Abstract
Networks from a wide range of physical, biological, and social systems have been recently described as “small-world” and “scale-free.” However, studies disagree whether ecological networks called food webs possess the characteristic path lengths, clustering coefficients, and degree distributions required for membership in these classes of networks. Our analysis suggests that the disagreements are based on selective use of relatively few food webs, as well as analytical decisions that obscure important variability in the data. We analyze a broad range of 16 high-quality food webs, with 25–172 nodes, from a variety of aquatic and terrestrial ecosystems. Food webs generally have much higher complexity, measured as connectance (the fraction of all possible links that are realized in a network), and much smaller size than other networks studied, which have important implications for network topology. Our results resolve prior conflicts by demonstrating that although some food webs have small-world and scale-free structure, most do not if they exceed a relatively low level of connectance. Although food-web degree distributions do not display a universal functional form, observed distributions are systematically related to network connectance and size. Also, although food webs often lack small-world structure because of low clustering, we identify a continuum of real-world networks including food webs whose ratios of observed to random clustering coefficients increase as a power–law function of network size over 7 orders of magnitude. Although food webs are generally not small-world, scale-free networks, food-web topology is consistent with patterns found within those classes of networks.
Food webs, which depict networks of trophic relationships in ecosystems, provide complex yet tractable depictions of biodiversity, species interactions, and ecosystem structure and function. Although food web studies have long been central to ecological research (1–3), there has been controversy over whether there are regularities in food-web structure worth explaining (4). Early analyses of topological properties of empirical food webs emerged from research on ecological diversity–stability relationships (e.g., refs. 5 and 6) and typically used low resolution, species-poor (S < 20) depictions of food webs (e.g., refs. 7 and 8). Dramatic improvements in data (e.g., refs. 9–11) led to the successful description and modeling of general food-web properties among ecosystems (12), including how food-web properties vary with species richness, resolution, and sampling effort (e.g., refs. 13–15).
Research on food-web structure is one example of an expanding range of “real-world” network topology studies (16). In particular, complex systems research across many disciplines has resulted in renewed interest in the study of “small-world” network topology (17) inspired by the “six degrees of separation” sociology experiment by Milgram (18). The two characteristics required of small worlds are (i) high clustering compared with a random graph, with neighbors of a node much more likely to be connected to each other than in a random graph, and (ii) small path length compared with a regular lattice, with the average shortest path length (“characteristic path length”) among all pairs of nodes increasing logarithmically with the number of nodes, similar to what is seen in random graphs (17). Types of networks examined for these topological properties include social, technological, economic, and biological networks (see refs. 16 and 19 for reviews).
Researchers have explicitly examined several food webs for small-world properties, with conflicting results and interpretations. Studies have generally agreed that food webs display short path lengths, consistent with small-world topology (20–23). However, early research using low-quality (i.e., poorly resolved, highly aggregated, low diversity) food-web data speculated that characteristic path lengths would increase with greater inclusion of species (20), as did a more recent study (23), contrary to recent findings that path lengths decrease with increasing food-web size and complexity measured as species richness and connectance (links/species2), respectively (21). Two recent studies that have examined clustering have come to opposite conclusions: a set of four food webs was described as having greater than random clustering, consistent with small-world topology (22), whereas an overlapping set of seven food webs was described as having similar to random clustering (23). Thus, with regard to small-world topology there has been little agreement about what empirical food web structure generally looks like.
A great deal of attention has been placed on the power–law, or “scale-free” distribution of node degrees of many small-world networks (24). Real-world networks display degree distributions that deviate from a Poisson distribution found for simple random graph models (25, 26). Many networks, including the world wide web, Internet domains and routers, scientific coauthors and citations, metabolic and protein networks, and phone–call networks display a scale-free degree distribution, with power–law exponents ranging from ≈1 to 2.5 (19). However, real-world networks can display other types of degree distributions, including “broad-scale” distributions that show a power–law regime with a sharp cutoff in the tail (e.g., movie–actor collaborations) and “single-scale” distributions with fast-decaying tails including exponential (e.g., Caenorhabditis elegans neural network) and Gaussian (e.g., acquaintances among a Mormon social group) distributions (27). As with small-world topology, there has been little agreement about what degree distributions in food webs look like, except that their distributions deviate from random. One previous study suggests that food webs are scale-free (22), whereas another study suggests that food webs display exponential degree distributions, and provides a quantitative formulation of that pattern in a universal functional form (23).
An early study (20) acknowledged limitations on drawing general conclusions about topology from the species-poor food webs available at that time. The more recent studies, which examine higher quality, more speciose food webs, have been limited by their use of relatively few (<8) data sets (21–23). Two of the studies used a simple and predictive “niche model” of food-web structure (12) to extend their results (21, 23). However, generalities derived from a model may obscure informative details of empirically observed topology, especially in ecology, where variability can be both extremely high and critically important.
In contrast to the limitations of the previous studies, we used a set of 16 high-quality food webs from a variety of ecosystems to develop the most comprehensive picture to date of whether food webs display small-world and scale-free structure (22), similar to topology of many other real-world networks (19), or whether food webs display other kinds of topology. Our use of a wider array of food webs also allows us to probe the relationship of the discussed metrics of food-web topology to basic measures of food web complexity including network size (S), links per species (L/S), and connectance (L/S2). We briefly discuss some potential ecological implications of observed food-web network structure.
Methods
The 16 food webs studied, two of which are variants of the same web, represent a wide range of species number, linkage densities, taxa, and habitat types (Table 1). There are five lakes or ponds, two streams, three estuaries, and five terrestrial ecosystems (temperate, desert, subtropical) represented. The food webs are described in more detail in their individual references (Table 1; refs. 9, 11, 15, and 28–37; see also ref. 38). Food webs consist of L directed trophic links between S nodes or “trophic species.” Trophic species, functional groups of taxa that share the same set of predators and prey within a food web, are a widely accepted convention in structural food-web studies that reduce methodological biases related to uneven resolution of taxa within and among food webs (12). Trophic species webs are constructed by aggregating taxa from the original “taxonomic food webs” into trophic species. Taxonomic to trophic species aggregation occurs for <10% of nodes in nine of the food webs studied (Table 1).
Table 1.
Topological properties of empirical and random food webs, listed in order of increasing connectance
| Ref. | Taxa | S | C(L/S2) | L/S | D | Dran | Cl | Clran | Cl/Clran | |
|---|---|---|---|---|---|---|---|---|---|---|
| Grassland | 15 | 75 | 61 | 0.026 | 1.59 | 3.74 | 3.63 | 0.11 | 0.03 | 3.7 |
| Scotch Broom | 28 | 154 | 85 | 0.031 | 2.62 | 3.11 | 2.82 | 0.12 | 0.04 | 3.0 |
| Ythan Estuary 1 | 29 | 134 | 124 | 0.038 | 4.67 | 2.34 | 2.39 | 0.15 | 0.04 | 3.8 |
| Ythan Estuary 2 | 30 | 92 | 83 | 0.057 | 4.76 | 2.20 | 2.19 | 0.16 | 0.06 | 2.7 |
| El Verde Rainforest | 31 | 156 | 155 | 0.063 | 9.74 | 2.20 | 1.95 | 0.12 | 0.07 | 1.4 |
| Canton Creek | 32 | 108 | 102 | 0.067 | 6.83 | 2.27 | 2.01 | 0.02 | 0.07 | 0.3 |
| Stony Stream | 32 | 112 | 109 | 0.070 | 7.61 | 2.31 | 1.96 | 0.03 | 0.07 | 0.4 |
| Chesapeake Bay | 33 | 33 | 31 | 0.071 | 2.19 | 2.65 | 2.40 | 0.09 | 0.09 | 1.0 |
| St. Marks Seagrass | 34 | 48 | 48 | 0.096 | 4.60 | 2.04 | 1.94 | 0.14 | 0.11 | 1.3 |
| St. Martin Island | 35 | 44 | 42 | 0.116 | 4.88 | 1.88 | 1.85 | 0.14 | 0.13 | 1.1 |
| Little Rock Lake | 11 | 182 | 92 | 0.118 | 10.84 | 1.89 | 1.77 | 0.25 | 0.12 | 2.1 |
| Lake Tahoe | * | 800 | 172 | 0.131 | 22.59 | 1.81 | 1.74 | 0.14 | 0.13 | 1.1 |
| Mirror Lake | * | 586 | 172 | 0.146 | 25.13 | 1.76 | 1.72 | 0.14 | 0.15 | 0.9 |
| Bridge Brook Lake | 36 | 75 | 25 | 0.171 | 4.28 | 1.85 | 1.68 | 0.16 | 0.19 | 0.8 |
| Coachella Valley | 37 | 30 | 29 | 0.312 | 9.03 | 1.42 | 1.43 | 0.43 | 0.32 | 1.3 |
| Skipwith Pond | 9 | 35 | 25 | 0.315 | 7.88 | 1.33 | 1.41 | 0.33 | 0.33 | 1.0 |
“Taxa” refers to the number of compartments in the original food web, which can range from ontogenetic stages (e.g., largemouth bass juveniles) to nonphylogenetic categories (e.g., detritus, seeds) to highly aggregated taxa (e.g., microbes). S refers to trophic species, C refers to connectance, L refers to trophic links. D refers to characteristic path length, and Cl refers to the clustering coefficient. Dran and Clran refer to the mean D and Cl for 100 random webs.
Unpublished data held by N.D.M.
We measured three properties central to recent network topology research for the 16 food webs: (i) characteristic path length (D), the average shortest path length between all pairs of species (21), (ii) clustering coefficient (Cl), the average fraction of pairs of species one link away from a species that are also linked to each other (17), and (iii) cumulative degree distribution, the fraction of trophic species P(k) that have k or more trophic links (both predator and prey links). We treated trophic links as undirected when calculating path length and clustering because effects can propagate through the web in either direction, through changes to both predator and prey species (17). For 100 random webs with the same C and S as each empirical web, we also calculated mean D and Cl. We rejected random webs that had nodes or groups of nodes disconnected from the main web because our empirical data lacks disconnected subwebs or species (12).
Results
The 16 food webs range in size from 25 to 172 trophic species, connectance (L/S2) ranges from 0.026 to 0.315, and links per species ranges from 1.59 to 25.13 (Table 1). The average connectance over all 16 webs is 0.11 (SD = 0.09), similar to mean connectance values reported for other reliable sets of community food webs (ref. 39: 5 webs, mean C = 0.11, SD = 0.03; ref. 36: 50 webs, mean C = 0.10, SD = 0.04). Characteristic path lengths range from 1.33 to 3.74, generally decreasing with increasing connectance (21). Empirical food webs display similar, slightly longer (for 13 of 16 webs analyzed) path lengths compared with random webs (Table 1).
A comparison of the clustering coefficients of empirical food webs to that of counterpart random webs (Table 1, Cl/Clran) gives ratios ranging from 0.3 to 3.8. Only five food webs, the four very low connectance Grassland, Scotch Broom, and Ythan Estuary 1 and 2 webs, plus Little Rock Lake, display clustering that is twice or more that of random webs (Cl/Clran = 2.1 to 3.8). Eleven webs have Cl/Clran <1.5, with six of those webs displaying equal or lower clustering than random webs (Cl/Clran = 0.3 to 1.0). The largest clustering coefficient ratio, 3.8 for the large version of the Ythan Estuary web, is lower than ratios for other biological networks. The substrate and reaction graphs for Escherichia coli display clustering coefficient ratios of 12.3 and 6.6 (40) and the C. elegans neural network has a ratio of 5.6 (17).
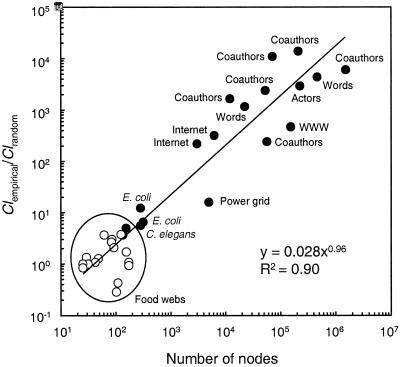
Across 34 biological and nonbiological network data sets including the current food webs, the clustering coefficient ratio increases as an approximate power–law function, specifically a linear function of the size of the network (Fig. 1, refs. 17, 22, 40–47). A 1:1 ratio occurs in networks with ≈40 nodes. The nonbiological networks displayed, which range in size from 4,941 to 1,520,251 nodes (19) have clustering ratios that range from 16 for the power grid (17) to ≈14,000 for neuroscience coauthorship (43). Biological networks, especially ecological networks, have relatively few nodes compared with nonbiological networks and clustering coefficients that are much closer to random.
Figure 1.
Log–log plot of the clustering coefficient ratios (empirical/random web values) as a function of size of the network. Open circles represent data from 16 trophic food webs from the current analysis. Dark circles represent data from previous studies of 18 scale-free small-world networks summarized in ref. 19: 2 taxonomic food webs (22); E. coli substrate and reaction graphs (40); C. elegans neural network, movie actors, and power grid (17); 4 science coauthorship data sets (41, 42); 2 math and science coathorship data sets (43); low and high estimates for Internet domains (44, 45); world wide web sites (46); and concurrence and synonomy of words (47, 44).
The linear relationship of the clustering ratio to network size arises because clustering in random networks should be equal to connectance (C = L/S2), because the likelihood that one node is connected to another node is the same as the likelihood that two neighbors of a node are connected. This is demonstrated by the near identity of Clran with C for the 34 networks of Fig. 1 (linear regression; slope = 1.04, r2 = 0.996, P < 0.001). However, observed clustering appears to scale linearly with L/S for the same networks across 7 orders of magnitude (linear regression; slope = 0.003, r2 = 0.22, P = 0.005). Thus, the ratio of observed to random clustering should increase approximately linearly with S, because (L/S)/(L/S2) = S. The relationship of observed clustering to L/S depends on analyzing the full complement of networks. Observed clustering in 18 food webs (Fig. 1) scales linearly with connectance (linear regression; slope = 0.89, r2 = 0.61, P < 0.001) but not with S or L/S, consistent with clustering in food webs being similar to random. Observed clustering in the 16 other real-world networks (Fig. 1) does not scale with S, L/S, or C.
Following ref. 27, we analyzed cumulative rather than density distributions of food-web degree data. A previous study of food-web density degree distributions included degree bins with a frequency of 0 that were ignored when scale-free functions were fit to the data (22). The use of cumulative distributions avoids the problem of arbitrary exclusion of null bins and gives a more accurate picture of the shape of the distribution in small, noisy data sets. We examined the cumulative distribution for each food web separately rather than pooling data across food webs as done in another study (23) to make sure we illuminated informative detail and variation.
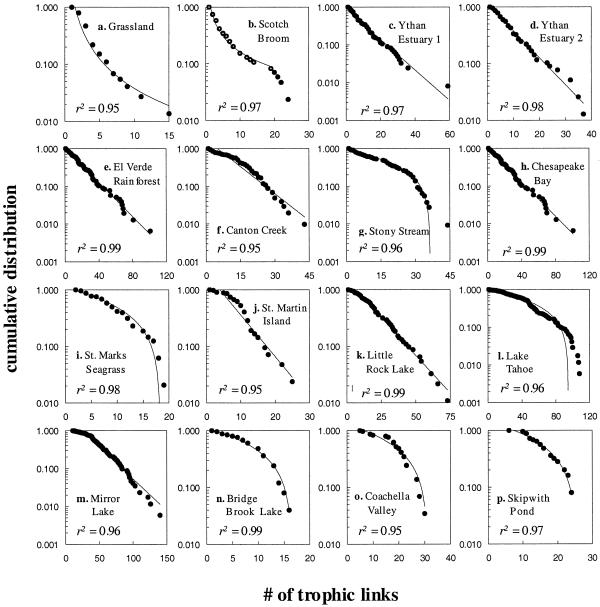
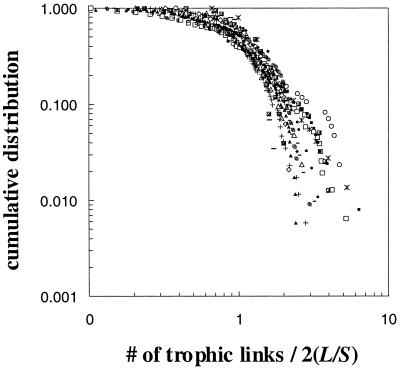
All 16 of the trophic food webs display cumulative degree distributions that differ from a Poisson distribution expected in random networks (Fig. 2). The two least connected food webs display scale-free or broad-scale degree distributions, with power–law behavior over the whole range of the Grassland web (exponent = 1.71) and part of the range of the Scotch Broom web (exponent = 0.92) (Fig. 2). The Scotch Broom web displays an exponential drop-off in the tail. The remaining 14 food webs display single-scale distributions. Eight food webs have data consistent with an exponential distribution (Ythan Estuary 1 and 2, El Verde Rainforest, Canton Creek, Chesapeake Bay, St. Martin Island, Little Rock Lake, and Mirror Lake) and six webs, generally those with relatively high connectance, have data consistent with a uniform distribution (Stony Stream, St. Marks Seagrass, Lake Tahoe, Bridge Brook Lake, Coachella Valley, and Skipwith Pond). One of the uniform distribution food webs, Lake Tahoe, displays a tail well fit by a Gaussian distribution. An overlay of normalized food-web data from all 16 webs clearly demonstrates the general trend that distribution tails drop off faster than expected for scale-free networks (Fig. 3). The normalized data also display a large amount of scatter that reinforces the lack of a universal functional form of degree distributions in food webs (Fig. 3).
Figure 2.
Linear-log plots of the cumulative distributions of links per species (both predator and prey links) in 16 food webs. Webs are ordered by increasing connectance (see Table 1). Lines and r2 values show the fit to the data of the best of three simple models: power–law distribution (upward curved line), exponential decay (straight line), or uniform distribution (downward curved line). No food web is well fit by a Poissonian or Gaussian distribution.
Figure 3.
Log–log overlay plot of the cumulative distributions of links per species in 16 food webs. The link data are normalized by the average number of links/species in each web. If the distributions followed a power law, the data would tend to follow a straight line. The overlay also displays a significant amount of scatter in the data.
Discussion
An increasingly wide array of information, social, physical, and biological networks (19), including food webs (22), have been described as small-world networks with short path lengths and high clustering. Our analyses suggest that most food webs do not display typical small-world topology, corroborating and extending a more empirically limited study (23). Characteristic path lengths of food webs are short and only slightly longer than random (20–23), consistent with small-world topology and observations for most real-world networks (19). However, only 5 of 16 trophic food webs analyzed, four of which are the lowest connectance webs examined, unambiguously display the much greater than random clustering expected for small-world topology, as also reported for the taxonomic versions of four of the same webs (22).
The apparent deviation of most food webs from small-world clustering topology is related not only to connectance but to the small size of food webs (101 to 102 nodes) compared with most other networks (102 to 107 nodes). When network size is taken into account, food webs fit into a predictable continuum of clustering, with increasingly greater than random clustering observed in larger networks. A well-known property of networks is that clustering is generally independent of network size (19). Our results show that observed clustering coefficients across a wide variety of real-world networks increase as a linear function of links per node, but are independent of network size and connectance. The specific relationship between observed clustering and links per species only emerges when food web data are combined with non-food-web data. These findings suggest that the expectation of high clustering in small-world topology (17) is generally inapplicable to small networks with relatively few links per species.
As with small-world topology, the degree distributions of food webs appear similar to other real-world networks in some respects and different in others. We observed degree distributions that deviated from random (see also refs. 22 and 23), as observed for other networks (19). The deviation of food-web degree distributions from distributions in random networks for prey and predator links considered separately (“generality” and “vulnerability,” in ref. 20) was first reported in a set of seven trophic food webs (12) reanalyzed in this study for nondirected degree distributions. A simple one-dimensional “niche model” has been found to successfully predict nonrandom distributions and other topological properties (12, 21, 23).
Although the shapes of food-web degree distributions deviate from random, they also differ from scale-free, power–law distributions observed in many other networks (19). Our analysis of 16 food webs shows that food webs display a variety of functional forms, including power–law, partial power–law, exponential, and uniform degree distributions. This finding contradicts two prior contrasting claims: (i) food webs display scale-free degree distributions (22), and (ii) food webs display exponential degree distributions, which, when normalized, can be expressed in a universal functional form (23). What is responsible for the discrepancies between these studies and the current study? The first study (22) looked at only four food webs, two of which were variants of the same web, and analyzed density distributions, which tend to obscure the role of empty bin data thus altering the shapes of the distributions. The study also dismissed a non power–law distribution observed for one of their webs as an artifact of poor taxonomic resolution. This dismissal is not supported by our study, which examines 16 webs with variable resolution (38) that does not correlate with the presence of power–law degree distributions. Also, a reanalysis of cumulative distributions for the deaggregated, taxonomic versions of the 16 food webs did not substantively alter our results (data not shown). The second study (23) used cumulative degree distributions, but failed to report the results from one (Ythan Estuary) of the seven food webs analyzed and pooled the results for the remaining six food webs. Thus, both studies were limited to a few food webs, dismissed data sets from consideration, and made analytical decisions that obscured important variability in the data.
Although we find that degree distribution in food webs does not follow a universal functional form, we do find a general systematic relationship between the shape of the degree distribution of a food web and its network complexity measured as connectance. Food webs with relatively high connectance typically display uniform distributions, webs with middle connectance tend to have exponential distributions, and webs with very low connectance display power–law or partial power–law distributions. The size of the food web also plays a role in the form that the degree distribution takes. The tendency toward uniform degree distributions in high connectance food webs may occur because networks with relatively few nodes and high connectance have relatively high average degree. This minimizes the difference between the average (2L/S) and the maximum possible (2S) degree, cutting off distribution tails that would include nodes with much higher than average degree, as often seen in large, low-connectance, small-world networks (e.g., Fig. 1; non-food-web networks have mean nodes = 140,000, C = 0.01, and L/S = 1,400). Conversely, sparsely connected food webs with much greater differences between average (2L/S) and maximum (2S) degree can display extremely heterogeneous scale-free distributions as seen in many real-world networks. The lowest connectance food web, Grassland (C = 0.026), displays a power–law distribution and has S = 61 and L/S = 1.59, which differ by a factor of C−1 = 38.4. In contrast, the highest connectance food web, Skipwith Pond (C = 0.315), displays a uniform distribution and has S = 25 and L/S = 7.88, which differ by a factor of only 3.2.
Both low- and high-connectance food webs are unusual, and their more extreme connectances may sometimes be artifacts of particular food-web collection or assembly procedures. The two lowest connectance webs (C ≈ 0.03), Grassland and Scotch Broom, the only webs that display complete or partial power–law degree distributions, are “source webs” constructed by following food chains upward from one or a few basal species. In addition, these webs ignore co-occurring generalists such as spiders and birds and focus on specialist parasitoid insects whose immature stages develop on or within a single insect host, ultimately killing the host. Such parasitoids tend to be linked to very few other species, resulting in webs with low C (15). The two highest connectance webs (C ≈ 0.3), Coachella Valley and Skipwith Pond, which display uniform distributions, are small webs dominated by omnivores, taxa that feed at multiple trophic levels. Such taxa tend to be generalists with links to many other species, resulting in high C. In Coachella Valley, the high levels of omnivory are an artifact of high node aggregation (only 1 of 30 taxa are identified at the species level). Many food webs are less skewed toward specialists or generalists and thus have connectances closer to 0.1 and may be more likely to display exponential degree distributions. However, even webs with intermediate levels of connectance do not always display exponential distributions.
The relative lack of power–law degree distributions in food webs also may relate to how ecosystems assemble and evolve compared with other networks. Networks that display scale-free degree distributions probably emerge via a set of mechanisms that differ from those that produce networks with broad- and single-scale distributions (27). A simple “scale-free model” for networks has been proposed that produces networks with power–law degree distributions (24). This model incorporates two generic mechanisms thought to be common to many real-world networks: (i) growth of the network by addition of nodes and links at each time step and (ii) preferential attachment of new nodes to existing nodes with a high number of links. Although these assumptions may be useful for describing evolution of the world wide web or citation networks (but see refs. 48 and 49), they appear less appropriate for ecosystems and some other types of networks. Alternate models that remove either of the assumptions (24, 50) or incorporate “aging” (some nodes cannot accept new links) or “cost” (a maximum number of links per node) (27) eliminate scale-free topology and produce broad- or single-scale degree distributions.
In ecosystems, both assumptions of the scale-free model may be problematic. The simple growth assumption is violated because there are both additions and losses of nodes (species) at ecological and evolutionary time scales by means of immigration, emigration, speciation, and extinction. The net effect of such changes can be expansion, contraction, or no change of species richness (as well as trophic links) within an ecosystem over time. An elaboration of the scale-free model suggests that in evolving networks with both node addition and removal, a scale-free topology can emerge as long as the overall network size does not systematically decrease (e.g., ref. 51). However, this still suggests that ecosystems undergoing no net change or contraction of species richness will not produce scale-free distributions.
Whether new species preferentially link to highly connected species already in the food web is unclear. Data from a wide range of studies suggest that generalists (species with a large number of links to prey) are more likely than specialists to consume invasive species, as observed for insect herbivores of invasive plants (52, 53) and parasitoids of invasive insect herbivores (54). This observation supports the preferential attachment hypothesis. However, we can also hypothesize that an invasive species will be more likely to establish successfully if it has few consumers. Initial data from some Hawaiian food webs suggest that there are more successfully established alien parasitoids than alien herbivores or plants, which may be attributable to parasitoids typically having fewer consumers than species at lower trophic levels (J. Memmott, personal communication). Regarding predation links to resource species, there is little data to suggest whether invasive species have any tendency to consume species that already have a large number of consumers. Theoretically, competitive exclusion causes overlapping niches to repulse each other (55), reducing the average number of consumers preying on resource species. However, a species may have many consumers because it is relatively abundant, which may make it more likely to be preyed on by new species. Empirically, a balance of these two mechanisms is suggested by the success of the niche model (12, 21, 23), which randomly distributes niches free of repulsion or attraction (12).
A general understanding of the structure of food webs from a variety of ecosystems is useful for developing a more universal understanding of biological and nonbiological network topology. In particular, although food webs generally display far less clustering than expected in small world networks, we show how low clustering in food webs represents an extreme in a previously unrecognized continuum based on the number of nodes within networks. Also, the small size and relatively high connectance of most food webs compared with other real-world networks has illuminated which types of network topology can be expected to display various kinds of degree distributions. From a more applied, conservation-minded perspective, an understanding of food web topology can be used to explore and predict functional responses of ecosystems to structural changes. The most dramatic structural change that many ecosystems face is human-driven biodiversity loss. Structural analyses can provide another tool for exploring how robust or fragile ecosystems are to species loss (38, 56), as assessed for Internet and world wide web topology (57) and metabolic and protein networks (58, 59). Such analyses can also more generally provide evidence of whether and what aspects of topology and network complexity drive robustness (38).
Acknowledgments
We thank Jane Memmott for helpful discussions and Mark Newman and two anonymous reviewers for useful comments on a draft of the paper. This work has been supported by the Santa Fe Institute and National Science Foundation Grants DEB/DBI-0074521 “Effects of Biodiversity Loss on Complex Communities: A Web-Based Combinatorial Approach” (to J.A.D.), DEB-0083929 “Scaling of Network Complexity with Diversity in Food Webs” (to N.D.M.), and DUE-9950461 “Instructional Environmental Science Computer Lab” (to N.D.M. and R.J.W.).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.May R M. Ecology. 1986;67:1115–1126. [Google Scholar]
- 2.Pimm S L, Lawton J H, Cohen J E. Nature (London) 1991;350:669–674. [Google Scholar]
- 3.Levin S A. Ecology. 1992;73:1943–1967. [Google Scholar]
- 4.Paine R T. Ecology. 1988;69:1648–1654. [Google Scholar]
- 5.MacArthur R H. Ecology. 1955;36:533–536. [Google Scholar]
- 6.May R M. Stability and Complexity in Model Ecosystems. Princeton: Princeton Univ. Press; 1973. [PubMed] [Google Scholar]
- 7.Rejmanek M, Stary P. Nature (London) 1979;280:311–313. [Google Scholar]
- 8.Cohen J E, Briand F. ) Proc Natl Acad Sci USA. 1984;81:4105–4109. doi: 10.1073/pnas.81.13.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warren P H. Oikos. 1989;55:299–311. [Google Scholar]
- 10.Winemiller K O. Ecol Monogr. 1990;60:331–367. [Google Scholar]
- 11.Martinez N D. Ecol Monogr. 1991;61:367–392. [Google Scholar]
- 12.Williams R J, Martinez N D. Nature (London) 2000;404:180–183. doi: 10.1038/35004572. [DOI] [PubMed] [Google Scholar]
- 13.Martinez N D. Science. 1993;260:242–243. doi: 10.1126/science.260.5105.242. [DOI] [PubMed] [Google Scholar]
- 14.Martinez N D. Am Nat. 1994;144:935–953. [Google Scholar]
- 15.Martinez N D, Hawkins B A, Dawah H A, Feifarek B P. Ecology. 1999;80:1044–1055. [Google Scholar]
- 16.Strogatz S H. Nature (London) 2001;410:268–275. doi: 10.1038/35065725. [DOI] [PubMed] [Google Scholar]
- 17.Watts D J, Strogatz S H. Nature (London) 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 18.Milgram S. Psychol Today. 1967;2:60–67. [Google Scholar]
- 19.Albert R, Barabási A-L. Rev Mod Phys. 2002;74:47–97. [Google Scholar]
- 20.Schoener T W. Ecology. 1989;70:1559–1589. [Google Scholar]
- 21.Williams R J, Berlow E L, Dunne J A, Barabási A-L, Martinez N D. Proc Natl Acad Sci USA. 2002;99:12913–12916. doi: 10.1073/pnas.192448799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montoya J M, Solé R V. J Theor Biol. 2002;214:405–412. doi: 10.1006/jtbi.2001.2460. [DOI] [PubMed] [Google Scholar]
- 23.Camacho J, Guimerà R, Amaral L A N. Phys Rev Lett. 2002;88:228102. doi: 10.1103/PhysRevLett.88.228102. [DOI] [PubMed] [Google Scholar]
- 24.Barabási A-L, Albert R. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 25.Erdös P, Rényi A. Publ Math Inst Hung Acad Sci. 1960;5:17–61. [Google Scholar]
- 26.Bollobás B. Random Graphs. London: Academic; 1985. [Google Scholar]
- 27.Amaral L A N, Scala A, Barthélémy M, Stanley H E. Proc Natl Acad Sci USA. 2000;97:11149–11152. doi: 10.1073/pnas.200327197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Memmott J, Martinez N D, Cohen J E. J Anim Ecol. 2000;69:1–15. [Google Scholar]
- 29.Huxham M, Beany S, Raffaelli D. Oikos. 1996;76:284–300. [Google Scholar]
- 30.Hall S J, Raffaelli D. J Anim Ecol. 1991;60:823–842. [Google Scholar]
- 31.Waide R B, Reagan W B, editors. The Food Web of a Tropical Rainforest. Chicago: University of Chicago Press; 1996. [Google Scholar]
- 32.Townsend C R, Thompson R M, McIntosh A R, Kilroy C, Edwards E, Scarsbrook M R. Ecol Lett. 1998;1:200–209. [Google Scholar]
- 33.Baird D, Ulanowicz R E. Ecol Monogr. 1989;59:329–364. [Google Scholar]
- 34.Christian R R, Luczkovich J J. Ecol Model. 1999;117:99–124. [Google Scholar]
- 35.Goldwasser L, Roughgarden J. Ecology. 1993;74:1216–1233. [Google Scholar]
- 36.Havens K. Science. 1992;257:1107–1109. doi: 10.1126/science.257.5073.1107. [DOI] [PubMed] [Google Scholar]
- 37.Polis G A. Am Nat. 1991;138:123–155. [Google Scholar]
- 38.Dunne J A, Williams R J, Martinez N D. Ecol Lett. 2002;5:558–567. [Google Scholar]
- 39.Martinez N D. Am Nat. 1992;139:1208–1218. [Google Scholar]
- 40.Wagner A, Fell D A. Proc R Soc London Ser B. 2001;268:1803–1810. doi: 10.1098/rspb.2001.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newman M E J. Proc Natl Acad Sci USA. 2001;98:404–409. doi: 10.1073/pnas.021544898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newman M E J. Phys Rev E. 2002;64:016131. [Google Scholar]
- 43. Barabási, A.-L., Jeong, H., Néda, Z., Ravasz, E., Schubert, A. & Vicsek, T. (2001) http://www.arXiv.org/abs/cond-mat/0104162.
- 44.Yook S H, Jeong H, Barabási A-L, Tu Y. Phys Rev Let. 2001;86:5835. doi: 10.1103/PhysRevLett.86.5835. (4). [DOI] [PubMed] [Google Scholar]
- 45.Pastor-Satorras R, Vázquez A, Vespignani A. Phys Rev Let. 2001;87:258701. doi: 10.1103/PhysRevLett.87.258701. (4). [DOI] [PubMed] [Google Scholar]
- 46.Adamic L A. Proc. European Conf. Digit. Libr. (ECDL) London: Springer; 1999. pp. 443–452. [Google Scholar]
- 47.Cancho R F I, Solé R V. Proc R Soc London Ser B. 2001;268:2261–2265. [Google Scholar]
- 48.Kleinberg J, Kumar S R, Raghavan P, Rajagopalan S, Tomkins A. Proc Int Conf Combinatorics Computing LNCS. 1999;167:1–17. [Google Scholar]
- 49.Adamic L A, Huberman B A. Science. 2000;287:21150. [Google Scholar]
- 50.Barabási A-L, Albert R, Jeong H. Physica A. 1999;272:173–187. [Google Scholar]
- 51.Dorogovtsev S N, Mendes J F F. Phys Rev E. 2001;63:056125. doi: 10.1103/PhysRevE.63.056125. [DOI] [PubMed] [Google Scholar]
- 52.Connor E F, Faeth S H, Simberloff D, Opler P A. Ecol Entom. 1980;5:205–211. [Google Scholar]
- 53.Strong D R, Lawton J H, Southwood T R E. Insects on Plants: Community Patterns and Mechanisms. Oxford: Blackwell Scientific; 1984. [Google Scholar]
- 54.Cornell H V, Hawkins B A. Am Nat. 1993;141:847–865. doi: 10.1086/285512. [DOI] [PubMed] [Google Scholar]
- 55.Begon M, Harper J L, Townsend C R. Ecology: Individuals, Populations and Communities. 3rd Ed. Oxford: Blackwell Science; 1996. [Google Scholar]
- 56.Solé R V, Montoya J M. Proc R Soc B. 2001;268:2039–2045. doi: 10.1098/rspb.2001.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Albert R, Jeong H, Barabási A-L. Nature (London) 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 58.Jeong H, Tombor B, Albert R, Oltvai Z N, Barabási A-L. Nature (London) 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 59.Jeong H, Mason S P, Barabási A-L, Oltvai Z N. Nature (London) 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]