fluorometric studies of reporter constructs encoding luciferase are typically performed on tissue homogenates, often of pooled populations of embryos. For studying dynamic gene activity in individual cells, green fluorescent protein (GFP) has distinct advantages. However, limitations in standard detection techniques and light scattering from tissue make it challenging to use GFP for quantitative analysis of gene expression in many systems (1, 2). Even in sea urchin embryos, model organisms noted for their optical transparency, losses in fluorescence intensity can make internal structures of the embryo invisible. Thus, ongoing efforts to establish networks of genes and, specifically, comparisons of the activities of promoter/enhancer regions of genes transcribed during sea urchin embryogenesis (3, 4) often rely on in situ hybridization or biochemical analyses of homogenates of several embryos. Gene-expression studies in living embryos carried out with GFP reporters offer the advantage of longitudinal studies with single-cell resolution, but the available technology does not permit its use for quantitative measurements (5). Here we describe methods to accomplish this objective, developed for use in gene-expression studies in sea urchin embryos, but easily applicable to many other subjects as well.
Confocal laser scanning microscopy (CLSM) is well suited for imaging live specimens, because it allows the study of cellular processes with high spatial (≈0.1-μm) and temporal (on the order of seconds) resolution. However, several difficulties must be overcome to use CLSM for quantitation of gene-expression levels in living embryos. First, scatter from living tissue dictates that laser excitation and fluorescence collection efficiencies vary with the depth of the fluorescent protein in the sea urchin embryo. Second, the orientation of different embryos on the coverslip varies relative to the microscope objective, thereby complicating comparisons of regions within an embryo or between different embryos. Finally, injected DNA integrates mosaically into the sea urchin genome (in ≈75% of eggs the DNA is incorporated into the nucleus of a blastomere by the fourth cleavage stage) (6, 7).
Some prior work involving quantitative imaging of indicator dyes was facilitated by the ratiometric analysis of fluorescence intensities at two different wavelengths. For example, the ratio of fluorescence from a β-lactamase substrate and product was monitored in vivo, thereby permitting direct assay for the regional distribution of enzyme activity (8). Accurate ratiometric indicators of intracellular calcium have shown similar advantages (9). Previous studies on sea urchin embryos measured GFP production relative to fluorescence from rhodamine-labeled dextran to measure protein synthesis as a function of cell cycle (10). These results suggested the use of ratiometry to make in vivo imaging of GFP and related reporters more quantitative.
We built on these previous approaches to develop CLSM data collection and analytical methods for the determination of gene-expression levels in living sea urchin embryos (Fig. 1 a–f). Ratiometry between a synthetic dye, Texas red (TR)-labeled dextran, and GFP provides a quantitative in vivo assay for reporter gene activation. TR was coinjected with the well characterized hatching enzyme (HE) cis-regulatory system (11) fused to a GFP reporter gene. The dye diffuses homogeneously throughout the embryos and serves as an internal fluorescent standard. Custom-built image-processing scripts correct the observed GFP signal based on the changes in TR-fluorescence intensity with depth. Finally, embryo lysis protocols and fluorometric methods were developed to correlate in vivo fluorescence intensity with absolute numbers of GFP molecules. By this approach, gene-expression levels may now be determined with dramatically improved accuracy and speed, thereby accelerating the analysis of numerous reporter gene constructs for functional genomics studies.
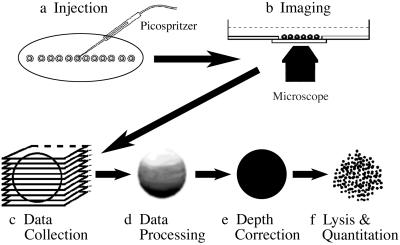
Figure 1.
Method of GFP quantitation. (a) Microinjection of newly fertilized sea urchin embryos. Embryos first are rowed in a straight line onto a poly-L-lysine-coated coverslip. (b) Confocal microscopy at high magnification. (c) Collection of fluorescence data for both GFP and TR-labeled dextran throughout the embryo. (d) Input of data, showing large attenuation of signal with depth, into correction algorithms. (e) Output of depth-corrected fluorescence data showing considerable signal enhancement. (f) Lysis of the same Krox-GFP mRNA-injected embryos for GFP quantitation in solution using a fluorometer.
Materials and Methods
Sea Urchin Eggs and Embryos.
Gametes from Strongylocentrotus purpuratus were harvested, and the eggs were rowed onto plates and fertilized just before microinjection (12). The plates were prepared by boring a 20-mm-diameter hole into the center of a Petri dish and covering the hole with a 25-mm-diameter, 170-μm-thick glass coverslip sealed with vacuum grease. The dimensions of the coverslip allowed imaging with a high-magnification, high-numerical aperture objective. A poly-L-lysine solution (molecular mass, 150–300 kDa; 50 μg/ml in 10 mM Tris, pH 8.0, Sigma P1399) was applied to the coverslip for 10 min at room temperature, after which the plates were rinsed with doubly distilled water and air-dried in a dust-free place (13).
Because S. purpuratus embryos typically hatch 18 h postfertilization (hpf), most imaging was performed 14–18 hpf while the embryos were still immobilized on the coverslip within the fertilization membrane. To image later stages, embryos were deciliated through brief exposure to 2× seawater.
Microinjection.
S. purpuratus embryos were microinjected (Fig. 1a) and cultured as described (12). However, a freely diffusing internal fluorescent standard, TR-labeled dextran (neutral; molecular weight, 10,000; Molecular Probes), was coinjected with either DNA or mRNA. Depth-dependent losses in GFP fluorescence then were corrected by using the corresponding TR intensity. In parallel experiments, HE-GFP DNA (14) and Krox-GFP mRNA were injected into freshly fertilized eggs from the same sea urchin. Primers were designed to amplify a 182-bp fragment at the 5′ region of the SpKrox1 message: 5′-AAGCTG- GGTACCCTAAAGAAATTCCCCGATTGAAAC-3′ (forward primer with KpnI site) and 5′-CTGCTGGACGTCCCGTTCGGGGTCCTTGACCTC-3′ (reverse primer with AatII site). The Krox gene (15) contains a good Kozak sequence, and the start codon was placed in frame with the coding sequence for a “bright” GFP mutant, S65T. This construct increased the GFP translation–initiation efficiency ≈3-fold (data not shown), thus facilitating quantitation efforts.
Both DNA and RNA injection solutions had a total volume of 10 μl. Solution A was 50 ng of HE-GFP DNA/250 ng of carrier DNA/0.65 mM TR dextran/0.12 M KCl/15% glycerol. Solution B was 1.5 μg of Krox-GFP mRNA/0.65 mM TR-labeled dextran/0.12 M KCl/15% glycerol. Centrifugation (16,000 × g for 5 min) was performed to pellet any precipitate that could clog the injection needle.
Imaging with CLSM.
To maintain S. purpuratus embryos at an optimum culture temperature, an aluminum stage fitted with a Peltier cooling device was attached to the universal stage adapter of an Axiovert 100 M inverted microscope configured for CLSM (LSM 5 PASCAL, Zeiss). This imaging chamber held the Petri dish and maintained the seawater at 10–15°C, such that the embryos continued to develop normally during repeated confocal imaging over several hours.
The embryos (≈80 μm in diameter) were imaged by CLSM using a ×40, 0.9-numerical aperture Plan-Neofluar multiimmersion lens (Zeiss) with coverslip correction (Fig. 1b). Using water as the immersion medium best matched the indices of refraction inside the embryo. The pinhole was kept at ≈100 μm (1 Airy unit), and identical detector gain (typically 525 V, red channel; 650 V, green channel; 215 V, transmitted light) and offset settings (−0.1, red; −0.1, green; −0.05, transmitted light) were maintained throughout each experiment, allowing direct comparison of different data sets. Laser power (argon ion, 20% power, 488 nm, and helium-neon, 80% power, 543 nm) was monitored periodically to ensure consistency between experiments. Virtually no crosstalk was observed between channels (green, 505–530-nm band-pass filter; red, 585-nm long-pass filter). Typically, embryos were imaged at scan speed 8 (1.76 μsec per voxel, less than 1 sec per image) in 1-μm-thick z sections for a total of 91 slices.
Data Processing.
The confocal images for each embryo (Fig. 1c) were resized (roughly 220 × 220 voxels per optical section) and saved as 8-bit .tif files by using commercially available software (LSM 5 IMAGE EXAMINER, Zeiss). These images (Fig. 1d) were read into in-house interactive data language (Research Systems, Boulder, CO) scripts. Commercial programs such as IMAGESPACE (Molecular Dynamics) interpolate data to maintain the dynamic range of intensity values within a specimen and preserve the integrity of deep structures. We developed algorithms to allow absolute comparisons of intensity values within embryos.
An array was created of voxels with red intensity exceeding a threshold value and contiguous with eight other red voxels (comprising a 3 × 3 square). The threshold was subtracted from each red intensity, and a red mean was determined for each slice. In the deepest 20 μm of each embryo, a second-order polynomial was fit to the mean red intensities to minimize the effects of lower signal-to-noise ratios. Altogether, this constituted the depth profile for the embryo.
As a first step toward depth-correcting the GFP fluorescence, the green channel was filtered by using a threshold value (typically 4) of twice the dark noise. Then the green intensities were divided by the depth profile at that slice and multiplied by the maximum red mean intensity to normalize the data. By this method, barely discernible GFP expressed deep within the embryo was amplified relative to GFP expressed in more optically transparent parts of the embryo (Fig. 1e). Finally, our scripts integrated the corrected in vivo GFP fluorescence intensity over the entire embryo to allow comparisons between analogously injected embryos. The corrected TR and GFP fluorescence intensity data were output as .tif files, and three-dimensional renderings were performed by using AMIRA 2.3 (TGS Template Graphics Software, San Diego).
Strategy of Correlating Fluorescence Intensity with Number of GFP Molecules.
For quantification purposes, the total corrected GFP fluorescence intensity was determined for mRNA-injected embryos, because they contained larger amounts of GFP. To correlate in vivo fluorescence intensity with the number of GFP molecules, mRNA-injected embryos were imaged individually by CLSM and then lysed in aliquots of 20 embryos for analysis with a fluorometer. The in vivo fluorescence intensity obtained by CLSM was related to the number of GFP molecules per lysed embryo. This correlation then was applied to fluorescent embryos injected with HE-GFP DNA.
Embryo Lysis and GFP Measurement with a Fluorometer.
After CLSM, mRNA-injected embryos were removed carefully from the coverslip in minimal volume (≈10 μl) by using a mouth pipette and placed inside a 0.6-ml Eppendorf tube on ice. An equal volume of 2× lysis buffer was added, for a total volume of 20–30 μl. The 2× lysis solution contained 3 mg/ml BSA, 100 mM Tris⋅HCl (pH 8.0), 300 mM NaCl, 20% glycerol, 1 mM EGTA, 0.2% Triton X-100, and 0.2% Nonidet P-40. The detergents punctured the cell and nuclear membranes and released the GFP into solution (Fig. 1f). While lysing, the embryos were stored at −20°C to prevent further GFP production or degradation.
Before analysis, tubes containing 20 embryos were microcentrifuged for 1 h at 16,000 × g at 4°C to remove cell debris. Each embryo solution was transferred by pipette to a 1-cm path length small-volume fluorescence cuvette (WG-16/10F-Q-10-1, Wilmad, Buena, NY). Samples were measured in 20-μl aliquots to avoid creating air bubbles in the detection path. Maximum gain (950 V), slit settings (10-nm excitation, 20-nm emission), and instrument response (8 sec) on the fluorescence spectrophotometer (F-4500, Hitachi, Tokyo) allowed nanomolar detection of the fluorophore. The samples were excited at 475 nm (near the 488-nm absorption maximum for both GFP and fluorescein), and a 500-nm long-pass filter minimized excitation light at the detector. GFP fluorescence was measured from 500 to 700 nm, and the absolute protein concentration was determined through comparison to purified S65T GFP standards. The published quantum yield of 0.65 for S65T GFP (16, 17) was confirmed relative to a 5 nM fluorescein stock solution (0.1 M NaOH; quantum yield, 0.95).
Results and Discussion
GFP Quantitation.
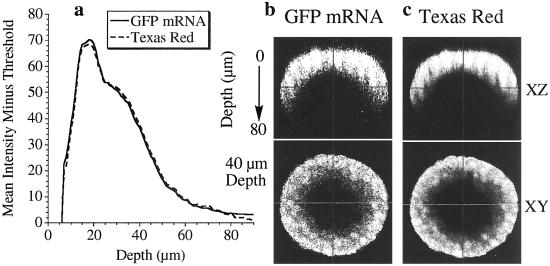
Fig. 2 presents orthogonal views of a uniformly labeled sea urchin embryo; the regional differences in signal intensity illustrate the intrinsic difficulty of imaging through a sea urchin embryo, where there are considerable losses in fluorescence intensity with depth. By coinjecting HE-GFP constructs with a TR-labeled dextran that diffuses uniformly throughout the embryos, it is possible to correct the GFP fluorescence intensity for the depth at each confocal slice. Injection of the Krox-GFP mRNA (see Materials and Methods) produces large amounts of fluorescent protein homogeneously throughout the embryos. Importantly, Krox-GFP mRNA typically produces 5-fold more total GFP than embryos expressing HE-GFP. Uniform distribution of TR and Krox-GFP is confirmed by comparing fluorescence intensities within and between blastomeres in many different embryos. Virtually identical depth dependence of the TR and GFP fluorescence intensities is achieved by adjusting the photomultiplier tube gain and offset appropriately for the red and green channels (Fig. 2 a–c). In this way, it was possible to compensate for differences in red- and green-light sensitivity between the selected photomultiplier tubes. The GFP data exhibit lower signal-to-noise (Fig. 2b), primarily because of lower concentrations of this fluorophore in early blastula-stage embryos.
Figure 2.
Depth profiles of GFP and TR evenly distributed throughout 18-hpf sea urchin embryos. (a) Overlaid plots of mean intensity (minus threshold) vs. depth for GFP and TR are virtually identical. (b) An XZ cross section through the embryo shows marked attenuation of GFP intensity in the bottom half of the embryo. An XY cross section (40-μm depth) shows fairly homogeneous GFP distribution in the embryo. (c) An XZ cross section also shows marked attenuation of TR intensity in the bottom half of the embryo. An XY cross section (40-μm depth) shows even TR distribution in the embryo.
After imaging the mRNA-injected embryos by CLSM, the embryos are lysed and the GFP quantified by using a fluorometer calibrated with fluorescein and GFP standards. It is important to arrest GFP production immediately after imaging and to preserve the fluorescent protein during lysis. The lysis conditions were tested by using purified recombinant S65T GFP (18) and found to maintain the integrity of the GFP fluorescence, as measured with a fluorometer. During lysis, the increasing GFP fluorescent signal was monitored until saturation was achieved (data not shown). It was found that typical sea urchin embryo lysis protocols involving repeated freeze–thaw cycles at −80 and 37°C result in rapid and irreversible protein degradation. By use of a nonionic detergent lysis procedure, as described in Materials and Methods, the CLSM data were correlated with the number of GFP molecules measured by fluorometry. This method permits the quantification of GFP for any orientation of the embryos on the coverslip and for a broad range of gene-expression levels.
Data Analysis.
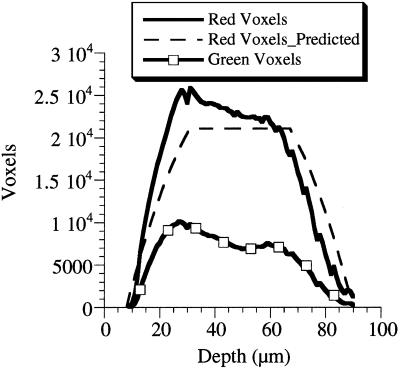
Plots of TR voxels vs. depth (Fig. 3) confirm that the appropriate number of voxels is distributed roughly in the shape of a spherical blastula (80-μm diameter) with a 36-μm diameter blastocoel in the center. Deviations from predicted values in the bottom quarter of the embryo (sections, 60–80 μm) show that fluorescence data from ≈10% of the voxels are never collected. Although our depth-correction algorithms compensate for attenuated signal, no efforts were made to compensate for lost signal in these voxels, where no fluorescence was detected. Critically, the filtering procedures succeeded in removing the majority of the halo caused by Rayleigh scatter at the outer and inner (blastocoel) surfaces of the embryo (Fig. 4 a and b). Voxels in the halo were discarded, and these intensities in the red and green channels were ignored during subsequent data processing.
Figure 3.
Plot of voxels vs. depth (in micrometers) showing agreement between the distribution of filtered voxels and the shape of the embryo. Stray voxels have been removed by a linear threshold and 3 × 3 median filter. The TR voxel distribution with depth (solid line) closely matches calculated predictions (dashed line) based on a spherical 80-μm diameter sea urchin embryo with a central 36-μm diameter blastocoel. DNA (HE-GFP) is incorporated mosaically into cells, as shown by the more irregular GFP-voxel distribution (squares).
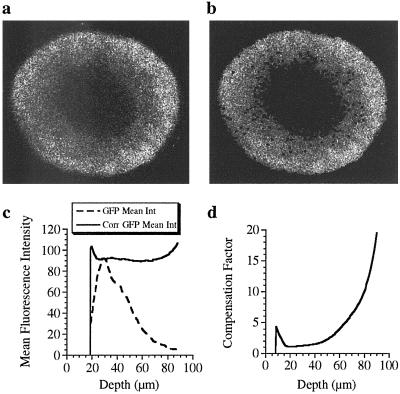
Figure 4.
CLSM image showing TR fluorescence collected halfway (40 μm) through a living sea urchin embryo. (a) Raw data show considerable light scatter at both outer and inner (blastocoel) surfaces. (b) Data filtered by a linear threshold and 3 × 3 median filter show well defined edges. The red intensity threshold was found empirically to decrease linearly with depth: threshold = (2 × dark noise) + constant − (slope × slice). Under standard TR loading and imaging conditions, a constant equal to 16 and slope equal to 0.2 gave a discreet outer boundary with minimal loss of signal within the embryo. Thus, for the deepest slice (80 μm), the threshold for the red channel equaled twice the mean dark noise. (c) Plot of depth-corrected GFP mean intensity (Int) vs. depth (in micrometers). Homogeneous distribution of GFP throughout the embryo is not reflected in the raw data (green mean). Dividing the green mean by the depth profile (for TR) results in a linear distribution of GFP mean intensity with only minor edge effects at shallow and deep slices. (d) Plot of signal compensation vs. depth (in micrometers). The compensation factor, typically 20 at the deepest slice, is determined by dividing the corrected GFP intensity by the raw GFP intensity at each depth.
Correction of the highly depth-attenuated GFP signal from mRNA-injected embryos occurred with only mild edge effects (≈10% peak error) at shallow and deep slices (Fig. 4c). Divergence of the depth-corrected mean from a horizontal line (Fig. 4c, measurements at 18- and 90-μm depths) highlights small errors in filtering fluorescence at the extremities of the embryo, where the data are particularly influenced by out-of-plane fluorescence and laser scatter. These effects lead to slight fluorescence overcompensation at the extremities. Because this error is minimal, it was possible to obtain accurate determinations for gene expression in sea urchin embryos at these dye and GFP concentrations.
Photobleaching was determined to be only a minor source of error. Photobleaching of TR and GFP is measured (5 and 10%, respectively) for a typical 91-slice z stack by repeated imaging of the same embryos. The degree of bleaching is proportional to the number of images taken (i.e., the dwell time of each laser on the sample). In the present experiments, where the embryos were imaged consistently from shallow to deep slices, the deepest slices are attenuated by the longest laser-dwell times. The depth-correction algorithms, in fact, compensate for most of the photobleaching, because the TR and GFP bleach at similar rates and are exposed to identical laser-dwell times.
Standard deviations of ±20% are typical for measurements of GFP intensity per voxel when comparing embryos (Table 1). Most of this variation arises from trying to image and analyze (i.e., filter and compensate for) differently injected embryos under identical imaging conditions and threshold parameters. In general, data collection errors are minimized when all embryos are injected with similar volumes of sufficiently concentrated TR and nucleic acid. When performed carefully, injection volumes vary by less than a factor of two between embryos (TR quantified per voxel, Table 1). Reproducible injections allow the use of consistent imaging parameters (photomultiplier tube gain, offset) throughout each experiment.
Table 1.
GFP expression in HE-GFP DNA- and Krox-GFP mRNA-injected embryos
| Embryo | Total GFP (×106) | GFP/voxel | TR/voxel | GFP/TR |
|---|---|---|---|---|
| DNA1* | 7.2 | 28 | 62 | 0.45 |
| DNA2† | 22 | 43 | 72 | 0.59 |
| DNA3‡ | 32 | 50 | 68 | 0.73 |
| DNA4 | 28 | 48 | 67 | 0.72 |
| DNA5 | 16 | 39 | 64 | 0.61 |
| RNA1§ | 96 | 100 | 41 | 2.4 |
| RNA2 | 85 | 91 | 63 | 1.4 |
| RNA3 | 82 | 83 | 40 | 2.1 |
The values presented are unitless but were collected with identical gain and offset settings on a detector with 8 bits (0–255) per channel.
Data correspond to embryo shown in Fig. 5a.
Data correspond to embryo shown in Fig. 5b.
Data correspond to embryo shown in Fig. 5c.
Data correspond to embryo shown in Fig. 5d.
Although these imaging methods are subject to continual optimization, already they represent a marked improvement over conventional CLSM techniques. Fig. 4d shows a typical fluorescence compensation vs. depth profile, where the compensation is the depth-corrected GFP fluorescence divided by the raw GFP data for each slice. The compensation factor has been observed to be as large as 50, depending on the dye-loading levels and the opacity of the 18-hpf sea urchin embryos. This enhancement will be particularly relevant in studies of tissue-specific gene expression in later-stage embryos (for example, in the gut), where GFP intensity will vary greatly depending on the orientation of the embryo.
Analyzing GFP Fluorescence in 18-hpf Embryos.
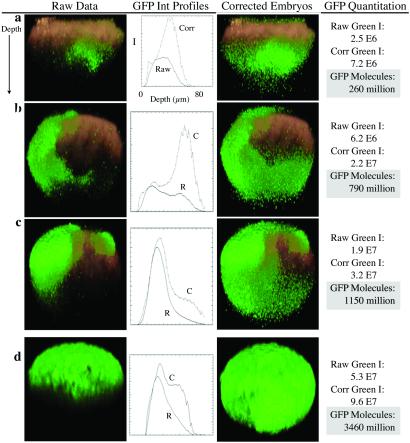
Multiplication by a compensating factor (i.e., depth correction) leads to improvement in fluorescence signal, most dramatically in the bottom third of the embryo (Fig. 5 a–d). Fig. 5 a–c represents HE-GFP DNA-injected embryos with increasing levels of gene expression, from top to bottom. The striking enhancement of the distal portion of each embryo is shown clearly in Fig. 5b. The location of the most intense gene expression shifts by 40 μm (slice 65 with correction vs. slice 25 for the raw data), and the integrated green intensity increases 3.5-fold from 6.2 × 106 to 2.2 × 107. The raw green intensity values vary almost 8-fold between Fig. 5a (2.5 × 106) and 5c (1.9 × 107), but after depth correction, the gap narrows to ≈4-fold. When comparing the distribution and number of fluorescent voxels, it appears that HE-GFP was incorporated at the two-cell stage in one blastomere for the embryo shown in Fig. 5c (6.3 × 105 voxels) and not until the four-cell stage in one blastomere for the embryo shown in Fig. 5a (2.6 × 105 voxels).
Figure 5.
Depth-corrected three-dimensional image reconstructions of HE-GFP DNA-injected (a–c) and Krox-GFP mRNA-injected (d) embryos. The embryos are not oriented on their animal–vegetal axis. Green voxels indicate areas of GFP expression. The boundaries of the embryo (shown in red) are delineated by TR. (a–c) Mosaic HE-GFP expression in three different embryos. Data processing results in clear signal enhancement in the bottom third of the embryos. (d) Representative embryo showing Krox-GFP translation and resulting GFP distribution. Depth correction restores signal at deeper slices. GFP quantitation is possible for all embryos by using depth-corrected GFP fluorescence intensities. The calculated GFP can be compared with predictions based on measured rates of sea urchin protein synthesis (19, 20): 6 pl injected × 150 fg/pl mRNA = 900 fg = 9 × 10−13 g of mRNA; 9 × 10−13 g of mRNA/2.8 × 105 g/mol = 3.2 × 10−18 mol of mRNA; and 3.2 × 10−18 × (6.02 × 1023 molecules per mol) = 1.9 × 106 mRNA molecules. mRNA translation rate = 140/h based on a spacing of 140 nucleotides per ribosome and a translational velocity of ≈1.8 codon⋅sec−1 at 15°C (19). Duration = 14 h, because GFP oxidation and chromophore formation take ≈4 h. Translation = (140/h) × 14 h = 1,960 GFPs per message at 18 h. 1,960 GFPs per mRNA × (1.9 × 106 mRNA) = 3.7 × 109 GFPs per embryo at 18 h. Predictions of GFP molecules in Krox-GFP-injected 18-hpf embryos represent an upper limit, because these calculations do not consider the degradation of mRNA or GFP.
To assess the strength of a particular promoter (in this case, the HE cis-regulatory system) in the face of mosaic incorporation and varying injection volumes, the corrected GFP intensity per voxel (GFP/voxel) is normalized by dividing by the TR intensity per voxel (TR/voxel) for each embryo (Table 1). A mean GFP/TR value of 0.62 ± 0.11 for HE-GFP indicates that the GFP intensities fall in a narrow distribution that can be compared between experiments and between different promoters.
Similar results are found for Krox-GFP mRNA-injected embryos but with higher GFP intensities (Table 1). The mean GFP fluorescence intensity of Krox-GFP mRNA-injected embryos is 8.8 (±0.7) × 107 (Table 1); embryo RNA1 corresponds to Fig. 5d and is a representative example. In lysates of these same embryos (18 hpf, injected with Krox-GFP mRNA, in aliquots of 20), a fluorometer identified 3.2 ± 0.3 × 109 GFP molecules per embryo. This value corresponds well with previous measurements of sea urchin protein synthesis, which predict an upper limit of 3.7 × 109 GFP molecules per embryo at 18 hpf; see Fig. 5 legend (19, 20). Thus, by dividing the fluorescence intensity by the number of GFP molecules, we conclude that each in vivo fluorescence intensity unit corresponds to 36 ± 7 GFP molecules. From these normalized data, GFPs per embryo was quantified as shown in the last column of Fig. 5 a–d.
Efforts were made to assess the degree to which the variability in the GFP/TR ratio in the mRNA-injected embryos (1.9 ± 0.5; Table 1) can be ascribed to artifacts of the imaging/data analysis vs. embryo–embryo variability in GFP expression. Sea urchin embryos were coinjected with varying concentrations of equimolar TR and fluorescein dextran. Although fluorescein photobleaches more readily than GFP and is slightly localized to the nucleus (unlike TR), the ratio of fluorescein to TR intensities varied little (1.55 ± 0.15) over a 2-fold concentration range typical for microinjected embryos (see Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). Thus, more than half the variation in GFP intensity observed in the mRNA-injected embryos must be due to biological variability.
Rigorous efforts to characterize GFP and color variants in vitro have identified many of the complications of using these reporter proteins to profile gene expression. A discrepancy in the published molar extinction coefficient of GFP (16, 17), reports of aggregation, and heretofore-unexplained single-molecule blinking behavior (21) raise questions about quantitative GFP measurements. However, it is evident from this study that measurements of bulk GFP (106–1010 molecules) can be performed reproducibly in embryos with acceptable standard error. In these experiments, there are 36 GFP molecules per intensity unit, 1–255 intensity units per voxel for an 8-bit detector, and therefore 36–9,200 GFP molecules per voxel. Because each voxel has a volume of ≈2 × 10−16 liter, the concentrations measured in vivo were typically 0.3–80 μM. Purified GFP was titrated linearly in this concentration range and quantified in solution by using a fluorometer. This indicates that aggregation and self-quenching phenomena are unlikely to compromise CLSM measurements at the GFP concentrations present within these living embryos.
It is worth noting that the relationship between the number of GFP molecules and the strength of a promoter cannot be defined absolutely unless we know the rates of GFP message and protein turnover within a particular cell. Experiments to determine these rates can be performed, in fact, to make direct measurements of gene activity. Although it is possible to assay more directly the mRNA levels with quantitative fluorescence PCR (3) or by whole-mount in situ hybridization, monitoring GFP provides high sensitivity and allows repeated measurements of gene expression in living embryos. In principle, the exercise of correlating fluorescence with mRNA data need only be performed once for a single embryonic stage with a given confocal microscope, microscope objective, laser power, and photomultiplier-tube voltage. Later experiments can compute GFP molecules from fluorescence intensity relative to this benchmark.
Extending Analyses to Developmental Biology.
Our future goals are to correct the fluorescence for both the depth and position of GFP within an embryo and to improve the spatial and kinetic resolution in measurements of gene expression. The analysis is simplest for the blastula-stage sea urchin embryos studied in this work; they are spherically symmetric, allowing correction of the GFP intensity relative to the depth dependence determined for TR. Subsequent image-processing algorithms will consider both the depth and position of the GFP within the embryo, and such changes are in progress for the consideration of 72-h sea urchin plutei, where the geometry and gene-expression patterns are more complex. In addition, it should be possible to measure localized (i.e., gut-specific) gene activity and tabulate GFP expression on a cell-by-cell or intracellular (nucleus vs. cytoplasm) basis. To determine gene expression within individual cells, the boundaries may be identified by using transmitted light images or fluorescent markers. Using less stable GFP constructs (≈1 h half-life) may exploit the high sensitivity of CLSM fluorescence detection to provide greater kinetic resolution of gene activity.
For CLSM experiments involving large specimens or applications requiring high-throughput or high temporal resolution, faster but similarly accurate GFP measurements can be achieved by compromising three-dimensional (spatial) resolution. For sea urchin embryos injected with TR and Krox-GFP mRNA, CLSM data were collected where the thickness of each optical section was varied from 1 to 10 μm. For the same section thickness (pinhole size), the total quantity of TR and GFP calculated for each embryo was found to vary inversely with the spacing between optical sections. This observation proves particularly useful in larger embryos where imaging GFP with fewer optical sections saves significant time. To test this principle and illustrate the broader utility of our depth-correction algorithms, we recently imaged 24-hpf starfish embryos (200-μm diameter, 3-μm sections) by CLSM. By using a ×40, 1.2-numerical aperture water-immersion, C-Apochromat lens (Zeiss), it was possible to section optically through the entire embryo and quantify GFP by using TR as a fluorescent standard.
Broad Applicability of Quantitative CLSM Technique.
Improvements in quantitative CLSM hold great promise for elucidating a wide variety of in vivo processes in molecular detail. GFP, DsRed (red fluorescent protein from Discosoma), and their color variants possess high quantum yields and have absorption and emission profiles that span the visible spectrum, are excited readily by conventional laser sources, and are spectrally resolvable with commercial filter sets (16, 22). In addition, a wide variety of fluorescent probes can be delivered inside living organisms and could be assayed by using confocal microscopy (23). These imaging methods will facilitate studies of embryonic development and biological processes in many model organisms including sea urchins, Caenorhabditis elegans, Drosophila, and zebrafish.
Supplementary Material
Acknowledgments
We thank Drs. Andrew Cameron and Chiou-Hwa Yuh for many helpful discussions; Carolina B. Livi for providing the Krox-GFP construct; Dr. Veronica Hinman for providing starfish embryos; and the California Institute of Technology Seismological Laboratory for computational facilities. This work was supported by the Helen Hay Whitney Foundation (to I.J.D.), National Aeronautics and Space Administration Earth System Science Fellowship Grant NGT5-30393 (to J.E.D.), and National Institutes of Health Grant HD37105 (to E.H.D. and S.E.F.).
Abbreviations
- CLSM
confocal laser scanning microscopy
- TR
Texas red
- HE
hatching enzyme
- hpf
hours postfertilization
References
- 1.Ferrer-Martinez A, Gomez-Foix A M. BioTechniques. 2002;32:62–66. doi: 10.2144/02321st01. [DOI] [PubMed] [Google Scholar]
- 2.Kam Z, Hanser B, Gustafsson M G L, Agard D A, Sedat J W. Proc Natl Acad Sci USA. 2001;98:3790–3795. doi: 10.1073/pnas.071275698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davidson E H, Rast J P, Oliveri P, Ransick A, Calestani C, Yuh C-H, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 4.Yuh C-H, Bolouri H, Davidson E H. Science. 1998;279:1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- 5.Pawley J B, editor. Handbook of Biological Confocal Microscopy. New York: Plenum; 1995. [Google Scholar]
- 6.Hough-Evans B R, Britten R J, Davidson E H. Dev Biol. 1988;129:198–208. doi: 10.1016/0012-1606(88)90174-1. [DOI] [PubMed] [Google Scholar]
- 7.Livant D L, Hough-Evans B R, Moore J G, Britten R J, Davidson E H. Development (Cambridge, UK) 1991;113:385–398. doi: 10.1242/dev.113.2.385. [DOI] [PubMed] [Google Scholar]
- 8.Zlokarnik G, Negulescu P A, Knapp T E, Mere L, Burres N, Feng L, Whitney M, Roemer K, Tsien R Y. Science. 1998;279:84–88. doi: 10.1126/science.279.5347.84. [DOI] [PubMed] [Google Scholar]
- 9.Hyrc K B, Bownik J M, Goldberg M P. Cell Calcium. 2000;27:75–86. doi: 10.1054/ceca.1999.0092. [DOI] [PubMed] [Google Scholar]
- 10.Terasaki M. Mol Biol Cell. 2000;11:897–914. doi: 10.1091/mbc.11.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei Z, Angerer L M, Gagnon M L, Angerer R C. Dev Biol. 1995;171:195–211. doi: 10.1006/dbio.1995.1271. [DOI] [PubMed] [Google Scholar]
- 12.Kirchhamer C V, Davidson E H. Development (Cambridge, U K) 1996;122:333–348. doi: 10.1242/dev.122.1.333. [DOI] [PubMed] [Google Scholar]
- 13.McClay D R, Wessel G M, Marchase R B. Proc Natl Acad Sci USA. 1981;78:4975–4979. doi: 10.1073/pnas.78.8.4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bogarad L D, Arnone M I, Chang C, Davidson E H. Proc Natl Acad Sci USA. 1998;95:14827–14832. doi: 10.1073/pnas.95.25.14827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W, Wikramanayake A H, Gonzalez-Rimbau M, Vlahou A, Flytzanis C N, Klein W H. Mech Dev. 1996;60:185–195. doi: 10.1016/s0925-4773(96)00614-4. [DOI] [PubMed] [Google Scholar]
- 16.Heim R, Tsien R Y. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 17.Patterson G H, Knobel S M, Sharif W D, Kain S R, Piston D W. Biophys J. 1997;73:2783–2790. doi: 10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ormö M, Cubitt A B, Kallio K, Gross L A, Tsien R Y, Remington S J. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 19.Davidson E H. Gene Activity in Early Development. New York: Academic; 1976. [Google Scholar]
- 20.Goustin A S, Wilt F H. Dev Biol. 1981;82:32–40. doi: 10.1016/0012-1606(81)90426-7. [DOI] [PubMed] [Google Scholar]
- 21.Dickson R M, Cubitt A B, Tsien R Y, Moerner W E. Nature (London) 1997;388:355–358. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 22.Matz M V, Fradkov A F, Labas Y A, Savitsky A P, Zaraisky A G, Markelov M L, Lukyanov S A. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 23.Haugland R P. Handbook of Fluorescent Probes and Research Chemicals. Eugene, OR: Molecular Probes; 1996. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.