Abstract
1,2-GG intrastrand cross-links formed in DNA by the enantiomeric complexes [PtCl2(R,R-2,3-diaminobutane (DAB))] and [PtCl2(S,S-DAB)] were studied by biophysical methods. Molecular modeling revealed that structure of the cross-links formed at the TGGT sequence was affected by repulsion between the 5′-directed methyl group of the DAB ligand and the methyl group of the 5′-thymine of the TGGT fragment. Molecular dynamics simulations of the solvated platinated duplexes and our recent structural data indicated that the adduct of [PtCl2(R,R-DAB)] alleviated this repulsion by unwinding the TpG step, whereas the adduct of [PtCl2(S,S-DAB)] avoided the unfavorable methyl-methyl interaction by decreasing the kink angle. Electrophoretic retardation measurements on DNA duplexes containing 1,2-GG intrastrand cross-links of Pt(R,R-DAB)2+ or Pt(S,S-DAB)2+ at a CGGA site showed that in this sequence both enantiomers distorted the double helix to the identical extent similar to that found previously for the same sequence containing the cross-links of the parent antitumor 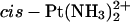 (cisplatin). In addition, the adducts showed similar affinities toward the high-mobility-group box 1 proteins. Hence, whereas the structural perturbation induced in DNA by 1,2-GG intrastrand cross-links of cisplatin does not depend largely on the bases flanking the cross-links, the perturbation related to GG cross-linking by bulkier platinum diamine derivatives does.
(cisplatin). In addition, the adducts showed similar affinities toward the high-mobility-group box 1 proteins. Hence, whereas the structural perturbation induced in DNA by 1,2-GG intrastrand cross-links of cisplatin does not depend largely on the bases flanking the cross-links, the perturbation related to GG cross-linking by bulkier platinum diamine derivatives does.
INTRODUCTION
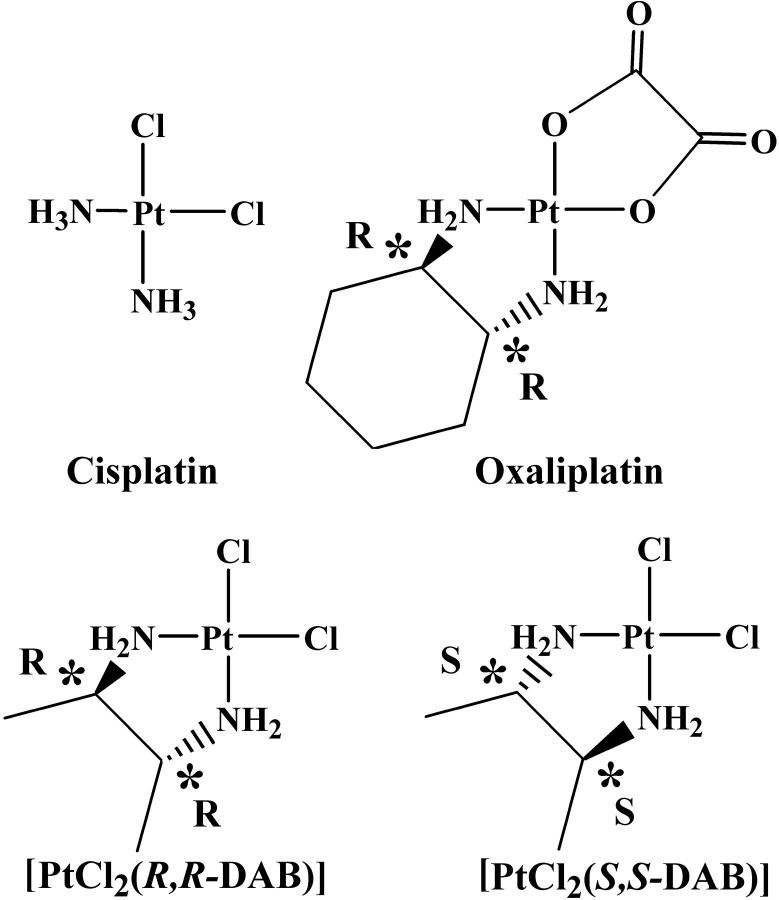
The activity of the antitumor drug cisplatin (cis-[PtCl2(NH3)2]) is related to its binding to DNA. The 1,2-GG intrastrand cross-links, formed as the major DNA adducts, are believed to play an important role in the mechanism underlying antitumor effects of this drug (Jamieson and Lippard, 1999). The ability to form the 1,2-GG cross-link as a major lesion has been also observed for a number of antitumor active analogs of cisplatin in which one or both NH3 ligands were replaced by primary amine(s) or by a diamine (Fig. 1). In this class of cytotoxic cisplatin analogs with a diamine ligand carrying chiral centers, such as [PtCl2(2,3-diaminobutane (DAB))] or [PtCl2(trans-1,2-diaminocyclohexane (DACH))] are of a particular interest. Enantiomeric pairs of these compounds have been shown to exhibit different biological activities. For example, the R,R isomer of [PtCl2(DACH)] has somewhat higher antitumor activity than the S,S isomer (Kidani et al., 1978; Noji et al., 1981), and the S,S isomer of [PtCl2(DAB)] has been found to be considerably more mutagenic than the R,R isomer (Fanizzi et al., 1987). Oxaliplatin, the R,R isomer of [Pt(oxalate)(DACH)], is currently in clinical use against metastatic colorectal cancer in which cisplatin is inactive (Raymond et al., 2002) and produces the same type of inter- and intrastrand DNA cross-links as cisplatin (Saris et al., 1996; Woynarowski et al., 2000).
FIGURE 1.
The antitumor compounds cisplatin and oxaliplatin, and the two enantiomers of [PtCl2(DAB)].
Since DNA is a chiral molecule, the fact that the optical isomers of platinum diamine complexes differ in their biological effects is a priori not surprising. The 1,2-GG-Pt intrastrand cross-link bends generally the DNA double helix toward the major groove (Jamieson and Lippard, 1999), and the contacts between the amine “spectator ligands” and the major groove can affect the adduct structure; the nature of these contacts would be chirality-dependent. However, convincing evidence for an influence of such contacts on the adduct structure has not yet been demonstrated. For instance, in the crystal structure of a 12-basepair (bp)-long DNA duplex cross-linked by Pt(R,R-DACH)2+ the DACH ligand forms a hydrogen bond with the O6 atom of the 3′-guanine, and it has been hypothesized that such hydrogen bonding could explain the differences in biological effects of the R,R and S,S isomers of [Pt(oxalate)(DACH)] (Spingler et al., 2001). On the other hand, the molecular structure of the same 12-bp duplex cross-linked with cisplatin is very similar, showing virtually identical bend and unwinding angles, although the NH…O6 hydrogen bond is not present (Takahara et al., 1995). Thus, the exact role of the contacts between the platinum ligands and DNA residues in determining the overall adduct structure is not yet clear.
The recognition and repair of the Pt-DNA cross-links by cellular proteins is crucial for the processing of these DNA lesions (Zamble and Lippard, 1999; Stehlikova et al., 2002; Brabec, 2002). A specific class of proteins that bind with high affinity to 1,2-GG adducts of platinum complexes are the high-mobility-group box (HMGB) proteins (Pil and Lippard, 1992). Vaisman et al. (1999) have investigated the binding of the ubiquitous HMGB1 protein to duplex DNA containing an AGGC sequence modified by different platinum complexes, and found that HMGB1 bound two times more strongly to DNA cross-linked by 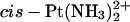 than to DNA cross-linked by Pt(R,R-DACH)2+. A similar result was obtained by Lippard et al. for the isolated A-domain of HMGB1 (HMGB1a) (Wei et al., 2001). However, when the affinity to Pt-cross-links within a TGGA sequence was probed, the HMGB1a domain discriminated by a factor of >15 between the
than to DNA cross-linked by Pt(R,R-DACH)2+. A similar result was obtained by Lippard et al. for the isolated A-domain of HMGB1 (HMGB1a) (Wei et al., 2001). However, when the affinity to Pt-cross-links within a TGGA sequence was probed, the HMGB1a domain discriminated by a factor of >15 between the 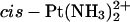 and Pt(R,R-DACH)2+ adducts (Wei et al., 2001). Recently, Malina et al. (2002) investigated DNA modified by the two isomers of [PtCl2(DAB)], a complex closely related to [PtCl2(DACH)] (Fig. 1)and determined the binding affinities of the two individual domains of HMGB1protein to Pt(R,R-DAB)2+, Pt(S,S-DAB)2+, and
and Pt(R,R-DACH)2+ adducts (Wei et al., 2001). Recently, Malina et al. (2002) investigated DNA modified by the two isomers of [PtCl2(DAB)], a complex closely related to [PtCl2(DACH)] (Fig. 1)and determined the binding affinities of the two individual domains of HMGB1protein to Pt(R,R-DAB)2+, Pt(S,S-DAB)2+, and 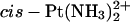 adducts formed at a TGGT sequence of a DNA duplex. They found that the A-domain bound ∼2 times more tightly to the Pt(R,R-DAB)2+ adduct than to the cisplatin adduct, whereas the Pt(S,S-DAB)2+ adduct was recognized ∼8 times less strongly than the cisplatin adduct. The B-domain of HMGB1 (HMGB1b) also bound more strongly to the Pt(R,R-DAB)2+ adduct than to the Pt(S,S-DAB)2+ adduct, but the difference was less pronounced (Malina et al., 2002). All these results indicate that the spectator ligands do affect the recognition of the 1,2-GG platinum cross-link, and that their influence is dependent on the nature of the bases flanking GG.
adducts formed at a TGGT sequence of a DNA duplex. They found that the A-domain bound ∼2 times more tightly to the Pt(R,R-DAB)2+ adduct than to the cisplatin adduct, whereas the Pt(S,S-DAB)2+ adduct was recognized ∼8 times less strongly than the cisplatin adduct. The B-domain of HMGB1 (HMGB1b) also bound more strongly to the Pt(R,R-DAB)2+ adduct than to the Pt(S,S-DAB)2+ adduct, but the difference was less pronounced (Malina et al., 2002). All these results indicate that the spectator ligands do affect the recognition of the 1,2-GG platinum cross-link, and that their influence is dependent on the nature of the bases flanking GG.
That the differential recognition by HMGB proteins of GG adducts formed by the enantiomers of the same platinum complex could originate from structural differences between the adducts was suggested by another study from the Brabec group in which the bending and unwinding angles of 20- to 23-bp DNA duplexes cross-linked at a central TGGT sequence by the R,R and S,S isomers of [PtCl2(DAB)] were investigated (Malina et al., 2000). The R,R isomer was shown to bend the double helix by 35 ± 2° and to cause an unwinding of 20 ± 2°, whereas for the S,S isomer these values were 24 ± 2° and 15 ± 3°, respectively. Under the same conditions, the cisplatin adduct formed with the same sequence bent the DNA helix by 33 ± 2° and unwound it by 15 ± 2° (Stehlikova et al., 2002). In an effort to contribute to understanding the effect of spectator ligands of platinum complexes on the structural perturbation of DNA due to the formation of the 1,2-GG intrastrand cross-links, we used molecular modeling techniques in this work to investigate the structures of adducts formed by Pt(R,R-DAB)2+, Pt(S,S-DAB)2+, or 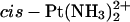 moieties with a 12-bp DNA duplex including a top strand with a central TGGT sequence (Scheme 1). The exact sequence of the duplex (designated subsequently TGGT) corresponded to the inner part of the duplexes studied previously (Malina et al., 2000). In addition, we have determined the bending and unwinding angles induced by 1,2-GG intrastrand cross-links of Pt(R,R-DAB)2+ or Pt(S,S-DAB)2+ formed in the duplexes having a top strand with a CGGA platinum binding site, by means of gel electrophoretic retardation (phasing) assay. Moreover, experiments with chemical probes of DNA conformation were carried out to characterize the distortion caused by Pt(DAB)2+ adducts bound to the CGGA sequence. In addition, we have probed the recognition of the adducts of Pt(R,R-DAB)2+ and Pt(S,S-DAB)2+ with CGGA by the A and B domains of the HMGB1 protein using the gel electrophoretic mobility-shift assay.
moieties with a 12-bp DNA duplex including a top strand with a central TGGT sequence (Scheme 1). The exact sequence of the duplex (designated subsequently TGGT) corresponded to the inner part of the duplexes studied previously (Malina et al., 2000). In addition, we have determined the bending and unwinding angles induced by 1,2-GG intrastrand cross-links of Pt(R,R-DAB)2+ or Pt(S,S-DAB)2+ formed in the duplexes having a top strand with a CGGA platinum binding site, by means of gel electrophoretic retardation (phasing) assay. Moreover, experiments with chemical probes of DNA conformation were carried out to characterize the distortion caused by Pt(DAB)2+ adducts bound to the CGGA sequence. In addition, we have probed the recognition of the adducts of Pt(R,R-DAB)2+ and Pt(S,S-DAB)2+ with CGGA by the A and B domains of the HMGB1 protein using the gel electrophoretic mobility-shift assay.
SCHEME 1.
Duplex TGGT cross-linked with Pt(DAB)2+.
EXPERIMENTAL PROCEDURES
Molecular modeling
The molecular mechanics calculations and molecular dynamics (MD) simulations used the SANDER module of the program AMBER6.0 (Case et al., 1999) implemented on a Beowulf cluster of Bi-athlon and Bi-xeon processors or on a Cray T3E computer. The parm98 force-field (Cheatham et al., 1999) was complemented with parameters describing Pt(II)-coordination by two guanines, as described previously (Herman et al., 1990; Elizondo-Riojas et al., 1997; Elizondo-Riojas and Kozelka, 2001). Density functional theory calculations on the model complex cis-[Pt(DAB)(1-Me-Guo)2]2+, (B3LYP hybrid functional, LANL2DZ pseudoorbital basis set for Pt and 3-21G* for the other atoms) were performed using the Gaussian98 program, (Frisch et al., 1998) to determine the atomic charges of the 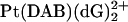 residue, and the Pt-N bond lengths. The MD simulator used a time step of 2 fs. The electrostatic energy was calculated using the particle-mesh Ewald method (Darden et al., 1993; Essman et al., 1995) with a 1-Å charge grid, cubic B-spline interpolation and sum tolerance of 10−6Å. During whole MD simulations, Lennard-Jones interactions were computed with a 9-Å cutoff distance, the nonbonded interactions list was updated every 10 steps, and the SHAKE algorithm (Ryckaert et al., 1977) was applied to all X-H bonds (tolerance criterion of 0.0005).
residue, and the Pt-N bond lengths. The MD simulator used a time step of 2 fs. The electrostatic energy was calculated using the particle-mesh Ewald method (Darden et al., 1993; Essman et al., 1995) with a 1-Å charge grid, cubic B-spline interpolation and sum tolerance of 10−6Å. During whole MD simulations, Lennard-Jones interactions were computed with a 9-Å cutoff distance, the nonbonded interactions list was updated every 10 steps, and the SHAKE algorithm (Ryckaert et al., 1977) was applied to all X-H bonds (tolerance criterion of 0.0005).
The starting structures were generated by adapting the NMR-based model of an intrastrand cisplatin-GG cross-link by Gelasco and Lippard. (PDB code 1A84) (Gelasco and Lippard, 1998) to our sequence. The choice was based on the similarity of the base sequence (the sequences differ only at positions 1,3, and 4). The Pt(DAB)2+ adducts were constructed by least-squares fitting of the Pt(DAB)2+ moiety (obtained from geometry optimization using B3LYP/Gaussian98) into the 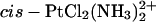 structure using the three-atom unit cis-PtN2 as reference. To neutralize the system, 20 sodium counterions were added to solute using the EDIT module of AMBER6.0; no counterions were placed at the phosphates of the platinated guanosines. The unit cell used for periodic boundary conditions applied in each MD simulation was composed of an ∼72 × ∼54 × ∼54 Å orthorhombic box ramdomly filled with 5864 to 5875 TIP3P water molecules. The protocol for energy minimization of starting structures and the subsequent MD simulations was as described previously (Elizondo-Riojas and Kozelka, 2001), except that the heating period consisted of two 10-ps steps, the first (0–150 K) with direct temperature scaling, and the second (150–300 K) using the Berendsen algorithm, and the subsequent equilibration period was extended to 87.5 ps. As previously, three Maxwell redistributions of velocities (see Supplementary Material, Table S1) were used to attenuate the influence of the initial configuration of the system on the dynamics. The production periods at a constant temperature of 300 K were 2–5 ns long. To avoid occasional openings of the terminal basepairs, their WC hydrogen bonds were reinforced by means of soft distance constraints of 10 kcal.mol−1 Å−2. Table S1 shows the whole MD protocol. The structures having αtγt conformation at the T5-G*6 step were generated by constraining the α and γ torsion angles to 180° with a harmonic constraint (k = 20 kcal.mol−1 rad−2) during 67.5 ps at the beginning of the equilibration period, followed by 20 ps of relaxation with the torsion constraints removed, and a production period of 2 ns.
structure using the three-atom unit cis-PtN2 as reference. To neutralize the system, 20 sodium counterions were added to solute using the EDIT module of AMBER6.0; no counterions were placed at the phosphates of the platinated guanosines. The unit cell used for periodic boundary conditions applied in each MD simulation was composed of an ∼72 × ∼54 × ∼54 Å orthorhombic box ramdomly filled with 5864 to 5875 TIP3P water molecules. The protocol for energy minimization of starting structures and the subsequent MD simulations was as described previously (Elizondo-Riojas and Kozelka, 2001), except that the heating period consisted of two 10-ps steps, the first (0–150 K) with direct temperature scaling, and the second (150–300 K) using the Berendsen algorithm, and the subsequent equilibration period was extended to 87.5 ps. As previously, three Maxwell redistributions of velocities (see Supplementary Material, Table S1) were used to attenuate the influence of the initial configuration of the system on the dynamics. The production periods at a constant temperature of 300 K were 2–5 ns long. To avoid occasional openings of the terminal basepairs, their WC hydrogen bonds were reinforced by means of soft distance constraints of 10 kcal.mol−1 Å−2. Table S1 shows the whole MD protocol. The structures having αtγt conformation at the T5-G*6 step were generated by constraining the α and γ torsion angles to 180° with a harmonic constraint (k = 20 kcal.mol−1 rad−2) during 67.5 ps at the beginning of the equilibration period, followed by 20 ps of relaxation with the torsion constraints removed, and a production period of 2 ns.
One structure was saved every 1 ps during the production period to constitute the trajectory file. The analysis of trajectories used the programs CURVES (Lavery and Sklenar, 1989) for the calculation of helical parameters (using the “global” helix axis) and the backbone torsion angles, and the CARNAL module of AMBER6.0 to determine average structures over the whole production period or for different conformational families. The averaged structures were submitted to a two-step energy minimization protocol before structural analysis (500 steps of in vacuo geometry optimization for hydrogen atoms only, followed by 1000 steps full-energy minimization using the GBSA module of AMBER6. The program VMD (Dalke et al., 1997) was used for superposition of molecules, visualization, and, coupled to the Raster3D software (Merrit and Bacon, 1997), for the figures. The xmgrace program (http://www.caida.org/tools/utilities/graphing/graph_xmgrace.xml) has been used to plot all graphs.
Materials
[PtCl2(R,R-DAB)] and [PtCl2(S,S-DAB)] were prepared and characterized as described (Fanizzi et al., 1987). Cisplatin was obtained from Sigma (Prague, Czech Republic). The stock solutions of platinum compounds were prepared at the concentration of 5 × 10−4 M in 10 mM NaClO4 and stored at 4°C in the dark. The synthetic oligodeoxyribonucleotides were synthesized and purified as described previously (Brabec et al., 1992). The 20- to 23-bp oligonucleotide duplexes containing central sequences CGGA/TCCG (for nucleotide sequences, see Figs. 4–6) uniquely and site-specifically intrastrand cross-linked by Pt(R,R-DAB)2+, PtCl2(S,S-DAB)2+, or cisplatin were prepared and characterized as described previously (Malina et al., 2000, 2002). Expression and purification of the domains HMGB1a and HMGB1b were carried out as described (Stros, 2001). T4 DNA ligase and T4 polynucleotide kinase were purchased from New England Biolabs (Beverly, MA). Acrylamide, bis(acrylamide), urea, and NaCN were from Merck KgaA (Darmstadt, Germany). Dimethyl sulfate, KMnO4, diethyl pyrocarbonate (DEPC), KBr and KHSO5 were from Sigma (Prague, Czech Republic). [γ-32P]ATP was from Amersham (Arlington Heights, IL). ATP was from Boehringer (Mannheim, Germany).
FIGURE 4.
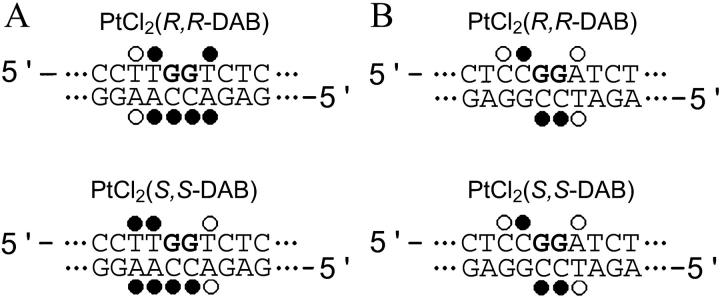
Summary of the reactivity of chemical probes with the TGGT(21) (left) and CGGA(21) (right) duplexes containing single, site-specific 1,2-GG intrastrand cross-link of Pt(R,R-DAB)2+ or Pt(S,S-DAB)2+. The reactivity of the chemical probes was evaluated via piperidine-induced specific strand cleavage at KMnO4-modified thymine residues, DEPC-modified adenine and guanine residues, and KBr/KHSO5-modified cytosine residues. (•) and (○) designate strong or weak reactivity, respectively. The data for the duplex TGGT were taken from our article published previously (Malina et al., 2000).
FIGURE 5.
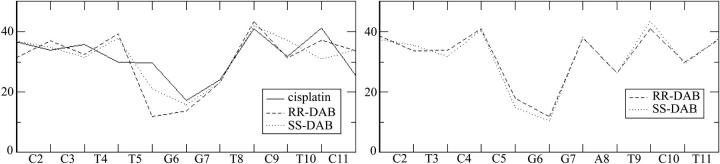
Twist angles calculated using the “global axis” option of CURVES (Lavery and Sklenar, 1988) for the time-averaged structures from MD simulations of adducts of duplexes TGGT (left) and CGGA (right).
FIGURE 6.
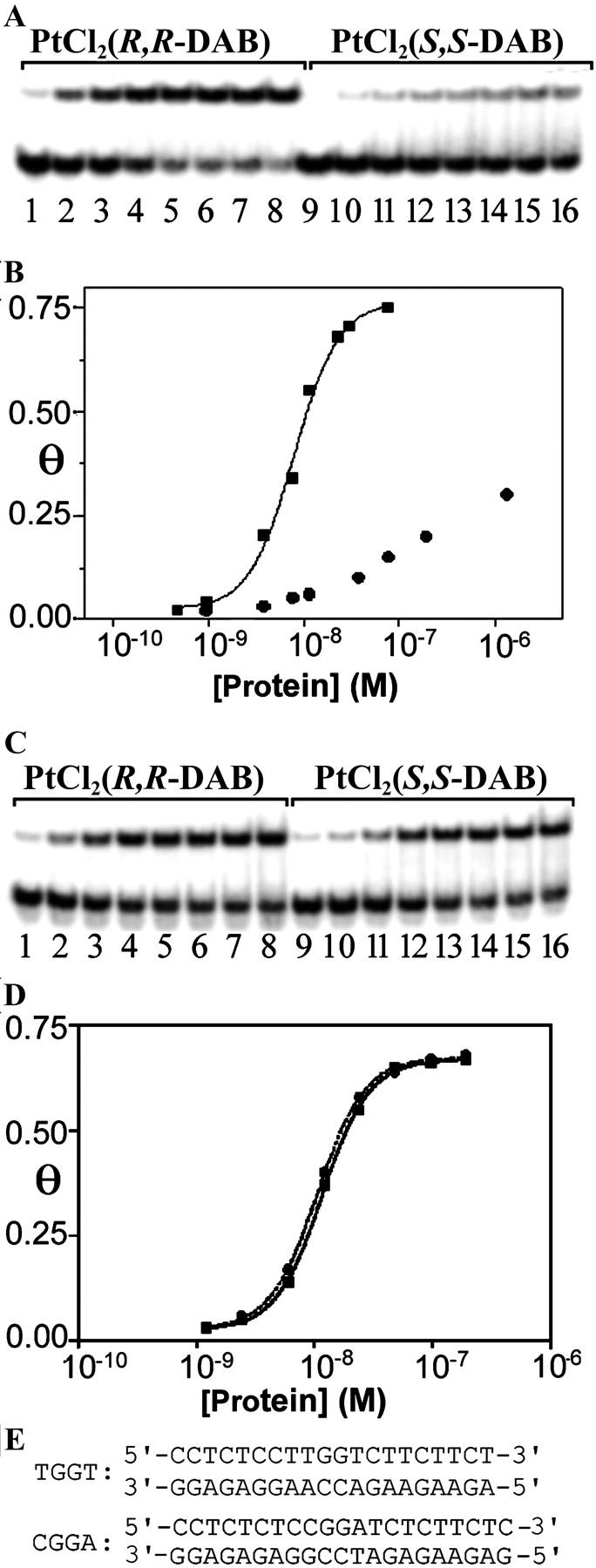
Gel mobility shift assay analysis of the titration of the duplexes TGGT and CGGA (for sequences, see E) containing the single 1,2-GG intrastrand cross-link of Pt(R,R-DAB)2+ or Pt(S,S-DAB)2+ with HMGB1a protein. (A and C) Autoradiograms of the gel mobility shift assay of the titration with HMGB1a (0.5 nM–1.4 μM). The concentration of the duplexes TGGT (A) and CGGA (C) was 10 nM. (B and D) Plots of the fraction of bound DNA (Θ) versus HMGB1a concentration with superimposed fits to the equation Θ = P/P+ KD(app) (P is the total protein concentration, and KD(app) is the apparent dissociation constant) (Dunham and Lippard, 1997; Lohman and Mascotti, 1992). DNA probes were the duplexes TGGT (B) or CGGA (D) (10 nM) containing the single 1,2-GG intrastrand CL of Pt(R,R-DAB)2+ (▪) or Pt(S,S-DAB)2+ (•). (E) The nucleotide sequences of the duplexes TGGT and CGGA used in the experiments demonstrated in A–D.
Ligation and electrophoresis of oligonucleotides
Details of these experiments were as described in previously published articles (Bellon and Lippard, 1990; Bellon et al., 1991; Kasparkova et al., 1996).
Chemical modifications
The modifications by KMnO4, DEPC, and KBr/KHSO5 were performed as described previously (Brabec et al., 1993). The strands of the duplexes were 5′-end-labeled with [γ-32P]ATP. In the case of the platinated oligonucleotides, the platinum complex was removed after reaction of the DNA with the probe by incubation with 0.2 M NaCN (pH 11) at 45°C for 10 h in the dark.
Electrophoretic mobility shift assays with HMGB1 domain proteins
Radioactively labeled 22-bp DNA probes (10 nM) were titrated with HMGB1a or HMGB1b proteins in 10-μl sample volumes in the buffer composed of 10 mM HEPES, pH 7.5, 10 mM MgCl2, 50 mM LiCl, 100 mM NaCl, 1 mM spermidine, 0.2 mg/ml bovine serum albumin, and 0.05% v/v Nonidet P40. For all gel mobility shift experiments, samples were incubated on ice for 1 h and made 7% in sucrose and 0.017% in xylene cyanol before loading on running, precooled (4°C), prerun (300 V, 1–2 h) 5% native polyacrylamide gels (29:1 acrylamide:bisacrylamide, 0.5× Tris-borate-EDTA buffer (45 mM Tris-HCl, 45 mM boric acid, and 1 mM EDTA, pH 8.3). Gels were electrophoresed at 4°C and 300 V for ∼1.5 h, dried, exposed to a molecular imaging plate, analyzed on a Fujifilm bioimaging analyzer, and the radioactivities associated with bands were quantitated with the AIDA image analyzer software. Other details have been published previously (Malina et al., 2002)
RESULTS
Structures of cisplatin adducts with GG sequences are little affected by flanking bases
An MD simulation of the duplex TGGT cross-linked with a 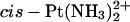 residue, explicitly solvated in a water bath (see Experimental Procedures) showed features similar to those seen in a previous simulation of a double-stranded decamer bearing a GG-Pt cross-link within a CGGA sequence (Elizondo-Riojas and Kozelka, 2001). Specifically, we observed the same lateral movement of the cross-linked GG dinucleotide with respect to the flanking basepairs, making the H2′ proton of the thymidine 5′ to the GG-Pt cross-link enter and quit the shielding cone of the 5′-guanine. The time-averaged kink angle (62 ± 13°) of the
residue, explicitly solvated in a water bath (see Experimental Procedures) showed features similar to those seen in a previous simulation of a double-stranded decamer bearing a GG-Pt cross-link within a CGGA sequence (Elizondo-Riojas and Kozelka, 2001). Specifically, we observed the same lateral movement of the cross-linked GG dinucleotide with respect to the flanking basepairs, making the H2′ proton of the thymidine 5′ to the GG-Pt cross-link enter and quit the shielding cone of the 5′-guanine. The time-averaged kink angle (62 ± 13°) of the 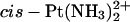 -TGGT adduct was similar to that determined previously for the
-TGGT adduct was similar to that determined previously for the 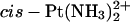 -CGGA adduct (60 ± 10) (Elizondo-Riojas and Kozelka, 2001). This similarity is in line with the finding of Stehlikova et al. (2002) that the bending and unwinding angles of XGGY sequences bound to
-CGGA adduct (60 ± 10) (Elizondo-Riojas and Kozelka, 2001). This similarity is in line with the finding of Stehlikova et al. (2002) that the bending and unwinding angles of XGGY sequences bound to 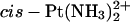 depend little on the nature of the bases X and Y.
depend little on the nature of the bases X and Y.
Structures of Pt(R,R-DAB)2+ and Pt(S,S-DAB)2+ adducts with TGGT are affected by the repulsion between the methyl groups of the DAB ligand and the 5′-thymine
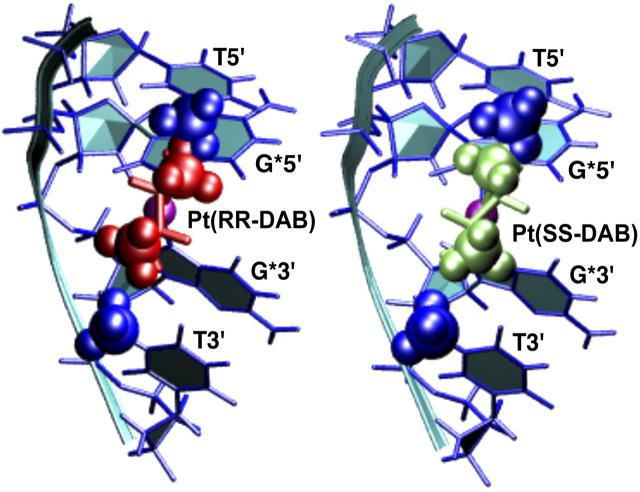
Replacing the 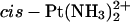 moiety in the time-averaged structure obtained from the MD simulation of the cisplatin-TGGT adduct by Pt(R,R-DAB)2+ or Pt(S,S-DAB)2+ results in clashes between the 5′-directed methyl group of the DAB ligand and the methyl group of the 5′-thymine of the TGGT tetranucleotide (Fig. 2). From Fig. 2, it appears clear that neither enantiomer of Pt(DAB)2+ bound to a TGGT sequence can accommodate the structure preferred by the cisplatin adduct. Possible ways to avoid this methyl-methyl repulsion include 1), untwisting or overtwisting the TG step; 2), displacements of the T and G bases along vectors perpendicular to the helix axis, i.e., modifying the slide and/or shift; 3), rotations of either base about such vectors, i.e., modifying roll and/or tilt; and 4), increasing the rise between the two bases. The differences in unwinding and kink angles observed experimentally between the adducts of Pt(R,R-DAB)2+ and Pt(S,S-DAB)2+ (Malina et al., 2000) indicated that the two structures obviate the methyl-methyl clashes differently. This is not surprising if one considers the different positioning of the DAB methyl groups (Fig. 2). It is interesting to note that no clashes are observed between DAB and thymine methyl groups at the 3′-side of the GG-Pt cross-link.
moiety in the time-averaged structure obtained from the MD simulation of the cisplatin-TGGT adduct by Pt(R,R-DAB)2+ or Pt(S,S-DAB)2+ results in clashes between the 5′-directed methyl group of the DAB ligand and the methyl group of the 5′-thymine of the TGGT tetranucleotide (Fig. 2). From Fig. 2, it appears clear that neither enantiomer of Pt(DAB)2+ bound to a TGGT sequence can accommodate the structure preferred by the cisplatin adduct. Possible ways to avoid this methyl-methyl repulsion include 1), untwisting or overtwisting the TG step; 2), displacements of the T and G bases along vectors perpendicular to the helix axis, i.e., modifying the slide and/or shift; 3), rotations of either base about such vectors, i.e., modifying roll and/or tilt; and 4), increasing the rise between the two bases. The differences in unwinding and kink angles observed experimentally between the adducts of Pt(R,R-DAB)2+ and Pt(S,S-DAB)2+ (Malina et al., 2000) indicated that the two structures obviate the methyl-methyl clashes differently. This is not surprising if one considers the different positioning of the DAB methyl groups (Fig. 2). It is interesting to note that no clashes are observed between DAB and thymine methyl groups at the 3′-side of the GG-Pt cross-link.
FIGURE 2.
Detail of the TGGT fragments from the time-averaged, energy-minimized model of the adduct of 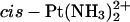 with duplex TGGT, where the
with duplex TGGT, where the 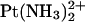 residue was replaced with Pt(R,R-DAB)2+ (left) and Pt(S,S-DAB)2+ (right), respectively. The methyl groups of the thymines (blue) and of the DAB ligand (green) are shown as CPK models. Clash is seen with the 5′-thymine but not with the 3′-thymine.
residue was replaced with Pt(R,R-DAB)2+ (left) and Pt(S,S-DAB)2+ (right), respectively. The methyl groups of the thymines (blue) and of the DAB ligand (green) are shown as CPK models. Clash is seen with the 5′-thymine but not with the 3′-thymine.
Both Pt(DAB)2+ adducts were simulated starting from the two structures whose details are shown in Fig. 2. In addition, for both adducts, we considered a variant in which the TpG step was switched from the canonical α−γ+ to the αtγt conformation. The αtγt conformation has been occasionally observed in oligonucleotides that are exposed to strain due to covalent (Sherman et al., 1985) or noncovalent (Djuranovic and Hartmann, 2003) modifications and has been reported to be connected with decreased twist (Djuranovic and Hartmann, 2003). Therefore, from the experimental observation that the TGGT adduct with Pt(R,R-DAB)2+ unwinds the double helix more than the adducts of cisplatin or of Pt(S,S-DAB)2+ (Malina et al., 2000; Stehlikova et al., 2002), one could expect that the Pt(R,R-DAB)2+-TGGT adduct unwinds the TpG step as a reaction to the methyl-methyl repulsion, and this unwinding could be stabilized by an α−γ+ → αtγt transition. Such transitions involve relatively high energy barriers (Varnai et al., 2002) and would not necessarily occur spontaneously during an MD simulation; we have therefore generated them using dihedral angle constraints and simulated the structures with the TpG step having α−γ+ and αtγt conformations separately. In line with the expectedly high energy barrier, all four simulations retained the starting α−γ+ or αtγt conformation.
Table 1 gives the time-averaged values of the twist and roll angles at the T5-G*6 step as well as the global twist per step and the global kink angle, as calculated with CURVES (Lavery and Sklenar, 1989). The extent to which these values fluctuate during the MD simulation is assessed as the standard deviation calculated over the ensemble of snapshots. Table 1 shows that the αtγt conformation indeed stabilizes smaller twist and less negative (or positive) roll at the TpG step. As a consequence, the global twist per step of the two structures having the αtγt conformation is smaller than that of the structures having a canonical α−γ+ conformation. The relationship between the global kink angle and the roll at the TpG step is less evident (since the global kink depends on a number of other local parameters), but the negative roll in the α−γ+ conformation seems to correlate with the smaller kink angle observed for these structures. It has to be noted that the kink values determined for solution structures of platinated DNA from molecular models are generally ∼2 times higher than those obtained from electrophoretic measurements. Possible sources for this discrepancy include experimental and computational errors, different conditions in a gel as compared to a diluted solution, and problems in defining the kink angle (Marzilli et al., 2001; Elizondo-Riojas and Kozelka, 2001; Spingler et al., 2001). We are concerned only with relative values here.
TABLE 1.
Twist and roll between the bases T5 and G*6, average twist per step and global kink angle, time-averaged over the MD simulations
| Adduct | Twist (T5-G*6) | Roll (T5-G*6) | Average twist | Kink |
|---|---|---|---|---|
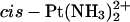 -TGGT -TGGT
|
31 (±8) | −10 (±6) | 31.4 (±1.1) | 62 (±13) |
| Pt(R,R-DAB)2+-TGGT | ||||
| α−γ+ at T5-G*6 | 30 (±11) | −19 (±10) | 31.4 (±1.1) | 59 (±14) |
| DAB equatorial (50%) | 30 (±11) | −18 (±10) | 31.4 (±1.2) | 57 (±14) |
| DAB axial (50%) | 30 (±11) | −19 (±9) | 31.5 (±1.0) | 61 (±13) |
| αtγt at T5-G*6 | 13 (±11) | 1 (±10) | 30.2 (±1.3) | 66 (±14) |
| DAB equatorial (37%) | 15 (±10) | −1 (±9) | 30.1 (±1.3) | 61 (±13) |
| DAB axial (63%) | 13 (±12) | 2 (±11) | 30.3 (±1.3) | 69 (±14) |
| Pt(S,S-DAB)2+-TGGT | ||||
| α−γ+ at T5-G*6 | 25 (±8) | −11 (±7) | 31.0 (±1.3) | 58 (±12) |
| DAB equatorial (56%) | 28 (±8) | −13 (±7) | 31.4 (±1.2) | 57 (±13) |
| DAB axial (44%) | 21 (±7) | −10 (±6) | 30.5 (±1.3) | 59 (±12) |
| αtγt at T5-G*6 | 7 (±9) | −1 (±8) | 28.8 (±1.1) | 70 (±13) |
| DAB equatorial (31%) | −3 (±7) | 7 (±6) | 27.4 (±1.2) | 80 (±9) |
| DAB axial (69%) | 12 (±6) | −4 (±6) | 29.5 (±1.1) | 65 (±11) |
Standard deviations in parentheses. Values in degrees, all calculated with the “global” helix axis using CURVES (Lavery and Sklenar, 1988).
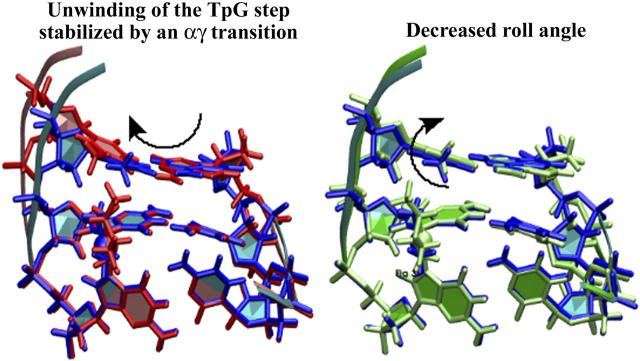
The previously reported biophysical measurements (Malina et al., 2000; Stehlikova et al., 2002), showing that the Pt(R,R-DAB)2+-TGGT adduct unwinds the double helix more strongly than the cisplatin-TGGT adduct but retains a similar kink angle, whereas the Pt(S,S-DAB)2+-TGGT adduct kinks the helix axis less but retains similar unwinding, can be thus plausibly explained by assuming that the TpG step retains the canonical α−γ+ conformation in the adduct of the S,S isomer but converts to αtγt in that of the R,R isomer. The corresponding models are shown in Fig. 3. The model that we propose for the Pt(R,R-DAB)2+-TGGT adduct was obtained by time-averaging the structures observed during the simulation “αtγt at T5-G*6” (see Table 1); that for the Pt(S,S-DAB)2+-TGGT adduct was obtained similarly from the corresponding simulation “α−γ+ at T5-G*6”. In Fig. 3, both structures are superimposed with the model for the 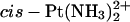 -TGGT adduct obtained from the same time-averaging procedure, so that the structural changes at the T5-G*6 step can be easily seen. To avoid the clash between the methyl group, the Pt(R,R-DAB)2+-TGGT adduct unwinds the T5-G*6 step, thus increasing the overall unwinding. On the other hand, the Pt(S,S-DAB)2+-TGGT adduct increases the negative roll of the T5-G*6 step, thus partially compensating the positive roll of the G*6-G*7 step and diminishing the overall kink angle.
-TGGT adduct obtained from the same time-averaging procedure, so that the structural changes at the T5-G*6 step can be easily seen. To avoid the clash between the methyl group, the Pt(R,R-DAB)2+-TGGT adduct unwinds the T5-G*6 step, thus increasing the overall unwinding. On the other hand, the Pt(S,S-DAB)2+-TGGT adduct increases the negative roll of the T5-G*6 step, thus partially compensating the positive roll of the G*6-G*7 step and diminishing the overall kink angle.
FIGURE 3.
TGG fragments of models for the duplex TGGT cross-linked with Pt(R,R-DAB)2+ (red, left) and Pt(S,S-DAB)2+ (green, right), both superimposed with the model for the adduct with 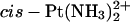 (blue). Structures were averaged over the production period of the MD simulations and subsequently energy-minimized using 1000 cycles of the conjugate-gradient minimizer of AMBER6.0. The structures were least-squares fitted using the Pt-G*pG* residue, which is nearly invariant in all three adducts. The arrows indicate how the two Pt(DAB)2+ adducts avoid the methyl-methyl clash.
(blue). Structures were averaged over the production period of the MD simulations and subsequently energy-minimized using 1000 cycles of the conjugate-gradient minimizer of AMBER6.0. The structures were least-squares fitted using the Pt-G*pG* residue, which is nearly invariant in all three adducts. The arrows indicate how the two Pt(DAB)2+ adducts avoid the methyl-methyl clash.
Hydrogen bonding involving the NH2 groups of DAB depends only marginally on DAB chirality
It has been suggested previously that stereoselective hydrogen bonding formed by the amino groups of the diamine ligand is responsible for the structural differences between DNA adducts formed by R,R and S,S isomers of Pt(DAB)2+ (Malina et al., 2000) and Pt(DACH)2+ (Spingler et al., 2001) complexes. Table 2 gives time-averaged N…O separations observed during our MD simulations, between either of the platinum-bound NH2 or NH3 groups and the proximate O6 atom of the adjacent guanine (G*6 at the 5′-side and G*7 at the 3′-side), and the O4 atoms of the next thymine (T5 at the 5′-side and T8 at the 3′-side). The distance from the 5′-directed NH2 group to the O2 atom of the phosphate 5′ to the GG-Pt cross-link is shown as well. It can be seen that the average distances characterizing the hydrogen bonding to the O6 atoms of either platinum-bound guanine are independent of DAB chirality, and unaffected by the α−γ+ → αtγt transition. The hydrogen atoms donated by the 5′-directed NH2 group of DAB to the 5′-phosphate and to the O4 atom of the 5′-thymine are very slightly dependent on the DAB chirality (R,R favoring the former and S,S the latter), and, above all, on the αγ conformation. In the canonical α−γ+ conformation, the NH2…OP hydrogen bond oscillates between direct bonding and bonding mediated by one water molecule, whereas in the αtγt conformation, there are two intervening water molecules most of the time. Conversely, the T5O4 atom forms predominantly a direct hydrogen bond in the αtγt conformation, whereas in the α−γ+ conformation it oscillates between direct and water-mediated bonding. The simulations further indicate a short and persistent hydrogen bond between the 3′-directed NH2 group of DAB and the 3′-thymine, independently of DAB chirality and αγ conformation.
TABLE 2.
Interatomic separations, and their fluctuations, between the two nitrogen atoms of the DAB (or NH3) ligand and potential hydrogen-bonding acceptors
| Adduct | N (5′)…T5O4 | N (5′)…G*6O6 | N (5′)…p6O2 | N (3′)…G*7O6 | N (3′)…T8O4 |
|---|---|---|---|---|---|
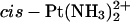 -TGGT -TGGT
|
4.9 (±0.8) | 4.2 (±0.3) | 4.3 (±1.0) | 3.4 (±0.2) | 3.3 (±0.4) |
| Pt(R,R-DAB)2+-TGGT | |||||
| α−γ+ at T5-G*6 | 5.2 (±1.0) | 3.9 (±0.4) | 4.6 (±1.2) | 3.2 (±0.2) | 3.1 (±0.3) |
| DAB equatorial (50%) | 5.4 (±1.0) | 4.0 (±0.3) | 4.6 (±1.3) | 3.3 (±0.2) | 3.1 (±0.2) |
| DAB axial (50%) | 5.0 (±1.0) | 3.8 (±0.4) | 4.6 (±1.1) | 3.2 (±0.2) | 3.1 (±0.3) |
| Pt(R,R-DAB)2+-TGGT | |||||
| αtγt at T5-G*6 | 3.7 (±0.7) | 4.0 (±0.3) | 7.3 (±1.3) | 3.1 (±0.2) | 3.2 (±0.3) |
| DAB equatorial (37%) | 3.8 (±0.7) | 4.1 (±0.3) | 7.2 (±1.2) | 3.2 (±0.2) | 3.1 (±0.2) |
| DAB axial (63%) | 3.6 (±0.7) | 4.0 (±0.3) | 7.4 (±1.4) | 3.1 (±0.1) | 3.2 (±0.4) |
| Pt(S,S-DAB)2+-TGGT | |||||
| α−γ+ at T5-G*6 | 4.7 (±0.8) | 3.9 (±0.4) | 5.1 (±1.3) | 3.3 (±0.2) | 3.2 (±0.3) |
| DAB equatorial (56%) | 5.0 (±0.6) | 4.1 (±0.3) | 4.5 (±1.3) | 3.3 (±0.2) | 3.1 (±0.3) |
| DAB axial (44%) | 4.3 (±0.7) | 3.7 (±0.4) | 5.8 (±0.9) | 3.3 (±0.2) | 3.2 (±0.3) |
| Pt(S,S-DAB)2+-TGGT | |||||
| αtγt at T5-G*6 | 3.4 (±0.4) | 3.7 (±0.4) | 8.9 (±0.9) | 3.2 (±0.2) | 3.2 (±0.3) |
| DAB equatorial (31%) | 3.3 (±0.3) | 3.5 (±0.3) | 9.6 (±0.8) | 3.2 (±0.2) | 3.2 (±0.3) |
| DAB axial (69%) | 3.5 (±0.5) | 3.9 (±0.4) | 8.6 (±0.7) | 3.2 (±0.2) | 3.2 (±0.3) |
Separation values, given in Å, were time-averaged over the MD simulations. Fluctuation values are given in parentheses as standard deviations. The phosphate 5′ to G*6 is labeled p6.
These observations indicate that the previously formulated hypothesis, that the adduct of Pt(R,R-DAB)2+ favors hydrogen bonding to the 3′-guanine whereas that of Pt(S,S-DAB)2+ instead forms a hydrogen bond with the 5′-guanine (Malina et al., 2000, 2002), is probably incorrect. Our simulations suggest that both adducts can form both hydrogen bonds simultaneously. The fact that in the crystal structure of Pt(R,R-DACH)2+, the DACH moiety forms a hydrogen bond only with the 5′-guanine and not with the 3′-guanine is probably due to the considerably smaller kink angle in the solid state. Although the absolute values of the kink angle determined by our force-field calculations may be overestimated, it is very likely that solution structures of Pt-GG adducts are more severely bent than solid-state structures (Ano et al., 1999; Marzilli et al., 2001; Elizondo-Riojas and Kozelka, 2001).
Conformational transitions of the DAB ligands
The five-membered ring of the Pt(DAB)2+ moieties can assume two stable conformations, one with equatorial orientations of the methyl groups and another where these groups are oriented axially. In an isolated [PtL2(DAB)] complex such as [PtCl2(DAB)], the preferred methyl groups orientation is expected to be equatorial. However, in a DNA adduct, interactions with the DNA double helix can affect such conformational equilibria. In fact, in the simulations of the Pt(R,R-DAB)2+and Pt(S,S-DAB)2+ adducts of TGGT, we observe equatorial-axial transitions (Supplementary Material, Fig. S1), suggesting that the energies of the two conformations are similar in the DNA adducts, and that the barrier is not very high. We checked this using DFT (B3LYP/3-21*) calculations on the model compound [Pt(DAB)(NH3)2]2+, for which we found an energy barrier of 8.7 kcal/mol separating the axial and equatorial DAB conformations, the latter being favored by 2.4 kcal/mol [J. Kozelka and O. Delalande, unpublished results]. As can be seen in Table 1, the twist and roll angles of the T5-G*6 step, and the global twist and kink angles are to some extent affected by the equatorial-axial transitions, as expected, since these transitions modify the position of the DAB methyl groups, and thus affect the methyl(DAB)-methyl(T) repulsion. The equatorial-axial transitions have, however, no influence on the average hydrogen-bonding lengths of the DAB ligand (Table 2). This confirms our conclusion that the orientation of the N-H bonds of DAB does not play a significant role in determining the adduct geometry.
The MD simulations indicate that the differential responses of the double helix to the chirality of the Pt(DAB)2+ moiety are characteristic of a TGG sequence
The main conclusion of our MD study is that the different kink and unwinding angles observed previously for the GG adducts of Pt(R,R-DAB)2+and Pt(S,S-DAB)2+ (Malina et al., 2000) are likely to be specific for sequences TGGN (N = any nucleotide). To check this conclusion, we have determined experimentally, in the second part of this work, the kink and unwinding angles of adducts of Pt(R,R-DAB)2+and Pt(S,S-DAB)2+ complexes bound to 20- to 23-bp DNA duplexes similar to those investigated previously (Malina et al., 2000) but having the inner TGGT-ACCA tetranucleotide replaced by CGGA-TCCG. These CGGA-containing adducts were also examined using chemical probes, similarly to the previous study on the TGGT-containing adducts (Malina et al., 2000). In addition, since the adducts of Pt(R,R-DAB)2+and Pt(S,S-DAB)2+ with the TGGT-sequence were previously found to be differentially recognized by both domains of HMGB1, an effect which was attributed to the different kink and/or unwinding angles of the adducts (Malina et al., 2002), we have compared the binding of both HMG domains of HMGB1 to the adducts of Pt(R,R-DAB)2+and Pt(S,S-DAB)2+ formed with the CGGA-containing 20-bp duplex, as described in the following paragraph.
Electrophoretic mobility assays, experiments with chemical probes, and molecular dynamics simulations indicate that adducts of Pt(R,R-DAB)2+ and Pt(S,S-DAB)2+ with the CGGA-sequence perturb the double helix in a way similar to that of cisplatin
As in the previous study (Malina et al., 2000), we used electrophoretic retardation as a quantitative measure of the extent of planar curvature to analyze bending and unwinding induced by the single, site-specific 1,2-GG intrastrand cross-link formed by Pt(R,R-DAB)2+ and Pt(S,S-DAB)2+ in the sequence CGGA. The oligodeoxyribonucleotide duplexes CGGA(20–23) (20–23 bp long, whose sequences were identical or similar to that of the duplex CGGA(21), i.e., 5′-CCTCTCTCCGGATCTCTTCTC/5′-GGAGAAGAGATCCGGAGAGAG; the 20-bp duplex had one terminal C·G pair deleted, whereas one additional T·A pair was added to the 3′ end in the 22-bp duplex and T·A and C·G pairs were added to the 3′ end in the 23-bp duplex) were used for the bending and unwinding studies. The ligation products of these unplatinated or Pt(DAB)2+-containing duplexes were analyzed on native polyacrylamide electrophoresis gel. Experimental details of these studies are given in our recent report (Malina et al., 2000). The results are summarized in Table 3 and compared to those obtained with the TGGT sequence (Malina et al., 2000). The DNA bending of 34 ± 2° toward the major groove and unwinding of 20 ± 2° due to the single, site-specific 1,2-GG intrastrand cross-link formed by Pt(R,R-DAB)2+ or Pt(S,S-DAB)2+ at the CGGA sequence were identical for both enantiomers. The direction of the bend was determined using the 33-bp duplex, which also contained, besides the single 1,2-GG intrastrand cross-link formed by either Pt-DAB enantiomer in the CGGA sequence, the (A·T)5 tract located “in phase” from the cross-link (the cross-linked basepair and the center of the A tract were separated by 11 bp), in the same way as in our recent articles (Kasparkova et al., 2002, 2003; Loskotova and Brabec, 1999).
TABLE 3.
DNA bending and unwinding induced by the 1,2-GG intrastrand cross-link within the CGGA and TGGT contexts
| CGGA
|
TGGT*
|
|||||
|---|---|---|---|---|---|---|
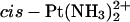 |
Pt(R,R-DAB)2+ | Pt(S,S-DAB)2+ | 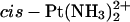 |
Pt(R,R-DAB)2+ | Pt(S,S-DAB)2+ | |
| Bend angle (°)† | 34 | 34 | 34 | 33 | 34 | 24 |
| Unwinding (°) | 19 | 20 | 20 | 15 | 20 | 15 |
Bending and unwinding were determined by the gel electrophoresis retardation assay.
TGGT data are taken from Malina et al. (2000).
Bend angle was toward the major groove.
Further studies focused on analysis of the distortion induced by the 1,2-GG intrastrand cross-link formed by Pt(R,R-DAB)2+ and Pt(S,S-DAB)2+ in the sequence CGGA by chemical probes of DNA conformation. The duplex CGGA(21) (vide supra) containing a single, site-specific cross-link of Pt(R,R-DAB)2+ or Pt(S,S-DAB)2+ was treated with several chemical agents used as tools for monitoring the existence of conformations other than canonical B-DNA. These agents included KMnO4, bromine, or DEPC as the probes of thymine, cytosine, or adenine and guanine residues, respectively. These probes react, under the conditions used, with base residues in single-stranded DNA and distorted double-stranded DNA, but not with the base residues in intact, double-stranded DNA (Bailly et al., 1994; Bailly and Waring, 1997; Brabec et al., 1993; Nielsen, 1990; Ross and Burrows, 1996). For this analysis, we used exactly the same methodology as in our recent studies dealing with DNA adducts of various antitumor platinum drugs so that the details of this experiment can be found in those articles (Brabec et al., 1993; Zehnulova et al., 2001). The results are schematically summarized in Fig. 4 A. The pattern and degree of reactivity toward the chemical probes was identical for the cross-links formed by both enantiomers, indicating a similar character of the conformational distortion. This is in contrast to the results of the analogous experiments carried out with the duplexes containing the cross-links in the sequence TGGT (Malina et al., 2000), where the pattern and degree of reactivity toward the chemical probes was different for the cross-links of the two isomers (Fig. 4 B), indicating a chirality-dependent character of the conformational distortion.
An additional check of the sequence impact was accomplished using molecular modeling. The starting structures that were used for the MD simulations of the adducts of Pt(R,R-DAB)2+ and Pt(S,S-DAB)2+ were mutated so that the platinated TG*G*T sequence became CG*G*A (the new DNA duplex is subsequently designated CGGA). MD simulations identical to those of the TGGT adducts were performed with the two CGGA adducts. Indeed, the structures of both adducts remained similar, with no appreciable differences at the TpG* step. This is demonstrated in Fig. 5, which shows a plot of the twist angles. Obviously, the patterns of twist angles observed in the adducts of TGGT are different, whereas those of the adducts of CGGA are virtually identical.
Neither HMG domain of HMGB1 differentiates between the 1,2-GG intrastrand adducts of Pt(R,R-DAB)2+ and Pt(S,S-DAB)2+ in the sequence CGGA
We demonstrated in our recent work (Malina et al., 2002) that different conformations of the 1,2-GG intrastrand cross-links formed in DNA by Pt(R,R-DAB)2+ and Pt(S,S-DAB)2+ are differently recognized by HMG-domain proteins. The domains A and B of HMGB1 protein (HMGB1a and HMGB1b, respectively) bound to the cross-link of Pt(R,R-DAB)2+ with a considerably higher affinity (∼10 times) than to the cross-link of Pt(S,S-DAB)2+. In these experiments, the 20-bp duplex was modified so that it contained a single, site-specific 1,2-GG intrastrand cross-link of either Pt(R,R-DAB)2+ or Pt(S,S-DAB)2+ formed at a central sequence TGGT in the pyrimidine-rich top strand (shown for HMGB1a in Fig. 6, A and B). Interestingly, the binding affinity of HMG-domain proteins that specifically recognize 1,2-GG intrastrand cross-link to DNA modified by cisplatin is modulated by the nature of the base pairs that immediately flank the platinated d(GpG) site (Dunham and Lippard, 1997). As the different recognition of the 1,2-GG intrastrand cross-links formed in DNA by the two enantiomers Pt(R,R-DAB)2+ or Pt(S,S-DAB)2+ by HMG-domain proteins was observed for the 1,2-GG intrastrand cross-link flanked by thymine residues (Malina et al., 2002), it was of great interest to examine whether HMG-domain proteins also discriminate between the 1,2-GG intrastrand cross-links of these enantiomers if the cross-link is flanked by other bases than two thymines. Hence, in the other experiments we used the 21-bp duplex, which was modified so that it contained a single, site-specific 1,2-GG intrastrand cross-link of either Pt(R,R-DAB)2+ or Pt(S,S-DAB)2+ formed at a central sequence CGGA in the pyrimidine-rich top strand.
HMGB1a and HMGB1b exhibited negligible binding to the nonmodified 21-bp duplex CGGA. As indicated by the presence of a shifted band whose intensity increases with increasing protein concentration, both HMGB1a and HMGB1b recognize the duplex containing the 1,2-GG intrastrand cross-link of either Pt(R,R-DAB)2+ or Pt(S,S-DAB)2+ (shown for HMGB1a in Fig. 6 C). Since only single-shifted bands are seen after incubation of the cross-linked CGGA duplex with either HMGB1a or HMGB1b, detailed titration studies were possible (the results of the titration of this duplex containing the 1,2-GG intrastrand cross-link of either Pt(R,R-DAB)2+ or Pt(S,S-DAB)2+ with HMGB1a are shown in Fig. 6, C and D). These titration data indicate that HMGB1a binds the probes containing the cross-link of both Pt(R,R-DAB)2+ or Pt(S,S-DAB)2+ with a relatively high affinity, which was similar to that of HMGB1a to the same probe, but containing the cross-link of Pt(R,R-DAB)2+ formed in the sequence TGGT (Fig. 6 A and see Malina et al., 2002). Most interestingly, the affinity of the cross-links formed by both enantiomers in the sequence CGGA was identical. The titration of the 22-bp duplex containing the 1,2-GG intrastrand cross-link of Pt(R,R-DAB)2+ or Pt(S,S-DAB)2+ formed at the sequence CGGA with HMGB1b revealed that this protein also binds to the intrastrand cross-link of these platinum compounds with the same affinity although with considerably less affinity (∼30 times) than HMGB1a (not shown).
DISCUSSION
Cross-links between DNA bases and the resulting DNA distortions are believed to be at the origin of the antitumor activity of cisplatin and other platinum complexes (Sherman and Lippard, 1987; Jamieson and Lippard, 1999). An interesting aspect of the structural deformations of DNA caused by each individual class of cross-links is dependence of these distortions on the bases flanking the cross-link. As for the 1,2-GG intrastrand cross-link caused by cisplatin as its most frequent DNA lesion, it has been shown recently that the most characteristic effects—the bending and unwinding of the double helix—are very little affected by the bases flanking the cross-link (Stehlikova et al., 2002).
Recently, Malina et al. (2000) found that the bend and unwinding angles accompanying the 1,2-GG cross-links formed in the sequence context of TGGT by the chiral cisplatin analog [PtCl2(DAB)] depend on the chirality of the asymmetric centers, and that the chirality also affects the recognition of the lesions by either domain of HMGB1 (Malina et al., 2002). In this work, we found that this structural differentiation originates from clashes of the 5′-oriented methyl group of the DAB ligand with the methyl group of the 5′-thymine. This discovery implied that this chiral differentiation should disappear in sequences XGG with X being different from T, and we have verified this prediction using gel electrophoresis measurements on single, site-specific [PtCl2(DAB)] adducts formed in the sequence CGGA of short oligonucleotide duplexes. Moreover, we present evidence that both domains of HMGB1 recognize the Pt(DAB)-GG cross-link within the CGGA sequence with the same affinity (Fig. 6, C and D), indicating that the differential protein recognition of the Pt(DAB)-GG cross-links in the TGGT context is also a result of the clashes between the methyl groups of the DAB ligand and the 5′-thymine.
Our results suggest that all platinum ethylenediamine complexes having a substituent at the carbon centers should form at the TGG sequences adducts whose structure and recognition are different from those of adducts at other XGG sequences. This conclusion is particularly interesting in the context of previous studies on adducts of the anticancer drug oxaliplatin ([Pt(oxalate)(R,R-DACH)], see Fig. 1). This drug is more cytotoxic than its S,S isomer, as concluded from results with the two enantiomers of its dichloro form (Kidani et al., 1978; Noji et al., 1981), and this has been explained by hydrogen bonding between the NH2 groups of the DACH ligand (whose positions and orientations are chirality-dependent) and the O6 atoms of the cross-linked guanines (Malina et al., 2000; Spingler et al., 2001). Since the DAB and DACH ligands are closely related, our finding that the hydrogen bonding from the NH2 groups of DAB are neither the origin of the different geometries observed for the DNA adducts of Pt(R,R-DAB)2+ and Pt(S,S-DAB)2+, nor the cause of their discrimination by HMGB1, probably applies to adducts of Pt(DACH)2+ as well. Our results to be published elsewhere (J. Malina and V. Brabec, unpublished data) indeed confirm that the bend and unwinding angles of the GG adducts of Pt(DACH)2+ at a TGGT sequence are chirality-dependent in a way similar to the Pt(DAB)2+ adducts, whereas at a CGGA sequence the difference disappears.
Finally, this work demonstrates both the power and the limits of molecular dynamics simulations. These simulations allowed us to identify the clashes between the 5′-directed methyl group of DAB and the 5′-thymine, and thus to correct the previous “hydrogen-bonding hypothesis” to explain the structural differences between TGGT-cross-links formed by [PtCl2(R,R-DAB)] and [PtCl2(S,S-DAB)] (Malina et al., 2000). However, it was necessary to use chemical intuition based on previous structural studies of nucleic acids structure to manipulate the MD simulations toward the plausible structural models. Simulations 10–100 times longer would probably have been needed to observe spontaneous transitions toward the αtγt conformation that seems to stabilize the adduct of the R,R enantiomer. Also, energy calculation for the time-averaged structures using the generalized Born approximation to account for solvent effect did not allow us to conclude that the adduct of the R,R-enantiomer favors the α−γ+ → αtγt transition at the TpG step, whereas the adduct of the S,S enantiomer does not. The successful assignment of a structural model to each adduct was therefore based on the comparison between kink and unwinding angles of the models with those determined experimentally. Thus, both computational and experimental methods were needed to explain the structural differences between the GG adducts of [PtCl2(R,R-DAB)] and [PtCl2(S,S-DAB)], and to conclude that they are specific to the TGG sequence.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
The authors acknowledge that their participation in the EC COST Chemistry Action D20 enabled them to exchange regularly the most recent ideas in the field of platinum anticancer drugs with several European colleagues. The authors are indebted to Drs. F. Fanizzi and G. Natile for providing us with samples of [PtCl2(R,R-DAB)] and [PtCl2(S,S-DAB)], as well as to S. Teletchéa and Dr. M.-A. Elizondo-Riojas for technical assistance with the molecular dynamics protocols.
This work was supported by the Association pour la Recherche sur le Cancer (grant P01/3/4297 to J.K. and scholarship to O.D.). This research was also supported by the Grant Agency of the Czech Republic (grant 305/05/2030) and the Grant Agency of the Academy of Sciences of the Czech Republic (grants A5004101 and S5004107).
References
- Ano, S. O., Z. Kuklenyik, and L. G. Marzilli. 1999. Structure and dynamics of Pt anticancer drug adducts from nucleotides to oligonucleotides as revealed by NMR methods. In Cisplatin: Chemistry and Biochemistry of a Leading Anticancer Drug. B. Lippert, editor. Verlag Helvetica Chimica Acta, Zürich. 247–291.
- Bailly, C., D. Gentle, F. Hamy, M. Purcell, and M. J. Waring. 1994. Localized chemical reactivity in DNA associated with the sequence-specific bisintercalation of echinomycin. Biochem. J. 300:165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly, C., and M. J. Waring. 1997. Diethylpyrocarbonate and osmium tetroxide as probes for drug-induced changes in DNA conformation in vitro. In Drug-DNA Interaction Protocols. K. R. Fox, editor. Humana Press, Totowa, NJ. 51–79. [DOI] [PubMed]
- Bellon, S. F., J. H. Coleman, and S. J. Lippard. 1991. DNA unwinding produced by site-specific intrastrand cross-links of the antitumor drug cis-diamminedichloroplatinum(II). Biochemistry. 30:8026–8035. [DOI] [PubMed] [Google Scholar]
- Bellon, S. F., and S. J. Lippard. 1990. Bending studies of DNA site-specifically modified by cisplatin, trans-diamminedichloroplatinum(II) and cis-[Pt(NH3)2(N3-Cytosine)Cl]+. Biophys. Chem. 35:179–188. [DOI] [PubMed] [Google Scholar]
- Brabec, V. 2002. DNA modifications by antitumor platinum and ruthenium compounds: their recognition and repair. Prog. Nucleic Acid Res. Mol. Biol. 71:1–68. [DOI] [PubMed] [Google Scholar]
- Brabec, V., J. Reedijk, and M. Leng. 1992. Sequence-dependent distortions induced in DNA by monofunctional platinum(II) binding. Biochemistry. 31:12397–12402. [DOI] [PubMed] [Google Scholar]
- Brabec, V., M. Sip, and M. Leng. 1993. DNA conformational distortion produced by site-specific interstrand cross-link of trans-diamminedichloroplatinum(II). Biochemistry. 32:11676–11681. [DOI] [PubMed] [Google Scholar]
- Case, D. A., D. A. Pearlman, J. W. Caldwell, T. E. Cheatham III, W. S. Ross, C. L. Simmerling, T. A. Darden, K. M. Merz, Jr., R. V. Stanton, A. L. Cheng, J. J. Vincent, M. Crowley, V. Tsui, R. J. Radmer, Y. Duan, J. Pitera, I. Massova, G. L. Seibel, U. C. Singh, P. K. Weiner, and P. A. Kollman. 1999. AMBER 6. University of California, San Francisco.
- Cheatham III, T. E., P. Cieplak, and P. A. Kollman. 1999. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 16:845–862. [DOI] [PubMed] [Google Scholar]
- Dalke, A., W. Humphrey, and J. Ulrich. 1997. VMD (Visual Molecular Dynamics), Theoretical Biophysics Group, University of Illinois and Beckman Institute, Urbana, IL.
- Darden, T. A., D. York, and L. G. Pedersen. 1993. Particle mesh Ewald: an Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. [Google Scholar]
- Djuranovic, D., and B. Hartmann. 2003. Conformational characteristics and correlations in crystal structures of nucleic acid oligonucleotides: evidence for sub-states. J. Biomol. Struct. Dyn. 20:771–788. [DOI] [PubMed] [Google Scholar]
- Dunham, S. U., and S. J. Lippard. 1997. DNA sequence context and protein composition modulate HMG-domain protein recognition of cisplatin-modified DNA. Biochemistry. 36:11428–11436. [DOI] [PubMed] [Google Scholar]
- Elizondo-Riojas, M.-A., F. Gonnet, P. Augé-Barrere-Mazouat, F. Allain, J. Bergès, R. Attias, J.-C. Chottard, and J. Kozelka. 1997. Molecular modeling of platinum complexes with oligonucleotides: Methodological lessons and structural insights. In Molecular Modeling and Dynamics of Bioinorganic Systems. L. Banci and P. Comba, editors. Kluwer Academic, Dordrecht, The Netherlands. 131–160.
- Elizondo-Riojas, M.-A., and J. Kozelka. 2001. Unrestrained 5 ns molecular dynamics simulation of a cisplatin-DNA 1,2-GG adduct provides a rationale for the NMR features and reveals increased conformational flexibility at the platinum binding site. J. Mol. Biol. 314:1227–1243. [DOI] [PubMed] [Google Scholar]
- Essman, U., L. Perera, M. Berkowitz, T. A. Darden, H. Lee, and L. G. Pedersen. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103:8577–8593. [Google Scholar]
- Fanizzi, F. P., F. P. Intini, G. Maresca, G. Natile, R. Quaranta, M. Coluccia, L. Di Bari, D. Giordano, and M. A. Mariggió. 1987. Biological activity of platinum complexes containing chiral centers on the nitrogen or carbon atoms of a chelate diamine ring. Inorg. Chim. Acta. 137:45–51. [Google Scholar]
- Frisch, M. J., G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, V. G. Zakrzewski, J. J. A. Montgomery, R. E. Stratmann, J. C. Burant, S. Dapprich, J. M. Millam, A. D. Daniels, and others. 1998. Gaussian 98. Gaussian, Inc., Pittsburgh, PA.
- Gelasco, A., and S. J. Lippard. 1998. NMR solution structure of a DNA dodecamer duplex containing a cis-diammineplatinum(II) d(GpG) intrastrand cross-link, the major adduct of the anticancer drug cisplatin. Biochemistry. 37:9230–9239. [DOI] [PubMed] [Google Scholar]
- Herman, F., J. Kozelka, V. Stoven, E. Guittet, J.-P. Girault, T. Huynh-Dinh, J. Igolen, J.-Y. Lallemand, and J.-C. Chottard. 1990. A d(GpG)-platinated decanucleotide duplex is kinked. An extended NMR and molecular mechanics study. Eur. J. Biochem. 194:119–133. [DOI] [PubMed] [Google Scholar]
- Jamieson, J. R., and S. J. Lippard. 1999. Structure, recognition, and processing of cisplatin-DNA adducts. Chem. Rev. 99:2467–2497. [DOI] [PubMed] [Google Scholar]
- Kasparkova, J., K. J. Mellish, Y. Qu, V. Brabec, and N. Farrell. 1996. Site-specific d(GpG) intrastrand cross-links formed by dinuclear platinum complexes. Bending and NMR studies. Biochemistry. 35:16705–16713. [DOI] [PubMed] [Google Scholar]
- Kasparkova, J., O. Novakova, N. Farrell, and V. Brabec. 2003. DNA binding by antitumor trans-[PtCl2(NH3)(thiazole)]. Protein recognition and nucleotide excision repair of monofunctional adducts. Biochemistry. 42:792–800. [DOI] [PubMed] [Google Scholar]
- Kasparkova, J., J. Zehnulova, N. Farrell, and V. Brabec. 2002. DNA interstrand cross-links of the novel antitumor trinuclear platinum complex BBR3464. Conformation, recognition by high mobility group domain proteins, and nucleotide excision repair. J. Biol. Chem. 277:48076–48086. [DOI] [PubMed] [Google Scholar]
- Kidani, Y., K. Inagaki, M. Iigo, A. Hoshi, and K. Kuretani. 1978. Antitumor activity of 1,2-diamminocyclohexane-platinum complexes against Sarcoma 180 ascites form. J. Med. Chem. 21:1315–1318. [DOI] [PubMed] [Google Scholar]
- Lavery, R., and H. Sklenar. 1988. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 6:63–91. [DOI] [PubMed] [Google Scholar]
- Lavery, R., and H. Sklenar. 1989. Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn. 6:655–667. [DOI] [PubMed] [Google Scholar]
- Lohman, T. M., and D. P. Mascotti. 1992. Thermodynamics of ligand-nucleic acid interactions. Methods Enzymol. 212:400–424. [DOI] [PubMed] [Google Scholar]
- Loskotova, H., and V. Brabec. 1999. DNA interactions of cisplatin tethered to the DNA minor groove binder distamycin. Eur. J. Biochem. 266:392–402. [DOI] [PubMed] [Google Scholar]
- Malina, J., C. Hofr, L. Maresca, G. Natile, and V. Brabec. 2000. DNA interactions of antitumor cisplatin analogs containing enantiomeric amine ligands. Biophys. J. 78:2008–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malina, J., J. Kasparkova, G. Natile, and V. Brabec. 2002. Recognition of major DNA adducts of enantiomeric cisplatin analogs by HMG box proteins and nucleotide excision repair of these adducts. Chem. Biol. 9:629–638. [DOI] [PubMed] [Google Scholar]
- Marzilli, L. G., J. S. Saad, Z. Kuklenyik, K. A. Keating, and Y. Xu. 2001. Relationship of solution and protein-bound structures of DNA duplexes with the major intrastrand cross-link lesions formed on cisplatin binding to DNA. J. Am. Chem. Soc. 123:2764–2770. [DOI] [PubMed] [Google Scholar]
- Merrit, E. A., and D. J. Bacon. 1997. Raster3D-photorealistic molecular graphics. Methods Enzymol. 277:505–524. [DOI] [PubMed] [Google Scholar]
- Nielsen, P. E. 1990. Chemical and photochemical probing of DNA complexes. J. Mol. Recognit. 3:1–24. [DOI] [PubMed] [Google Scholar]
- Noji, M., K. Okamoto, Y. Kidani, and T. Tashiro. 1981. Relation of conformation to antitumor activity of platinum(II) complexes of 1,2-cyclohexanediamine and 2-(aminomethyl)cyclohexyl- amine isomers against leukemia P388. J. Med. Chem. 24:508–514. [DOI] [PubMed] [Google Scholar]
- Pil, P. M., and S. J. Lippard. 1992. Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science. 256:234–237. [DOI] [PubMed] [Google Scholar]
- Raymond, E., S. Faivre, S. Chaney, J. Woynarowski, and E. Cvitkovic. 2002. Cellular and molecular pharmacology of oxaliplatin. Mol. Cancer Therap. 1:227–235. [PubMed] [Google Scholar]
- Ross, S. A., and C. J. Burrows. 1996. Cytosine-specific chemical probing of DNA using bromide and monoperoxysulfate. Nucleic Acids Res. 24:5062–5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert, J. P., G. Ciccotti, and H. J. C. Berendsen. 1977. Numerical integraton of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alcanes. J. Comput. Phys. 23:327–341. [Google Scholar]
- Saris, C. P., P. J. M. van de Vaart, R. C. Rietbroek, and F. A. Blommaert. 1996. In vitro formation of DNA adducts by cisplatin, lobaplatin and oxaliplatin in calf thymus DNA in solution and in cultured human cells. Carcinogenesis. 17:2763–2769. [DOI] [PubMed] [Google Scholar]
- Sherman, S. E., D. Gibson, A. H. J. Wang, and S. J. Lippard. 1985. X-ray structure of the major adduct of the anticancer drug cisplatin with DNA: cis-[Pt(NH3)2{d(pGpG)}]. Science. 230:412–417 [DOI] [PubMed] [Google Scholar]
- Sherman, S. E., and S. J. Lippard. 1987. Structural aspects of platinum anticancer drug interactions with DNA. Chem. Rev. 87:1153–1181. [Google Scholar]
- Spingler, B., D. A. Whittington, and S. J. Lippard. 2001. 2.4 A crystal structure of an oxaliplatin 1,2-d(GpG) intrastrand cross-link in a DNA dodecamer duplex. Inorg. Chem. 40:5596–5602. [DOI] [PubMed] [Google Scholar]
- Stehlikova, K., H. Kostrhunova, J. Kasparkova, and V. Brabec. 2002. DNA bending and unwinding due to the major 1,2-GG intrastrand cross-link formed by antitumor cis-diamminedichloroplatinum(II) are flanking-base independent. Nucleic Acids Res. 30:2894–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stros, J. 2001. Two mutations of basic residues within the N-terminus of HMG-1 B domain with different effects on DNA supercoiling and binding to bent DNA. Biochemistry. 40:4769–4779. [DOI] [PubMed] [Google Scholar]
- Takahara, P. M., A. C. Rosenzweig, C. A. Frederick, and S. J. Lippard. 1995. Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin. Nature. 377:649–652. [DOI] [PubMed] [Google Scholar]
- Vaisman, A., S. E. Lim, S. M. Patrick, W. C. Copeland, D. C. Hinkle, J. J. Turchi, and S. G. Chaney. 1999. Effect of DNA polymerases and high mobility group protein 1 on the carrier ligand specificity for translesion synthesis past platinum-DNA adducts. Biochemistry. 38:11026–11039. [DOI] [PubMed] [Google Scholar]
- Varnai, P., D. Djuranovic, R. Lavery, and B. Hartmann. 2002. α/γ Transitions in the B-DNA backbone. Nucleic Acids Res. 30:5398–5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, M., S. M. Cohen, A. P. Silverman, and S. J. Lippard. 2001. Effects of spectator ligands on the specific recognition of intrastrand platinum-DNA cross-links by high mobility group box and TATA-binding proteins. J. Biol. Chem. 276:38774–38780. [DOI] [PubMed] [Google Scholar]
- Woynarowski, J. M., S. Faivre, M. C. S. Herzig, B. Arnett, W. G. Chapman, A. V. Trevino, E. Raymond, S. G. Chaney, A. Vaisman, M. Varchenko, and P. E. Juniewicz. 2000. Oxaliplatin-induced damage of cullular DNA. Mol. Pharmacol. 58:920–927. [DOI] [PubMed] [Google Scholar]
- Zamble, D. B., and S. J. Lippard. 1999. The response of cellular proteins to cisplatin-damaged DNA. In Cisplatin: Chemistry and Biochemistry of a Leading Anticancer Drug. B. Lippert, editor. Verlag Helvetica Chimica Acta, Zürich. 73–110.
- Zehnulova, J., J. Kasparkova, N. Farrell, and V. Brabec. 2001. Conformation, recognition by high mobility group domain proteins, and nucleotide excision repair of DNA intrastrand cross-links of novel antitumor trinuclear platinum complex BBR3464. J. Biol. Chem. 276:22191–22199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.