Abstract
Whereas many physicochemical investigations have shown that among monovalent cations Na+ ion possesses minimal potential for DNA binding, biological assays have shown that Na+ ion (in contrast to K+ ion) plays a primary role in chromatin compaction and related processes. It is difficult to explain this inverse relationship between the compaction potentials of Na+ and K+ and their binding abilities. In this study we sought to resolve this contradiction and emphasize the phenomenological distinction between DNA compaction and DNA binding processes in the case of DNA compaction by monocations. Using polyethylene glycol solutions as a model of a crowded cell environment, we studied DNA compaction by alkali metal salts LiCl, NaCl, KCl, RbCl, and CsCl, and found that all of these monocations promote DNA compaction. Among these monovalent cations Na+ produces the greatest compaction and the ratio of K+ cand Na+ oncentrations for DNA compaction is ∼1.5–2. A comparative analysis of recent experimental results indicates that a higher binding activity of monocation generally corresponds to a low compaction potential of the corresponding monovalent ion. This inverse relation is explained as a result of partial dehydration of monocations in the compact state.
INTRODUCTION
DNA compaction in living cells is a process of extraordinary biological significance. During interphase, DNA exists in a rather compact state (compared to the elongated state in aqueous solution) and extra compaction is required during mitosis to realize cell division. Charge neutralization of DNA is a driving force of DNA compaction. The compaction of DNA in vivo is achieved by histone proteins, which neutralize ∼57% of the DNA negative charge (Morgan et al., 1987). The remaining charge of DNA is neutralized by other positively charged chemicals in cell fluid, where mono- and divalent cations comprise up to 1% of the cell weight. The most abundant monovalent cations in cells are K+ and Na+ ions, which are not only involved in DNA compaction but are also responsible for the regulation of other important biological processes. A dramatic difference in the functions of K+ and Na+ has been noted in a large number of biological reports (Strick et al., 2001). Stimulated by the biological importance of this subject, many in vitro investigations of DNA interaction with K+ and Na+ ions as well as other monovalent alkali cations (Li+, Rb+, and Cs+) have been performed by biophysicists (Bleam et al., 1980). However, almost all of these reports described DNA-monocation interaction without any consideration of the change in the higher-order structure of DNA, which is essential for understanding the role of monocations in DNA compaction in living cells.
Although DNA cannot be condensed by monocations in aqueous media (Wilson and Bloomfield, 1979), in a crowded environment DNA condensation is promoted by an increase in the concentration of monocations (Livolant and Amelie, 1996). Zimmerman has reviewed a number of experimental systems to demonstrate the promotion of molecular association in crowded environments (Zimmerman, 1993). In solutions of polyethylene glycol and salt, DNA undergoes polymer- and salt-induced (PSI) condensation (Post and Zimm, 1979; Grosberg et al., 1982). DNA compaction under such conditions can be considered as a model of DNA compaction in the protein-crowded environment inside living cells. According to an established model of Ψ-condensation, thermodynamically unfavorable contact between DNA and PEG decreases the available free space for coil DNA in solution and DNA undergoes a collapse transition at some critical concentration of PEG. The translational entropy of counterions and a hydration effect play important roles in this process. Despite the well-known role of the salt concentration in this process, there have been only a few investigations of the influence of the chemical nature of the monocation. Mayama and co-workers studied single-molecule compaction of DNA by comparing sodium and potassium chlorides in PEG solutions and noted a large difference in the temperature-dependencies of DNA compaction mediated by these two monocations (Mayama et al., 2000; Mayama and Yoshikawa, 2000). They showed that DNA compaction by both monocations is preferred at higher temperature; however, the slopes of these dependencies were different as a result of different thermodynamic parameters of DNA compaction. However, there has been no systematic investigation of the influence of the monocation nature on its ability to induce DNA compaction. To elucidate the compaction potentials of different alkali ions, we performed single-DNA observation to study the compaction of individual DNA molecules caused by LiCl, NaCl, KCl, RbCl, and CsCl in concentrated solutions of polyethylene glycol.
EXPERIMENTAL SECTION
Materials
Bacteriophage T4 DNA (165.5 kbp) was purchased from Nippon Gene (Tokyo, Japan). The fluorescent dye, 4,6′-diamidino-2-phenylindole (DAPI) was obtained from Wako Pure Chemical Industries (Osaka, Japan). Polyethylene glycols with molecular weights of 3000 and 10000 were purchased from Merck (Hohenbrunn, Germany). Lithium, sodium, potassium, rubidium, and cesium chlorides were purchased from Wako Pure Chemical Industries.
Methods
Samples for fluorescent microscopy were illuminated with 365 nm ultraviolet light, and fluorescence images of DNA molecules were observed using a Zeiss Axiovert 135 TV microscope equipped with a 100× oil-immersed lens and recorded on S-VHS videotape through a Hamamatsu SIT TV camera. All observations were carried out at room temperature. The population of DNA molecules in coil or compact states was determined by an analysis of at least 100 DNA molecules.
Sample solutions
Solutions for microscopic observations were prepared by mixing PEG 3000 or PEG 10000 in twice-distilled water until dissolution, adding T4 DNA with further mixing and incubation for 30 min, adding DAPI with further mixing and incubation for 30 min, and finally adding alkali chloride solution and letting the sample solution sit for 1–2 h before observations. The final solution contained 100 g/L PEG, 2 × 10−7 M DNA (in basepairs), 2 × 10−7 M DAPI and an appropriate amount of alkali chloride. All measurements were performed at room temperature.
RESULTS
The compaction of giant T4 DNA in PEG-salt systems was monitored by fluorescent microscopy at the level of single DNA chains. Fig. 1 illustrates DNA compaction in PEG 3000 with an increase in the NaCl concentration. Compaction of DNA was observed as a change in the conformation of DAPI-stained T4 DNA from a coil (Fig. 1 A, [NaCl] = 0.2 M) into a globule (Fig. 1 C, [NaCl] = 0.4 M). The compaction of giant T4 DNA proceeds as an all-or-none-type transition, and suggests the existence of only two distinct DNA states (coil and globule) and bimodality in the intermediate region (Fig. 1 B). Fig. 1, A″–C″, show the distributions of the T4 DNA long-axis length in these three characteristic regions. In the coil state, the DNA length shows a broad distribution, with an average value of ∼3 μm (Fig. 1 A″). In the coexistence region (Fig. 1 B″), DNA coils and globules can both be recognized, and during DNA compaction the ratio between these modes changes as in a first-order phase transition. The distribution of DNA lengths in the final globule conformation is very narrow, with an average length of ∼0.5 μm. (Fig. 1 C″) A detailed description of all-or-none DNA compaction in a PEG-salt system as observed by FM is given in the literature (Minagawa et al., 1994; Vasilevskaya et al., 1995).
FIGURE 1.
Fluorescent images of T4 DNA in coil (A) and compact (C) states, and the coexistence of these two states (B); intensity distributions of corresponding FM images (A′–C′) (image size 5 × 5 μm). Distributions of long-axis length of T4 DNA in the coil (A″, gray bars), coexistence (B″), and globule (C″, black bars) states. Images and distributions correspond to the observations and measurements in PEG 3000 solution at NaCl concentrations of 0.2 M (A series), 0.3 M (B series), and 0.4 M (C series).
Further, using the T4 DNA long-axis length distributions as shown in Fig. 1, A″–C″, we calculated the ratios of DNA in the coil and globule conformations. DNA compaction can be represented by a compaction curve as the dependence of the globule (compact DNA) fraction in solution on the concentration of the compaction agent, i.e., monovalent salt. Compaction curves for T4 DNA compaction in PEG 3000 solution induced by LiCl, NaCl, KCl, RbCl, and CsCl are shown on Fig. 2. All of these monocations collapsed DNA into globules and the concentrations of monovalent salts needed to induce DNA compaction varied greatly. In particular, the monocation with the highest compaction activity (Na+) generated compact DNA at half the concentration than the monocation with the lowest potential (Cs+). Similar microscopic measurements and analysis were performed in solutions of PEG with a higher molecular weight and the same PEG concentration as used in the above experiments. As with the PEG 3000 sample in Fig. 2, Fig. 3 shows compaction curves of T4 DNA in PEG 10000 solution. Comparison of Figs. 2 and 3 reveals that the concentrations of salts that were necessary to collapse DNA decreased when we used PEG with a higher molecular weight. In PEG 10000, the concentrations of salts required to collapse DNA were about half those with PEG 3000. These data are consistent with previous reports which have demonstrated that neutral polymers such as PEG with a higher molecular mass are more effective for DNA condensation (Vasilevskaya et al., 1995).
FIGURE 2.
T4 DNA compaction curves for LiCl (•), NaCl (▴), KCl (▾), RbCl (♦), and CsCl (◂) in PEG 3000 (100 g/L) shown as the dependence of the globule fraction (Fg) in the ensemble of DNA molecules on the concentration of monovalent salt.
FIGURE 3.
T4 DNA compaction curves for LiCl (•), NaCl (▴), KCl (▾), RbCl (♦), and CsCl (◂) in PEG 10000 (100 g/L) shown as the dependence of the globule fraction (Fg) in the ensemble of DNA molecules on the concentration of monovalent salt.
The potentials of monocations for DNA compaction in PEG 3000 and 10000 solutions are summarized in Fig. 4 as the correlation between monocations' concentrations necessary for a 50% transition from coil DNA to compact DNA calculated from corresponding compaction curves on Fig. 3. As shown, regardless of the PEG length, the order of compaction potential for metal chlorides with the exception of LiCl is as follows:
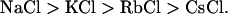 |
FIGURE 4.
Correlations between the concentrations of monovalent cation chlorides for T4 DNA compaction in a crowded environment of PEG 3000 (100 g/L) and PEG 10000 (100 g/L). The concentrations correspond to the point of 50% coexistence between DNA coil and DNA globule states on the corresponding compaction curves on Figs. 2 and 3.
Thus, sodium chloride has the highest activity among the salts studied. With an increase in the molecular mass of the monovalent metal ion, higher concentrations are required for DNA compaction. As shown in Fig. 4, there is a good correlation between the chemical potentials of monocations in solutions of PEG with different molecular weights. In solutions of PEG with a higher molecular weight, where monocations show higher chemical potentials, the differences between the chemical potentials of monocations are less pronounced. The increase in Na+ activity is rather steep compared to the moderate changes in activity between K+, Rb+, and Cs+. As shown in Fig. 4, Li+ demonstrates unique behavior in this set of monocations. Li+ ion shows a significantly lower DNA compaction potential than expected based on the trend for other ions, which suggests that monocations with a smaller atomic number are more active in DNA compaction. Furthermore, there is also no correlation between the DNA compaction activity of Li+ ion in PEG 3000 and PEG 10000 with respect to other monocations (Fig. 4). Li+ shows moderate chemical potential in PEG 3000 but is the weakest DNA compaction agent in PEG 10000.
DISCUSSION
The influence of the chemical nature of a monocation on its DNA compaction ability is clearly demonstrated by the observation of the collapse of single DNA molecules by fluorescent microscopy. However, more important conclusions arise when our results are compared with previous reports on DNA binding with monocations. The most important studies on DNA-monocation binding are summarized in Table 1, which shows the activity sequences, methods used in research, and the corresponding citations in our list of references. With the exception of LiCl, the sequences of salt activities are generally similar, and increase from Na+ toward ions with higher molecular weights. This dependence of the binding of monocations is usually explained from the viewpoint of a decreasing hydrated radius of monocations in aqueous solutions (Ivanov et al., 1973). Monocations with a lager hydrated radius are weaker at binding DNA than those with a smaller radius. It is recognized that the binding of the part of monocations to DNA can be specific in the minor or major grooves of B-form DNA (for review, see Hud and Polak 2001; McFail-Isom et al., 1999); however, there still exists some disagreement about exact location of monocations and distribution of bound monocations between different binding sites on DNA. Upon binding in DNA grooves, especially in minor groove, monocations become partially dehydrated (Howerton et al., 2001). Although the other monocations may be involved with DNA binding solely through electrostatic force, there may be some other possibilities with Li+ ion, and the position of Li+ ion in this monocation activity sequence seems to depend strongly on the detection method and experimental conditions. The specific properties of Li+ ion have been discussed previously (Wolf et al., 1977).
TABLE 1.
Binding potentials of monovalent cations with DNA determined by different methods
| Method | Sequence | Ref. |
|---|---|---|
| 23Na NMR | Cs+ > K+ > Li+ > Na+ | Bleam et al. (1980) |
| Electrophoresis of circular DNA | Cs+ > Rb+ > Li+ > K+ > Na+ | Anderson and Bauer (1978) |
| CD in aqueous solutions | Cs+ > Rb+ > Li+ > K+ > Na+ | Hanlon et al. (1975); Wolf and Hanlon (1975); Wolf et al. (1977) |
| CD in aqueous/alcohol solutions | Cs+ > Rb+ > K+ > Na+ > Li+ | Ivanov et al. (1973) |
| 23Na NMR relaxation | Cs+ ≈ Rb+ ≈ K+ > Na+ | Denisov and Halle (2000); Cesare Marincola et al. (2004) |
| Ion-exchange measurements of competition in binding with DNA fibers | Na+ ≈ K+ > Li+ | Korolev et al. (2001) |
| DNA compaction (PEG 10000) | Na+ > K+ > Li+ > Rb+ > Cs+ | This study |
When these previous reports (Table 1) are compared with our results on the DNA compaction potentials of monocations, dramatic differences are noted between the DNA binding and DNA compaction potentials of monovalent ions. Indeed, compaction potential appears to be inversely related to the binding activity of monocations. For example, sodium ion, which has been reported to have the lowest DNA binding potential, is the most effective monocation for DNA compaction.
In contrast to reports on DNA binding with monocations, the compaction potentials of monocations determined here correlate well with biological assays that describe the compaction of chromosomes. The concentration of Na+ ion fluctuates during the cell cycle, with peaks in the mitosis and synthesis phases, whereas the concentration of K+ does not change (Warley et al., 1983). Another report has demonstrated that Na+ ion nearly doubled the rate of mitosis in cells (Atkinson et al., 1983), probably due to the enhanced compaction of chromatin which facilitates cell division, whereas K+ ion had no effect. Thus, in real systems, sodium ion has a clear role as an effective component in DNA compaction.
For many years in the physical chemistry of DNA there was no clear discrimination between processes that involved chemical binding with coil DNA molecules in solution and changes in the conformation of DNA such as compaction. Since it has been widely recognized that changes in the conformation of DNA into higher density forms is the result of DNA negative charge neutralization (Bloomfield, 1991), the activity of cationic molecules in neutralizing DNA negative charge (through binding with DNA phosphates) has been associated with the potential to induce DNA phase separation processes, such as compaction, condensation, aggregation, and precipitation, also without clear distinctions between these different processes. Indeed, the activity of compounds for inducing DNA phase separation is generally determined by their DNA binding potential. In a well-known example, the higher cationicity of spermine4+ compared to spermidine3+, which evidently makes spermine bind more strongly to negatively charged DNA, determines the higher DNA compaction potential of spermine (Takahashi et al., 1997). In contrast, recent investigations on DNA compaction by weak cationic agents showed that under conditions of weak DNA-multication binding, there is not always a clear correlation between binding and compaction activities. Clear evidence of the lack of a binding-compaction correlation was seen in studies on DNA compaction by enantiomeric dications (Zinchenko et al., 2004). Although the dications studied showed no difference in their affinity for coil DNA, their compaction activities differed by 100-fold. The results of DNA compaction by metal chlorides in PEG clarify that in some cases the orders of relative binding and compaction activities can be even opposite. Therefore, with regard to DNA monomolecular compaction, it is important to consider morphological effects that result from the dramatic change in the molecular density of DNA.
The inverse relationship between compaction and binding activities can be explained as follows. It is well known that during DNA compaction the molecular volume of DNA molecules dramatically decreases and compact DNA represents an extremely tightly packed polymer chain that approaches the physical limits of molecular compaction. The different activities in elongated DNA and compact DNA correspond to the two very different DNA molecular environments. When studying the response of DNA to chemicals in solution, we always deal with elongated DNA, and under these conditions monovalent cations in free or DNA-bound states are substantially hydrated. This assumption has been used to explain the dependence of DNA-monocation binding. An ion with a smaller radius corresponds to stronger hydration, and the hydrated radius sequence for monocations is Li+ > Na+ > K+ > Rb+ > Cs+. As a result, the small size of a hydrated cation provides effective interaction with DNA due to higher Coulomb electrostatic potential (Rouzina and Bloomfield, 1996) and the possibility of being incorporated into the DNA minor groove (Bartenev et al., 1983) for necessary charge neutralization.
A different scenario must be realized in compact DNA. In general, we consider that DNA compaction is accompanied by dehydration of monocations and can proceed as through the decreasing of effective distance between DNA chains and through the changes in the parameters of DNA helix secondary structure. The distance between DNA helixes in a compacted DNA can largely vary as a function of PEG concentration from 25 Å to 38 Å (Maniatis et al., 1974) as well as a function of water activity from 18.8 Å to more than 30 Å (Suwalsky et al., 1969). The phenomenon of single molecular DNA compaction is expected to lead to higher order DNA structures with a high degree of packing of DNA segments, however the volume of DNA condensate can be also dependent on the nature of monocation. Additional dehydration of monocations can occur inside DNA grooves as a result of deformation of DNA secondary structure. It was reported that compact DNA in PEG solution demonstrates specific shape of circular dichroism spectra (the so called Ψ-spectra) assigned to severe distortion of typical B-form shape of DNA helix (Evdokimov et al., 1976); however, the detailed characterization of DNA secondary structure of Ψ-condensates has not been yet performed.
Thus, the effective size of the monocation in compact DNA is expected to be closer to the size of free ion without hydration shell, where the sequence of the monocation radius is Li+ < Na+ < K+ < Rb+ < Cs+. The correlation between DNA compaction potential and the ionic radius of a monocation is shown in Fig. 5, where the concentration of monocation needed to induce 50% DNA coil-globule transition in PEG 3000 and PEG 10000 solutions appears to be proportional to ionic radius of the monocation. These C50% concentrations in both cases linearly increase with a decrease in the radius of the alkali metal ion. Li+ ion also shows anomalous behavior in this case. To explain the exceptional case of lithium ion, we can suppose that a small lithium ion is significantly hydrated even in the compact DNA structure because its higher charge density and small radius leads to a higher hydration enthalpy.
FIGURE 5.
Correlations between the concentration of monocations needed to collapse 50% DNA into the globular state in PEG 3000 (•) and PEG 10000 (○) and the bare ionic radii of the monocations (Glusker, 1991).
In terms of thermodynamics, the change in free energy in DNA compaction can be considered as a sum of four different contributions: crowding effect (depletion forces), elasticity of DNA, effect of ions, and effect of hydration:
 |
where ΔGmix, ΔGela, ΔGion, and ΔGhyd are the change in the free energy of single DNA upon mixing with a linear polymer, the elastic contribution of a DNA chain, the contribution of a translational entropy of small ions, and free energy change due to hydration effects, respectively. It is expected that the first two terms are insensitive to the change in the chemical structure of monovalent cations. The ionic contribution relates to the change in the translational entropy of monocations and this contribution disfavors DNA compaction in a crowded environment. The hydration effect is a mixture of the entropic and enthalpic contributions, where the enthalpic component disfavors the compaction, whereas the entropic component, associated with a release of hydration water when monocarions are incorporated into compact DNA, favors this conformational transition. Thus, the third and forth terms in the above equation may tune the free energy of the DNA conformational transition depending on the chemical property of the monovalent ion.
CONCLUSION
Our investigation highlights the inverse relation between DNA compaction potentials of monovalent cations in a crowded environment and the previously reported DNA binding properties of different alkali ions. Monovalent cations that show higher binding activity for DNA had a lower DNA compaction potential. Therefore, the widely accepted correlation between cation binding and compaction potential is not an absolute. As for the biological aspects of this work, the noted high potential of Na+ cation in DNA compaction supports the notion that Na+ ion (contrary to K+) plays an important role in maintaining the compact higher order structure of DNA in vivo.
Acknowledgments
The authors are grateful to Prof. Y. Yoshikawa (Dept. of Food and Nutrition, Nagoya Bunri College, Nagoya, Japan) for helpful discussions. Z.A.A. thanks Japan International Science and Technology Exchange Center for fellowship No. P04154.
This work was supported in part by a grant-in-aid for the 21st Century COE “Center for Diversity and Universality in Physics” from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Anderson, P., and W. Bauer. 1978. Supercoiling in closed circular DNA: dependence upon ion type and concentration. Biochemistry. 17:594–601. [DOI] [PubMed] [Google Scholar]
- Atkinson, M. J., C. Cade, and A. D. Perris. 1983. Sodium and ouabain induce proliferation of rat thymic lymphocytes via calcium- and magnesium-dependent reactions. Cell Calcium. 4:1–12. [DOI] [PubMed] [Google Scholar]
- Bartenev, V. N., Eu. I. Golovamov, K. A. Kapitonova, M. A. Mokulskii, L. I. Volkova, and I. Y. Skuratovskii. 1983. Structure of the B-DNA cationic shell as revealed by an x-ray diffraction study of CsDNA. Sequence-specific cationic stabilization of B form DNA. J. Mol. Biol. 169:217–234. [DOI] [PubMed] [Google Scholar]
- Bleam, M. L., C. F. Anderson, and M. T. Record. 1980. Relative binding affinities of monovalent cations for double-stranded DNA. Proc. Natl. Acad. Sci. USA. 77:3085–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield, V. A. 1991. Condensation of DNA by multivalent cations: considerations on mechanism. Biopolymers. 31:1471–1481. [DOI] [PubMed] [Google Scholar]
- Cesare Marincola, F., V. P. Denisov, and B. Halle. 2004. Competitive Na+ and Rb+ Binding in the Minor Groove of DNA. J. Am. Chem. Soc. 126:6739–6750. [DOI] [PubMed] [Google Scholar]
- Denisov, V. P., and B. Halle. 2000. Sequence-specific binding of counterions to B-DNA. Proc. Natl. Acad. Sci. USA. 97:629–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimov, Y. M., T. L. Pyatigorskaya, O. F. Polyvtsev, N. M. Akimenko, V. A. Kadykov, D. Y. Tsvankin, and Y. M. Varshavsky. 1976. A comparative x-ray diffraction and circular dichroism study of DNA compact particles formed in water-salt solutions, containing poly(ethylene glycol). Nucleic Acids Res. 3:2353–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusker, J. P. 1991. Structural aspects of metal liganding to functional groups in proteins. Adv. Protein Chem. 42:1–75. [DOI] [PubMed] [Google Scholar]
- Grosberg, A. Yu., I. Ya. Erukhimovitch, and E. I. Shakhnovitch. 1982. On the theory of Ψ-condensation. Biopolymers. 21:2413–2432. [Google Scholar]
- Hanlon, S., S. Brundo, T. T. Wu, and B. Wolf. 1975. Structural transitions of deoxyribonucleic acid in aqueous electrolyte solutions. I. Reference spectra of conformational limits. Biochemistry. 8:1648–1660. [DOI] [PubMed] [Google Scholar]
- Howerton, S. B., C. C. Sines, D. VanDerveer, and L. D. Williams. 2001. Locating monovalent cations in the grooves of B-DNA. Biochemistry. 40:10023–10031. [DOI] [PubMed] [Google Scholar]
- Hud, N. V., and M. Polak. 2001. DNA–cation interactions: the major and minor grooves are flexible ionophores. Curr. Opin. Struct. Biol. 11:293–301. [DOI] [PubMed] [Google Scholar]
- Ivanov, V. I., L. E. Minchenkova, A. K. Schyolkina, and A. I. Poletayev. 1973. Different conformations of double-stranded nucleic acid in solution as revealed by circular dichroism. Biopolymers. 12:89–110. [DOI] [PubMed] [Google Scholar]
- Korolev, N., A. P. Lyubartsev, A. Rupprecht, and L. Nordenskiöld. 2001. Competitive substitution of hexammine cobalt(III) for Na+ and K+ ions in oriented DNA fibers. Biopolymers. 58:268–278. [DOI] [PubMed] [Google Scholar]
- Livolant, F., and L. Amelie. 1996. Condensed phases of DNA: structures and phase transitions. Prog. Polym. Sci. 21:1115–1164. [Google Scholar]
- Maniatis, T., J. H. Venable Jr., and L. S. Lerman. 1974. The structure of Ψ DNA. J. Mol. Biol. 84:37–64. [DOI] [PubMed] [Google Scholar]
- Mayama, H., and K. Yoshikawa. 2000. Thermodynamics in folding transition of DNA. Macromol. Symp. 160:55–60. [Google Scholar]
- Mayama, H., T. Iwataki, and K. Yoshikawa. 2000. Thermodynamics in the folding phase-transition of single T4DNA molecules in poly(ethylene glycol) solution. Chem. Phys. Lett. 318:113–117. [Google Scholar]
- McFail-Isom, L., C. Sines, and L. D. Williams. 1999. DNA structure: cations in charge? Curr. Opin. Struct. Biol. 9:298–304. [DOI] [PubMed] [Google Scholar]
- Minagawa, K., Y. Matsuzawa, K. Yoshikawa, A. R. Khokhlov, and M. Doi. 1994. Direct observation of the coil-globule transition in DNA molecules. Biopolymers. 34:555–558. [Google Scholar]
- Morgan, J. E., J. W. Blankenship, and H. R. Mattheus. 1987. Polyamines and acetylpolyamines increase the stability and alter the conformation of nucleosome core particles. Biochemistry. 26:3643–3649. [DOI] [PubMed] [Google Scholar]
- Post, C. B., and B. H. Zimm. 1979. Internal condensation of a single DNA molecule. Biopolymers. 18:1487–1501. [Google Scholar]
- Rouzina, I., and V. A. Bloomfield. 1996. Competitive electrostatic binding of charged ligands to polyelectrolytes: planar and cylindrical geometries. J. Phys. Chem. 100:4305–4313. [DOI] [PubMed] [Google Scholar]
- Strick, R., P. L. Strissel, K. Gavrilov, and R. Levi-Setti. 2001. Cation-chromatin binding as shown by ion microscopy is essential for the structural integrity of chromosomes. J. Cell Biol. 155:899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwalsky, M., W. Traub, U. Shmueli, and J. A. Subirana. 1969. An x-ray study of the interaction of DNA with spermine. J. Mol. Biol. 42:363–373. [DOI] [PubMed] [Google Scholar]
- Takahashi, M., K. Yoshikawa, V. V. Vasilevskaya, and A. R. Khokhlov. 1997. Discrete coil-globule transition of single duplex DNAs induced by polyamines. J. Phys. Chem. B. 101:9396–9401. [Google Scholar]
- Vasilevskaya, V. V., A. R. Khokhlov, Y. Matsuzawa, and K. Yoshikawa. 1995. Collapse of single DNA molecule in poly(ethylene glycol) solutions. J. Chem. Phys. 102:6595–6602. [Google Scholar]
- Warley, A., J. Stephen, A. Hockaday, and T. C. Appleton. 1983. X-ray microanalysis of HeLa S3 cells. II. Analysis of elemental levels during the cell cycle. J. Cell Sci. 62:339–350. [DOI] [PubMed] [Google Scholar]
- Wilson, R. W., and V. A. Bloomfield. 1979. Counterion-induced condensation of deoxyribonucleic acid: a light-scattering study. Biochemistry. 18:2192–2196. [DOI] [PubMed] [Google Scholar]
- Wolf, B., S. Berman, and S. Hanlon. 1977. Structural transitions of calf thymus DNA in concentrated LiCl solutions. Biochemistry. 16:3655–3662. [DOI] [PubMed] [Google Scholar]
- Wolf, B., and S. Hanlon. 1975. Structural Transitions of transitions of deoxyribonucleic acid in aqueous solutions. II. The role of hydration. Biochemistry. 8:1661–1670. [DOI] [PubMed] [Google Scholar]
- Zimmerman, S. B. 1993. Macromolecular crowding effects on macromolecular interactions: some implications for genome structure and function. Biochim. Biophys. Acta. 1216:175–185. [DOI] [PubMed] [Google Scholar]
- Zinchenko, A. A., V. G. Sergeyev, V. A. Kabanov, S. Murata, and K. Yoshikawa. 2004. Stereoisomeric discrimination in DNA compaction. Angew. Chem. Int. Ed. 43:2378–2381. [DOI] [PubMed] [Google Scholar]







