Abstract
Feeding relationships can cause invasions, extirpations, and population fluctuations of a species to dramatically affect other species within a variety of natural habitats. Empirical evidence suggests that such strong effects rarely propagate through food webs more than three links away from the initial perturbation. However, the size of these spheres of potential influence within complex communities is generally unknown. Here, we show for that species within large communities from a variety of aquatic and terrestrial ecosystems are on average two links apart, with >95% of species typically within three links of each other. Species are drawn even closer as network complexity and, more unexpectedly, species richness increase. Our findings are based on seven of the largest and most complex food webs available as well as a food-web model that extends the generality of the empirical results. These results indicate that the dynamics of species within ecosystems may be more highly interconnected and that biodiversity loss and species invasions may affect more species than previously thought.
The mean distance between all nodes in a web (D) is perhaps the most familiar property of complex networks. For example, the “small world” phenomenon in large social networks is popularly termed “six degrees of separation” (1). Known as the characteristic path length, D quantifies the average number of links necessary for information or an effect to propagate along the shortest paths between nodes in a network. Networks are enjoying an increasing amount of interdisciplinary interest (2) as illustrated by examinations of D among film actor guilds, electrical power grids, neural networks, and the Internet (1–3). The first study to calculate D in food webs found small D in relatively simple webs with 3–33 taxa, but the empirical variation in the data prompted a call for much more research on path lengths in food webs (4).
A food web consists of L directed trophic links among S nodes or “trophic species.” Trophic links occur between consumer taxa (i.e., predators, parasites, herbivores, etc.) and the resource taxa (i.e., prey, hosts, plants, etc.) that they eat. Trophic species are functionally distinct network nodes composed of all taxa within a particular food web that share identical consumers and resources. Aggregating functionally similar taxa into trophic species is a convention within structural food-web studies that appears to reduce methodological biases in the data and emphasizes the topologically distinct aspects of food-web networks while downplaying phylogenetic and other differences among lumped taxa (see references in ref. 5).
Connectance (C) is the fraction of all possible links that are realized (L/S2) and represents a standard measure of food web complexity thought to be independent of S (6, 7). The distance (d) between every species pair in a web is averaged to compute D (1). Paths are treated as undirected because effects can propagate through the network in either direction. Species one link from a focal species (d = 1) are those that are a direct consumer or resource of the focal species. A species two links (d = 2) from a focal species lacks a direct trophic interaction with that species and does one or more of the following: (i) consumes a resource species of the focal species, (ii) supports a consumer species of the focal species, (iii) consumes a consumer of the focal species, or (iv) is a resource of a resource of the focal species. When defining D, self-self links are typically ignored in network analyses (1). Because self-self trophic links may be dynamically important in ecosystems, we define d for the self-self species pair the same as for any other species pair. This method also allows us to include the important ecological distinction (8) between cannibals, which have d = 1, and other species, which have d = 2. Our method alters D among our webs an average of <1% compared with employing the more standard convention.
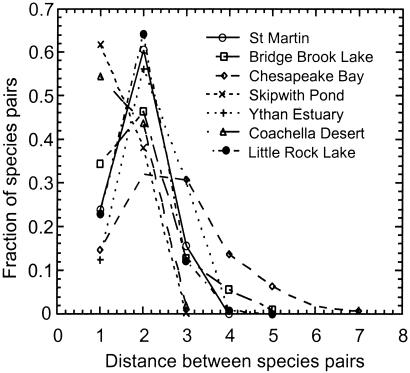
Although there are hundreds of food webs in the literature, the vast majority have been criticized for being incomplete, having too few species, and lacking a rigorous empirical base (4, 8–11). Therefore, we focused our analyses on seven of the largest, most comprehensive, and highest quality empirical food webs in the primary literature (Table 1; ref. 5). Three are from freshwater habitats: Skipwith Pond (12), Little Rock Lake (9), and Bridge Brook Lake (13). Two are from habitats at freshwater-marine interfaces: Chesapeake Bay (14) and Ythan Estuary (15). Two are from terrestrial habitats: Coachella Valley (8) and the island of St. Martin (16). Among these webs, D ranges between 1.40 and 2.71 and decreases with increasing connectance (Table 1, Fig. 1). On average, these values of D are 5% smaller than if we had not aggregated species into trophic species (data not shown). Distances between species pairs (d) are closely clustered around the mean, with very few widely separated species pairs (d > 3, Fig. 2). Across the seven webs, an average of 26% of species pairs interact directly (d = 1), and 80% and 97% of the species pairs are within two and three links of each other, respectively.
Table 1.
Properties of empirical and niche model food webs
| Food web | Taxa | S | C (L/S2) | Observed D | Model D | Error (model SD) |
|---|---|---|---|---|---|---|
| Skipwith Pond | 35 | 25 | 0.32 | 1.40 | 1.44 | 1.10 |
| Little Rock Lake | 181 | 92 | 0.12 | 1.90 | 1.88 | −0.94 |
| Bridge Brook Lake | 75 | 25 | 0.17 | 1.92 | 1.80 | −1.68 |
| Chesapeake Bay | 33 | 31 | 0.072 | 2.71 | 2.55 | −0.99 |
| Ythan Estuary | 92 | 78 | 0.061 | 2.20 | 2.32 | 2.06 |
| Coachella Valley | 30 | 29 | 0.31 | 1.47 | 1.45 | −0.68 |
| St. Martin Island | 44 | 42 | 0.12 | 1.92 | 1.98 | 0.99 |
Taxa, the original names for groups of organisms found in the primary reference. S, trophic species. The seven food webs address (i) primarily invertebrates in Skipwith Pond (12); (ii) pelagic and benthic species in Little Rock Lake (8), the largest food web in the primary literature; (iii) Bridge Brook Lake, the largest among a recent set of 50 Adirondak lake pelagic food webs (13); (iv) the pelagic portion of Chesapeake Bay emphasizing larger fishes (14); (v) mostly birds and fishes among invertebrates and primary producers in the Ythan Estuary (15); (vi) a wide range of highly aggregated taxa in the Coachella desert (9); and (vii) trophic interactions emphasizing Anolis lizards on the Caribbean island of St. Martin (16). Model D is the mean of 1,000 niche model trials. Errors between the niche model D and observed D are normalized by dividing the difference by the SD of the trial results. Ninety-five percent of normalized errors should be within 2 SD if the model is accurate (5).
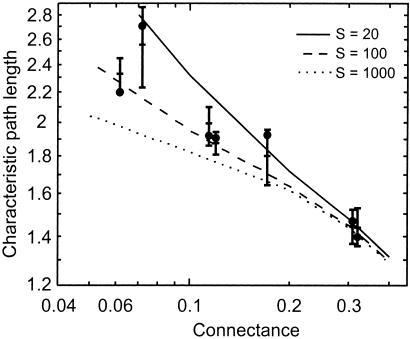
Figure 1.
Characteristic path length D of the seven empirical webs listed in Table 1 (●), error bars showing mean ± 2 SD for niche model webs with the same S and C as the empirical webs, and curves showing mean D vs. C for niche model webs with S = 20, 100, and 1,000. Log-log plot shows the approximate power-law relationship between mean D of niche model webs and C for all S. The seven empirical webs are, from left to right, Ythan Estuary (S = 78), Chesapeake Bay (S = 31), St. Martin Island (S = 42), Little Rock Lake (S = 92), Bridge Brook Lake (S = 25), Coachella Valley (S = 29), and Skipwith Pond (S = 25). Because the niche model is a stochastic model, previously described Monte Carlo techniques (5) were used to measure the mean and SD of the niche model predictions, and errors are normalized by the SD of the model prediction (Table 1). The mean normalized error is −0.02 model SD, which is very close to zero as expected when the model fits the data. The SD of the errors is 1.4, showing slightly greater variability of normalized error than the theoretically expected SD of 1 (5). A null model that randomly arranges trophic links while maintaining empirically observed S and C (5) fits the data much worse as indicated by a normalized error mean of −2.6 and SD of 5.4.
Figure 2.
Distributions of distances (d) between species pairs for the seven webs listed in Table 1. The histograms are normalized to show the fraction of species pairs at each distance.
Small values of D may be most surprising in Little Rock Lake because of its many species and the strong representation of both benthic and pelagic habitats. D had been hypothesized to increase beyond 2 in large food webs (4). Recent studies reinforce this expectation by asserting that D increases as log S (17) and by finding D = 3.4 in a Scotch Broom food web, which contains 154 taxonomic species and was suggested as the “best-defined web” for investigating topology (18). In addition, boundaries between habitats may be expected to result in food-web compartments (10) or clusters (1) whose presence would increase D well beyond that found in other webs with less representation of different habitats. The Little Rock Lake food web is more clustered than expected at random but the clusters are linked such that D remains small (18, 19, ‖). However, the small-world property of much greater than random clustering (1) appears absent among most food webs (17, 19), suggesting that predictions about D and other properties based on small-world assumptions may not apply to most food webs.
This theoretical limitation combined with limited empirical variation of diversity (S) and connectance (C) among available data leaves the systematic sensitivity of D to S and C in need of more study. Therefore, we explored this sensitivity with a recently described model of food web structure (5, 20). This “niche model” uses S and C as input parameters and successfully predicts a dozen food-web properties but was originally untested against D (5). We compared D of the seven empirical webs to that of webs generated by the niche model. The niche model constructs webs with the same S and C as the empirical webs by randomly arranging S species on a one-dimensional community niche. Each species eats all species within one contiguous section of the niche. The center of the section has a randomly chosen lower niche value than the consuming species, and the width of the section is randomly varied within the constraint that C in the synthesized web matches the input value. The niche model accurately and precisely predicts D for all seven empirical food webs (Fig. 1, Table 1).
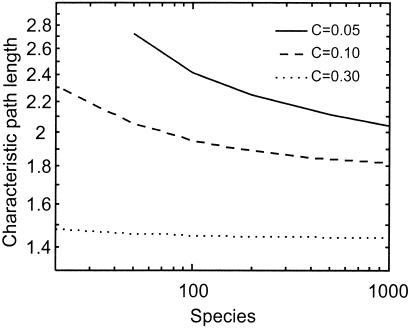
Given the success of the model, we used it to characterize the sensitivity of D to both S and C. Model results indicate that D is moderately sensitive to C and less sensitive to S (Figs. 1 and 3). For webs with constant S, D decreases by a factor of 2 with an order-of-magnitude increase of C in an approximate power law relationship whose slope is greater for smaller webs (Fig. 1). For webs with constant C, D surprisingly decreases with increasing S (Fig. 3). Other analyses employing outdated (7, 21) link-scaling assumptions asserted that D increases with S (4, 17). However, as S increases by two orders of magnitude, D decreases ≈5% in more complex webs (C > 0.20) and 15 to 50% in less complex webs (0.05 < C < 0.10).
Figure 3.
Sensitivity of D among niche model webs to S. Lines connect the means from 1,000 iterations for each level of S and C.
These findings show that even in high quality, species-rich food webs, species are generally linked by short chains. Eighty percent of species are connected by one or two links, which results in an average shortest path between species of approximately two. Thus, most species in a food web can be thought of as “local” to each other and can potentially interact with other species through at least one short trophic chain. Empirical studies show that short-chain indirect effects (path length = 2 or 3) can be as important as direct effects (path length = 1) and become evident nearly as quickly (22, 23). Combined with our results, this finding indicates that adding, removing, or altering species has the potential to rapidly affect many or most species in large complex communities.
Our findings should be tempered by the possibilities of either over- or underestimating D. Food webs may underestimate both the trophic and functional connectance of organisms in complex communities and thus overestimate the effective D and underestimate the potential for propagation of effects. This result may be partly due to scientists underreporting the actual number of trophic links present among species (11). More significantly, food webs depict only one of many types of interactions among species. Other nontrophic interactions include ecosystem engineering, facilitation, behavioral modification, and interference competition (24–27). If multiple types of interactions are accounted for, the ecological connectance among species should be higher than the trophic connectance we report. Thus, species may be ecologically closer than our results suggest because D decreases with increasing C. On the other hand, our results may overestimate the potential for propagation of effects because many food-web links may be “weak” and therefore unimportant in determining species dynamics and community structure and function (11, 28). This result suggests that food webs that include all trophic links overestimate functional connectance of species. However, recent theoretical and experimental studies show that many purportedly weak links are dynamically important (27, 29). Even if food webs overestimate functional connectance, empirical and model webs with low connectance (C < 0.08) still display short characteristic path lengths (D < 3) for all but the smallest and simplest webs (Fig. 1). Small empirical webs rarely have low connectance (4). These considerations show that our general conclusion of an average of two degrees of separation in complex food webs is theoretically robust and consistent with the best available data. However, two degrees may overestimate the size of ecological “worlds” when other interspecific interactions are taken into account.
Two degrees of separation in ecological worlds is particularly important because empiricists rarely observe strong effects between species to propagate further than three links (22, 24, 30, 31). For paths lengths ≤3, both theoretical and empirical studies have shown that shorter chain effects are not necessarily stronger than longer chains (e.g., refs. 22, 24, and 27). The most comprehensive review of experimentally demonstrated indirect effects suggests that both direct and indirect effects each account for ≈40% of the change in community structure due to manipulating species' abundances (24), with the remaining 20% of the variance left unexplained. Of the indirect effects, almost 95% were due to short chains (path length = 2 or 3; see appendixes in ref. 24). The percentage of variance explained because of direct and indirect effects appears independent of species richness (24), which is consistent with our result that D is insensitive to S. With increasing web size, the number of significant short-chain indirect effects per species increases, whereas the number of significant long-chain indirect effects remains small even as the number of longer paths grows rapidly (24).
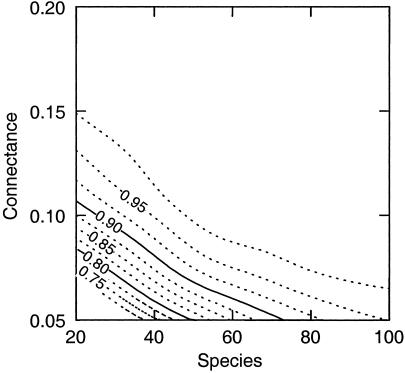
Other empirical studies have demonstrated strong “trophic cascades” in a variety of terrestrial and aquatic ecosystems (32–34), where significant effects often propagate two and sometimes three links from manipulated species. These studies confirm that path lengths with <3 links are frequently dynamically important. However, the paucity of demonstrated trophic cascades at long distances (path length > 3) is consistent with other empirical studies that suggest that species pairs with d ≥ 3 are functionally (or dynamically) disconnected. This paucity has several other plausible explanations. Our data and the niche model suggest that, in most webs, more than 95% of species pairs have d ≤ 3 (Fig. 4). The few empirical webs that are exceptions have relatively few species or are larger webs with unusually low connectance. Because d ≤ 3 chains are almost always present between species, attributing effects to longer chains may be particularly difficult because shorter effect chains must be excluded as responsible for the effects. The lack of long chain effects may also be due to aggregation of species (8, ∥) in these studies into trophic levels, which could conceal such effects (34). For example, manipulating the density of consumers of secondary carnivores could cause a positively responding plant species to compensate for a negatively responding plant species. More attention on disaggregated species within trophic levels could illuminate such possibilities (34).
Figure 4.
Fraction of species pairs with d ≤ 3 in niche model webs as a function of species number (S) and connectance (C). Adjacent lines designate isopleths that are 0.025 apart. Most empirical webs fall above and to the right of the 0.95 isopleth.
Together, the empirical studies of species manipulation effects suggest that “distant” (d > 3) species rarely influence each other. Therefore, finding D > 3 in food webs, as suggested in other food-web studies (4, 18) and found in other complex small-world networks (1, 35), would seriously challenge the popular ecological adage that “everything is connected to everything else.” D > 3 would have suggested that many if not most species within food webs are functionally isolated from one another. Therefore, our analyses may be the most systematic and thorough scientific corroboration of this ecological adage by demonstrating that all species within most ecological systems are potentially close neighbors. Overall, the robustness of short characteristic path length D in food webs to changes in size, complexity, and habitat suggests that the small world potential for widespread and rapid dispersion of effects (1) throughout a community of interacting organisms generally applies to ecosystems despite their lower than “small world” clustering (17, 19). Larger D may be found in food webs that span more distinct habitat boundaries (e.g., those between terrestrial and aquatic ecosystems). Larger D may also be methodologically generated by focusing on “source” webs. By definition, such webs bias web structure by restricting membership to direct and indirect consumers of only one or very few resource species found in a community (36). As a result of ignoring these consumers' links to excluded resources, source webs tend to have unusually low connectance (19), which our model shows is associated with larger D. We excluded source webs, including the Scotch Broom web focused on in another topology study (18), from analysis because of this bias. The Scotch Broom web is built up from a single shrub species and has C = 0.016 (37) and D = 3.4 (18).
Mechanisms potentially responsible for small D among food webs are currently unknown and deserve further investigation. Our findings suggest that such mechanisms are closely related to observed levels of connectance (6, 7) and other factors that generate web topology (5). Evolution of feeding capabilities has been suggested as responsible for the observed levels of C (6). Mechanisms associated with population dynamics may also be responsible as suggested by findings that increasing the number of weak interactions may increase dynamic stability and facilitate coexistence (7, 27, 29).
Within habitats as well as among habitats, attention to trophic paths between species could help conservation managers by suggesting whether and how species affect each other. However, more research on the variation of effect strengths between species is needed to enable managers to prioritize the attention given to species within 1, 2, and 3 links of a species of concern (e.g., endangered or invasive species). Without such research, our finding that almost everything is connected to everything else only slightly reduces the topologically conceivable effects to consider. Overall, our results specify how biodiversity loss (38), species invasions, and changes in populations have the topologically and dynamically demonstrated potential to affect many more co-occurring species than is often appreciated (30). The degree to which this potential is realized deserves much additional research.
Acknowledgments
The comments of Bruce Menge and several anonymous reviewers greatly helped the manuscript. This work was supported by the Santa Fe Institute (J.A.D.), National Science Foundation Grants DEB-0083929 (to R.J.W., A.-L.B., and N.D.M.) and DUE-9950461 (to N.D.M. and R.J.W.), and a Biological Informatics Postdoctoral Fellowship to J.A.D. (DEB/DBI-0074521).
Abbreviations
- D
characteristic path length
- d
shortest distance between two species
- S
number of trophic species
- L
number of trophic links
- C
connectance
Footnotes
Williams, R. J., Martinez, N. D., Berlow, E. L., Dunne, J. A. & Barabási, A.–L. (2001) Two Degrees of Separation in Complex Food Webs, Santa Fe Institute Working Paper 01-07-036.
References
- 1.Watts D J, Strogatz S H. Nature (London) 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 2.Strogatz S H. Nature (London) 2001;410:268–275. doi: 10.1038/35065725. [DOI] [PubMed] [Google Scholar]
- 3.Albert R, Jeong H, Barabási A-L. Nature (London) 1999;401:130–131. [Google Scholar]
- 4.Schoener T W. Ecology. 1989;70:1559–1589. [Google Scholar]
- 5.Williams R J, Martinez N D. Nature (London) 2000;404:180–183. doi: 10.1038/35004572. [DOI] [PubMed] [Google Scholar]
- 6.Martinez N D. Am Nat. 1992;139:1208–1218. [Google Scholar]
- 7.Warren P H. Trends Ecol Evol. 1994;9:136–141. doi: 10.1016/0169-5347(94)90178-3. [DOI] [PubMed] [Google Scholar]
- 8.Martinez N D. Ecol Monog. 1991;61:367–392. [Google Scholar]
- 9.Polis G A. Am Nat. 1991;138:123–155. [Google Scholar]
- 10.Pimm S L, Lawton J H, Cohen J E. Nature (London) 1991;350:669–674. [Google Scholar]
- 11.Paine R T. Ecology. 1988;69:1648–1654. [Google Scholar]
- 12.Warren P H. Oikos. 1989;55:299–311. [Google Scholar]
- 13.Havens K. Science. 1992;257:1107–1109. doi: 10.1126/science.257.5073.1107. [DOI] [PubMed] [Google Scholar]
- 14.Baird D, Ulanowicz R E. Ecol Monogr. 1989;59:329–364. [Google Scholar]
- 15.Hall S J, Raffaelli D. J Anim Ecol. 1991;60:823–842. [Google Scholar]
- 16.Goldwasser L, Roughgarden J. Ecology. 1993;74:1216–1233. [Google Scholar]
- 17.Camacho J, Guimerà R, Amaral L A N. Phys Rev Lett. 2002;88:228102–228105. doi: 10.1103/PhysRevLett.88.228102. [DOI] [PubMed] [Google Scholar]
- 18.Montoya J M, Solé R V. J Theor Biol. 2002;214:405–412. doi: 10.1006/jtbi.2001.2460. [DOI] [PubMed] [Google Scholar]
- 19. Dunne, J. A., Williams, R. J. & Martinez, N. D. (2002) Proc. Natl. Acad. Sci. USA, 12917–12922. [DOI] [PMC free article] [PubMed]
- 20.Camacho J, Guimerà R, Amaral L A N. Phys. Rev. E. 2002. http://link.asp.org/abstract/PRE/v65/e030901 , e-Print Archive, http://link.asp.org/abstract/PRE/v65/e030901. . [DOI] [PubMed] [Google Scholar]
- 21.Martinez N D. Science. 1993;260:242–243. doi: 10.1126/science.260.5105.242. [DOI] [PubMed] [Google Scholar]
- 22.Abrams P, Menge B A, Mittelbach G G, Spiller D, Yodzis P. In: Food Webs: Integration of Pattern and Dynamics. Polis G A, Winemiller K O, editors. New York: Chapman & Hall; 1995. pp. 371–396. [Google Scholar]
- 23.Menge B A. Am Nat. 1997;149:801–823. doi: 10.1086/286025. [DOI] [PubMed] [Google Scholar]
- 24.Menge B A. Ecol Monog. 1995;65:21–74. [Google Scholar]
- 25.Bertness M D, Leonard G H, Levine J M, Schmidt P R, Ingraham A. Ecology. 1999;80:2711–2726. [Google Scholar]
- 26.Jones C G, Lawton J H, Shachak M. Oikos. 1994;69:373–386. [Google Scholar]
- 27.Berlow E L. Nature (London) 1999;398:330–334. [Google Scholar]
- 28.Power M E, Tilman D, Estes J A, Menge B A, Bond W J, Mills L S, Daily G, Castilla J C, Lubchenco J, Paine R T. Bioscience. 1996;46:609–620. [Google Scholar]
- 29.McCann K S. Nature (London) 2000;405:228–233. doi: 10.1038/35012234. [DOI] [PubMed] [Google Scholar]
- 30.Strong D R. Ecology. 1992;73:747–754. [Google Scholar]
- 31.Schoener T W. In: Mutualism and Community Organization: Behavioral, Theoretical and Food-Web Approaches. Kawanabe H, Cohen J E, Iwasaki M, editors. New York: Oxford Univ. Press; 1993. pp. 365–411. [Google Scholar]
- 32.Brett M T, Goldman C R. Science. 1997;275:384–386. doi: 10.1126/science.275.5298.384. [DOI] [PubMed] [Google Scholar]
- 33.Pace M L, Cole J J, Carpenter S R, Kitchell J F. Trends Ecol Evol. 1999;14:483–488. doi: 10.1016/s0169-5347(99)01723-1. [DOI] [PubMed] [Google Scholar]
- 34.Schmitz O J, Hambäck, Beckerman A P. Am Nat. 2000;155:141–153. doi: 10.1086/303311. [DOI] [PubMed] [Google Scholar]
- 35.Jeong H, Tombor B, Albert R, Oltvai Z, Barabási A-L. Nature (London) 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 36.Hawkins B A, Martinez N D, Gilbert F. Acta Oecol-Int J Ecol. 1997;18:575–586. [Google Scholar]
- 37.Memmott J, Martinez N D, Cohen J E. J Anim Ecol. 2000;69:1–15. [Google Scholar]
- 38.Dunne J A, Williams R J, Martinez N D. Ecol Lett. 2002;5:558–567. [Google Scholar]