Abstract
Although the mechanical behavior of tendon and bone has been studied for decades, there is still relatively little understanding of the molecular basis for their specific properties. Thus, despite consisting structurally of the same type I collagen, bones and tendons have evolved to fulfill quite different functions in living organisms. In an attempt to understand the links between the mechanical properties of these collageneous structures at the macro- and nanoscale, we studied trimeric type I tropocollagen molecules by atomic force microscopy, both topologically and by force spectroscopy. High-resolution imaging demonstrated a mean (± SD) contour length of (287 ± 35) nm and height of (0.21 ± 0.03) nm. Submolecular features, namely the coil-pitch of the molecule, were also observed, appearing as a repeat pattern along the length of the molecule, with a length of ∼8 nm that is comparable to the theoretical value. Using force spectroscopy, we established the stretching pattern of the molecule, where both the mechanical response of the molecule and pull-off peak are convoluted in a single feature. By interpreting this response with a wormlike chain model, we extracted the value of the effective contour length of the molecule at (202 ± 5) nm. This value was smaller than that given by direct measurement, suggesting that the entire molecule was not being stretched during the force measurements; this is likely to be related to the absence of covalent binding between probe, sample, and substrate in our experimental procedure.
INTRODUCTION
Connective tissues in animals are the ensemble of a varied cell population distributed among protein fibers, embedded in a viscous ground substance, the extracellular matrix. The role of these connective tissues is not only to bind cells together, but also influences their behavior, development, and polarity, both directly and via the host of growth and regulatory factors that are concentrated within the matrix. Three main types of extracellular fibers can be identified in connective tissue: collagen, reticular, and elastic. One of their common characteristics is that they are formed indirectly by fibroblasts, the cells generating protein subunits, which later interact with each other within the matrix to form mature fibers. Among these, collagen is the major structural protein and is the most abundant in the human body. The collagen molecule consists of three left-handed coiled polypeptide chains, two α1-chains and one α2-chain intertwined in an overall right-handed coil, tropocollagen (Ramachandra and Karthan, 1955). The basic unit of each of the polypeptide chains consists of the repeating sequence, Glycine-X-Y, where X is often proline and Y hydroxyproline. It is the frequent occurrence of proline and hydroxyproline groups in the polypeptide chain that causes the collagen α-chains to be tightly packed into a triple helical conformation. Several collagen-related diseases occur in humans, osteoporosis and tendinitis being common examples. Both are linked to the mechanical properties of collagen and its higher order, structurally related forms: collagen fibrils and fibers, and the macroscopic tissues, tendon, and bone.
There have been many studies of collagen fibers using a range of experimental techniques, but more recently atomic force microscopy (AFM) has been used to study this protein under physiological conditions, unattainable by precursor techniques such as x-ray diffraction crystallography or transmission electron microscopy (TEM). The ability of AFM to image as well as to perform force measurements has enabled experimentalists to compare results obtained using molecular scale samples to those obtained at the macroscale using bulk techniques. Early studies involving AFM and collagen fibers were initially carried out by Chernoff et al. (1992), who imaged collagen fibers under dry conditions but were unsuccessful in observing the characteristic D-banding as initially proposed by Schmitt and co-workers in 1942 (Schmitt et al., 1942) based on TEM. This work was later pursued successfully by Baselt et al. (1993) and Revenko et al. (1994), who presented AFM images of collagen fibers displaying the characteristic D-banding pattern. Recently, Gutsmann et al. (2003) published a detailed account of both topological and mechanical behavior of the collagen fibers, isolated from rat tail tendon. One result supported the view that the fibers had the topology of a hollow tube and they suggested that the collagen fiber is composed of a hard shell with a less dense core, giving both flexibility and elasticity to the fiber. In a later publication, Gutsmann et al. (2004) presented some spectroscopic force measurements performed on rat tail collagen fibers. The aim of that study was to link the stretching pattern of the fiber with the D-banding pattern of the fiber, initially proposed by Schmitt et al. (1942). The stretching pattern that was obtained while pulling on the collagen fiber proved to be too complex to identify single characteristic events. Nevertheless, two features were identified with ruptures at 22 nm and 78 nm; these were hypothesized as being related to a possible repeat in the fiber structure, though all of these were not identified by AFM imaging.
There have been few investigations of collagen at the level of the single triple helical tropocollagen molecule structure using AFM. Mertig et al. (1997) presented an analysis of an in vitro generated two-dimensional collagen network formation. In their studies, they managed to isolate single collagen monomers that were prepared on a mica surface and observed that the monomer did not undergo denaturation as it bound the mica surface. More recently, Sun et al. (2002) used optical tweezers to characterize the mechanical behavior of collagen, but more specifically its flexibility. In their study, they used procollagen, the precursor form of the collagen monomer. Procollagen has cysteine groups at both its N- and C-termini, which enables covalent binding to the surface-probe or beads, though not with controlled orientation. They found that the monomer had a persistence length of 14.5 nm and a contour length of 309 nm.
The aims of our study were to use AFM to investigate type I collagen at the single molecule level to understand the fundamental mechanical behavior of this protein, and to correlate these findings with high-resolution topography imaging.
MATERIAL AND METHODS
Materials
In all experiments, purified soluble type I collagen monomer solution was used from a stock solution: 1 mg/ml in 0.1 M acetic acid (type I rat tail collagen, Sigma-Aldrich, Gillingham, UK) with the stock solution being kept at 4°C until use. Depending on the experiment performed, 1:100 or 1:1000 dilutions in phosphate buffer saline (PBS) were performed with the resulting solutions maintained at 4°C on ice during subsequent preparations.
Imaging collagen by AFM
For single molecule imaging, a droplet 40 μl of the 1:1000 solution was deposited on a mica substrate (mica disks; Agar Scientific, Stansted, UK) that had been freshly cleaved. The solution was left to incubate for 10 min, avoiding droplet evaporation by keeping a water droplet at the side of the sample but not in contact so as to avoid sample dilution. The sample was then rinsed, firstly using PBS and then using ultra high quality (UHQ) water to avoid any salt crystal formation. Finally the sample was dried using a gentle stream of dry N2. The imaging experiments were carried out using a Multimode-Nanoscope IV (Veeco, Santa Barbara, CA), equipped with an E scanner and NSC tips-D lever (MikroMasch, Tallinn, Estonia), with the following characteristics in air: ∼28 kHz resonant frequency and ∼0.35 N/m nominal spring constant. The samples were imaged in dry conditions in tapping mode, with the minimum amplitude set point selected to avoid either damaging or altering the sample on the surface. To do so, the probe was brought into contact with a false-engagement (the probe touches the surface and immediately retracts but remains very close), and then the amplitude set point was slowly reduced until the probe remade contact with the surface. This ensures that there is the minimal force applied by the probe onto the sample.
Single molecule pulling experiments
For single molecule force measurements, a single droplet of 80 μl of the 1/100 solution was deposited on a gold-coated glass slide, previously stored in liquid nitrogen. The solution was left to incubate for 5 min, avoiding evaporation. The sample was then gently rinsed using PBS. Finally a droplet of 150 μl PBS was deposited on the sample, before starting the measurements. Force measurements were performed on two different instruments: a Multimode-Nanoscope IV with a Picoforce attachment (Veeco) and a Molecular Force Puller, MFP1 (Asylum Research, Santa Barbara, CA). For consistency, the same type of probe was used on both the instruments during all of the experiments: Microlever-D tips lever (MikroMasch) with the following characteristics in buffer: ∼3.5 kHz resonant frequency and ∼0.03 N/m nominal spring constant. The spring constant of the levers was calibrated by performing a thermal tune of the lever in buffer conditions (fitting of the Brownian motion of the lever) (Hutter and Bechhoefer, 1993; Walters et al., 1996). Typical values of the spring constant varied from 0.030 N/m to 0.055 N/m, but the resonant frequency was consistently Fres= 3.5 kHz. The deflection sensitivity of the detector was calibrated by performing a force curve on a bare mica substrate in dry conditions. During the force measurement cycles, a typical load of <5 nN was applied at a constant loading rate of 1.8 μm/s. Series of 100 curves were recorded at a single location on the sample surface, before the probe was moved to a new location with up to 1000 force distance curves being accrued in each experiment. Finally, after the experiment, the samples were imaged using a new probe to evaluate surface coverage by collagen.
The resulting force-distance curves were subsequently analyzed using all the force curves that showed a stretching event and were fitted with the wormlike chain elasticity model (Bustamante et al., 1994) to determine the contour length of the molecule. Once the contour length had been established a frequency plot of the entire data set is produced, to establish the mean of the distribution (Lo: effective contour length). All numerical data are quoted as the mean ± SD of the data set.
RESULTS AND DISCUSSION
Single molecule imaging
Surface coverage of the substrate
The ideal approach to the study of mature tropocollagen would be to use covalent binding to a coated surface, as performed in related experiments (Rounsevell et al., 2004). However, producing recombinant collagen in eukaryotic cells with a defined sequence modification is difficult and collagen, being insoluble after enzymatic cleavage of procollagen, cannot easily be chemically modified. Thus, we took the simple approach of binding the collagen to the analysis surface noncovalently by physisorption. Since the strength of this interaction via van der Waals forces is weak, desorption of the adsorbate can easily occur in a buffer environment due to ionic screening lessening binding and to the substantial, though controlled, forces applied to the molecules during the AFM scanning process. This means that imaging collagen monomers in buffer solution in the absence of covalent linkage to the substrate was not possible; nevertheless, results were obtained when collagen was imaged under dry conditions.
The first step toward successful imaging of collagen monomers involved the deposition of collagen onto the mica surface with a low surface coverage to single out individual monomers. This implied adjusting both the concentration of the solution used as well as the corresponding incubation times. Fig. 1 presents two images obtained from solutions that were, respectively: a), 1 μg/ml for an incubation time of 10 min; b), 10 μg/ml for an incubation time of 5min; and c), 1 μg/ml for an incubation time of 10 min, the solution left to rest at room temperature for 8 h before incubation.
FIGURE 1.
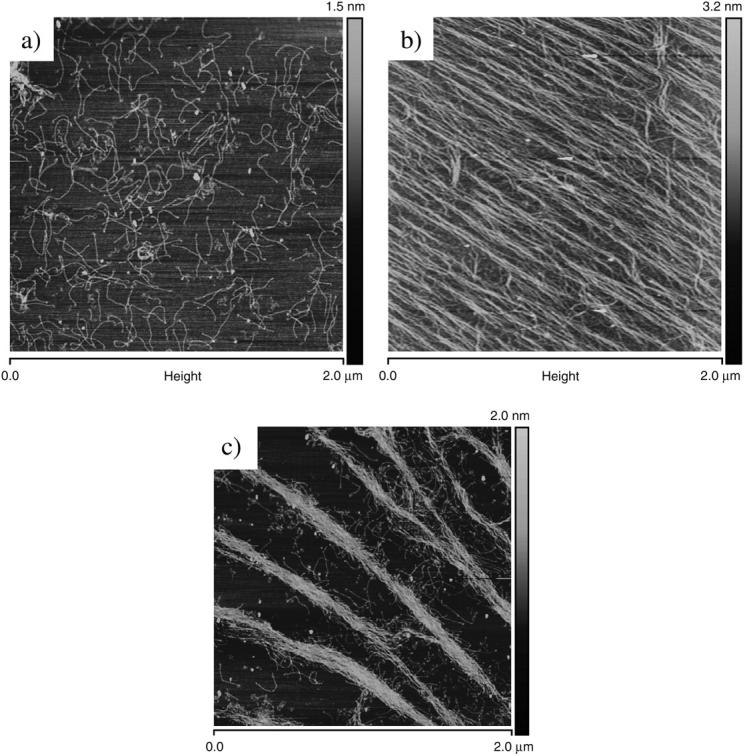
Topographical images (height; tapping mode in air) of type I collagen monomers on mica substrate: (a) low surface coverage, 1 μg/ml solution; (b) high surface coverage, 10 μg/ml solution; and (c) 1 μg/ml for an incubation time of 10 min, solution left at room temperature for 8 h before incubation.
At low surface coverage concentration, collagen monomers are distributed randomly on the mica surface (Fig. 1 a); aggregates were not seen and no particular molecular orientation observed. At much higher concentration, the collagen monomers are densely packed and seem to follow a generic orientation (Fig. 1 b). This overall orientation was found to be consistent over a very large area of the sample, as images could be recorded with offset of a few millimeters and still present the same pattern and orientation. It could be suggested that this effect is not due to the steric forces occurring during the rinsing (no flow was used), as successive sample preparation implying a rinsing in different directions was not linked to this overall orientation. Furthermore, the orientation of the collagen monomers in Fig. 1 b was not related either to the scanning direction of the AFM probe, as different scanning angles did not alter the orientation. Nevertheless, a drying orientation could occur as a result of the recession of the solvent base. Considering the in vivo process of collagen molecular aggregation, another possible explanation for the formation of this network would be that they are formed by electrostatic interactions between side chains exposed to solvent and available for intermolecular interactions. As there is no intrinsic dipole moment in the triple helix (Bella et al., 1994, 1995). Though the mechanism by which collagen molecules aggregates to form a fibrillar network is still not well understood, the presence of the orientation could be the first step toward molecular aggregation and a possible fibrillogenesis. For reference, fibrillogenesis usually takes place under low pH (∼4.2) and low salt concentrations, resulting in the formation of collagen fibers with the characteristic D-banding (Christiansen et al., 2000). Jiang et al. (2004), have recently published similar results demonstrating that the assembly of collagen monomers into ultrathin highly anisotropic ribbon structures is dependent on the pH of the buffer. In this publication, Jiang and his co-workers observed very similar patterns to that presented in Fig. 1 b, when the pH of the buffer was 7.5. This result was confirmed by considering Fig. 1 c. In this case, a low-concentration solution of collagen molecules was left to rest for a period of 8 h at room temperature. The resulting preparation enables molecular aggregation to occur. In this figure, a general orientation of the molecule is again observed but this time, the molecules have formed ribbons or prefibrillar structures which would support a case for fibrillogenesis to occur under defined conditions (Jiang et al., 2004).
Topographic features of collagen monomers
Topographic features of single type I collagen monomers, as presented in Fig. 1 a, were analyzed and found to have the following characteristics (Table 1). First, the contour length of the monomer, 287 ± 35 nm, corresponds to the value quoted in literature from collagen monomers with comparable numbers of residues. Second, the height of the monomer is smaller, 0.21 ± 0.03 nm, than the theoretical radius of the monomeric collagen triple helix (∼1.5 nm), whereas the measured width is much larger than predicted, 8.3 ± 0.7 nm (note that no probe deconvolution was performed). This deviation from predicted dimensions is commonly found and has been reported widely for DNA (Fritz et al., 1995; Schabert and Rabe, 1996). In our study, collagen was imaged in a dry environment where the molecule tends to collapse onto a surface, due to either the load applied by the mode of imaging (tapping mode in this case) or the binding forces between the monomer and substrate (stronger van der Waals forces).
TABLE 1.
Topographic analysis of type I collagen by direct AFM measurement
| Type1 collagen monomer | Values ± SD | N |
|---|---|---|
| Length (nm) | 287 ± 35 | 20 |
| Height (nm) | 0.21 ± 0.03 | 20 |
| Width (nm) | 8.3 ± 0.7 | 20 |
| Substrate roughness (nm) | <0.01 (mica) | — |
Note: Data were derived from images in which probe deconvolution was not performed.
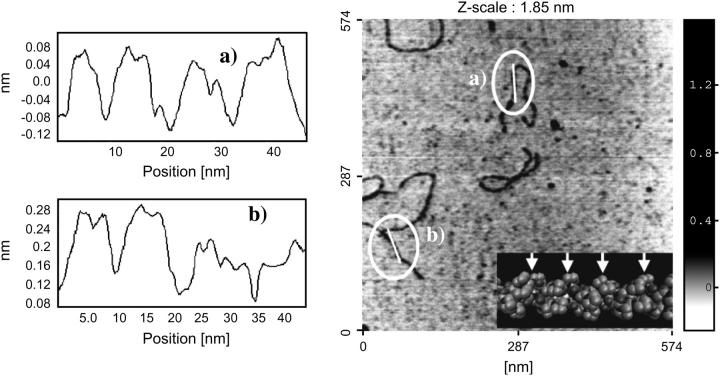
It was also noted that there was a repetitive pattern observable along the length the collagen monomer; Fig. 2 shows examples of line scans along the long axis of collagen. The line scans exhibit a series of peaks occurring regularly with a consistent width: ∼8 nm (n = 20). A possible explanation for this repeat pattern could be that the probe was following the coil-pitch of the collagen molecule. Indeed, the theoretical value of this coil-pitch is known to be 85.5 Å from x-ray analysis (Beck and Brodsky, 1998). However these results should be treated with some caution as similar experiments, carried out on DNA to study the coil-pitch of the double strand, have resulted in observation of apparent axial features (Hansma and Hansma, 1993; Han et al., 1995; Zhang et al., 1996) though proof that they relate to true molecular features is lacking. It is known that the AFM can generate probe-sample artifacts that would convolute real features of the sample with the end of the tip itself (typically 10 nm for tips used in this experiment) resulting in a false impression of the features in either width and/or length (Morris et al., 1999). This tends to occur when the repeat pattern has similar dimensions to that the tip itself. In our experiments, measurements of the coil-pitch induced repeats were taken in directions perpendicular and parallel to the scanning axis, which suggest that these observed repeats could be real surface features on the collagen molecule.
FIGURE 2.
(Right) Topographical image (height; tapping mode in air) of type I collagen monomers. The insert is a three-dimensional representation of the collagen monomer itself, with the arrows marking the coil-pitch of the three left-handed coils. (Left) example line-scan spectra obtained for locations a and b, respectively, from the right-hand side image.
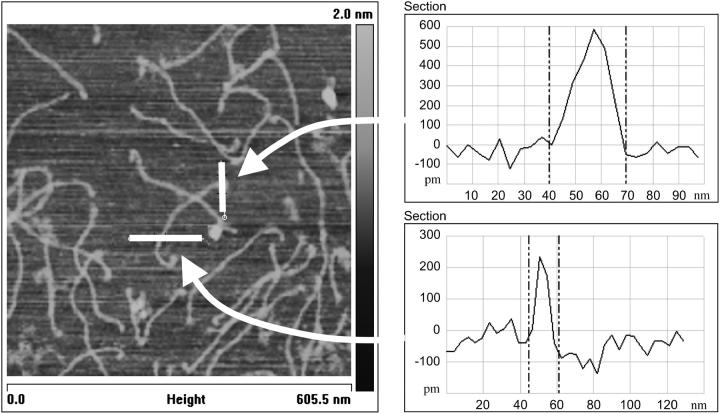
Finally, some of monomers (<10%) show a much wider end when compared to the rest of the molecule, as presented in Fig. 3. The increase in width between a middle section of the monomer and its end can be up to threefold. One reason for this occurrence may be the way that the collagen monomer is produced. Type I collagen monomers originate from procollagen; after formation by cells of the procollagen triple helix, proteolytic enzyme cleavage occurs to enable collagen molecules to assemble into fibers (Olsen, 1991), and, during this process, the amino and the carboxylic termini are cleaved, leaving the triple helix uncapped at both its ends. Therefore, it is possible that the triple helix could fray, resulting in a much wider end of the monomer, or it could be simply that the free end of the monomer refolds back on itself. Recent x-ray fiber diffraction experiments performed on the axial structure of the C-terminal telopeptide region (Orgel et al., 2000) supports this latter possibility; these showed the occurrence of a hairpin conformation with the C-terminus folded back onto the triple helix.
FIGURE 3.
(Left) Topographical image (height; tapping mode in air) of type I collagen monomers. The white marker lines are the location where the width of the monomer was measured and presented on the right-hand side. (Right) Line-scan spectra obtained from cross section of the monomer.
Single molecule mechanics
Stretching pattern of tropocollagen
Having successfully imaged and characterized the topology of type I collagen monomers, it was then possible to investigate their mechanical properties. To do so, Force (F) versus extension curves (z) were recorded on a sample of type I collagen monomers deposited onto glass freshly coated with gold (Rounsevell et al., 2004). The protein concentration and incubation times were those that were previously described (Fig. 1 a) and the force measurements were performed in PBS buffer (pH 7.4).
Fig. 4 a, shows a typical force versus distance curve with a complex structure containing features similar to that of those that Gutsmann et al. (2004) reported when studying the stretching pattern of macromolecular complexes of collagen (i.e., fibers consisting of several thousands of molecules) originating from rat tail tendon.
FIGURE 4.
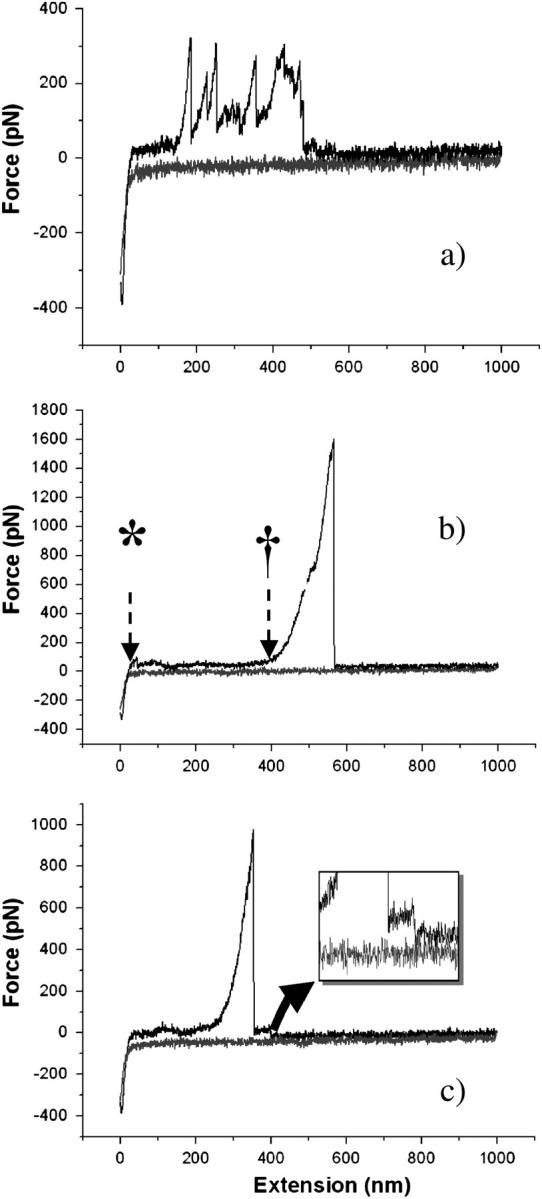
Force-distance curves obtained by pulling on type I collagen monomers. (a) Stretching pattern obtained when several monomers bind to the tip. (b) Stretching pattern obtained when a single monomer binds to the tip. Marker (*) indicates when the tip leaves the sample surface. Marker (†) indicates when the molecule starts being stretched. The lack of deflection between the two markers indicates that the monomer is being straightened with such a low force that the cantilever cannot detect it. (c) Stretching pattern presenting a plateau after the pull-off indicating the desorption or peel-off of the molecule from the surface. (Inset) Zoom on plateau after the pull-off.
In most cases, the force curves present a reproducible stretching pattern: the stretching peak occurs once the AFM probe has been retracted to a given distance rather than as soon as the probe leaves the sample surface (asterisk, Fig. 4 b). During this phase, there is little or no force exerted by the probe onto the monomer as no deflection can be detected. Thus, one may assume that the monomer undergoes a straightening during this phase that requires forces that are too weak to be detected with the current AFM setup. Once the monomer is fully extended, a load begins to be applied by the probe and the molecule starts to be stretched (dagger, Fig. 4 b). After the stretching phase, snap-off of the molecule from the probe or of the molecule from the substrate occurs.
To understand fully the mechanical properties of collagen, it is essential to know whether a single monomer or a series of monomers are being stretched at the same time. In common with the majority of AFM “pulling” experiments no specific covalent coupling between the AFM tip and collagen was used in our studies; thus there is no control over the binding process and hence no guarantee that single molecules are being examined. Nevertheless, by considering the stretching pattern of repeated experiments, one can differentiate between single and multiple stretching events. As mentioned earlier, two overall types of stretching pattern are observed: a unique stretching peak similar to those observed by Sun et al. (2002) or a complex series of stretching peaks as observed by Gutsmann et al. (2004). Thus, comparing our results with the above, one can consider a single stretching peak as being the mechanical response of a single monomer and a multiple stretching peak pattern to be the convoluted response of multiple monomers attached onto the probe.
Substrate binding and contour length
To understand, the mechanical behavior of the molecule, several parameters have to be assessed. First, it was noticed that the binding affinity of the collagen to the AFM tip was high compared to similar single molecule force measurement experiments. Typically, stretching events were detected in >26% of the force distance curves (picoforce, 26.3%; MFP, 27.4%).
A second point, as illustrated by Fig. 4 c, involves sample desorption. This phenomenon occurs when a plateau appears in the retraction curve after the pull-off of the probe (Conti et al., 2001). In our experiments, this was found to be uncommon (<1%) suggesting that seldom desorption of the molecule from either the tip or the substrate occurs during the stretching process.
To explain the mechanical behavior of the monomer, as expressed by the peaks present in the force-distance curves illustrated in Fig. 4, it was necessary to analyze them quantitatively using a classical entropic treatment such as the wormlike chain (WLC) model (Bustamante et al., 1994). Similar studies involving the characterization of the mechanical properties of DNA (Baumann et al., 1997; Wang et al., 1997) and titin (Rief et al., 1997; Tskhovrebova et al., 1997) suggested that this approach could also be applied to the collagen molecule.
The WLC model is often used to describe the mechanical behavior of a protein as it is being stretched or extended, by establishing a relation between the extension of the protein itself and the entropic force induced by the extension. The WLC can be written as follows:
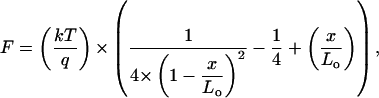 |
where F is the entropic force, x the extension, q the persistence length, and Lo is the contour length of the monomer. This model was successfully applied to all data that showed a stretching pattern, as presented in Fig. 5. The resulting values of the contour lengths were then plotted in a frequency plot and the most probable contour length was then extracted. Fig. 6 presents the frequency plots obtained after fitting data obtained in two experiments involving the same sample but performed on different instruments (see Materials and Methods). The resulting frequency plots were then fitted with a Gaussian distribution to define the mode of the distribution, thus giving the value of the contour length (Fig. 6). In our experimental set, the value of the contour length obtained after the Gaussian fitting are, respectively: Lo(a) = (206.5 ± 8.0) nm and Lo(b) = (198.3 ± 5.6) nm; a and b subscript referring, respectively, to the results obtained by the picoforce and MFP. The values of contour length obtained by force measurements are smaller than the topologically measured contour length of the monomer, which is: Lo(topo) = (287 ± 35) nm (vide supra). If the monomer had been covalently bound to the both the substrate and the tip, it would be expected that fitting of the WLC model to the force-distance curves would give the contour length of the entire molecule and any associated chemical linkers, provided that the monomer was elastic over its entire length. The frequency plots also present a tail in the region where the contour length is larger than the length of the monomer itself. This can be understood by the fact that several monomers may have bound together and the ensemble was being stretched by the probe. During a force measurement, the monomer is attached to the probe and substrate by physisorption and it is not possible to know where the probe is binding the molecule along its length. Thus, the resulting contour length is really a measure of the effective length of the monomer being stretched: Lo(eff) = (202.4 ± 5.0) nm, with the effective contour length corresponding to the average of Lo(a) and Lo(b). Thus, the mechanical properties obtained by this approach can only be representative of the actual length of the monomer stretched rather than the entire molecule; for a more complete experimental evaluation of collagen single molecule mechanics, further work will be required utilizing chemical cross-linking methods to ensure the entire molecule is extended (Hinterdorfer et al., 2002). Sun et al. (2002) used the endogenous cysteine residues in procollagen, though this limits observations to the precursor protein (2002). Manipulating the mature form might be problematic as it would require extensive molecular biology to insert specific binding tags into such a complex molecule as collagen—both termini would require differential tagging on one of the components of the triple helix at a location that would be perfectly exposed after proteolytic processing of the procollagen; a more fruitful approach may be to chemically modify the mature triple helix, though the extensive amino acid repeats in the collagen primary sequence would present other challenges.
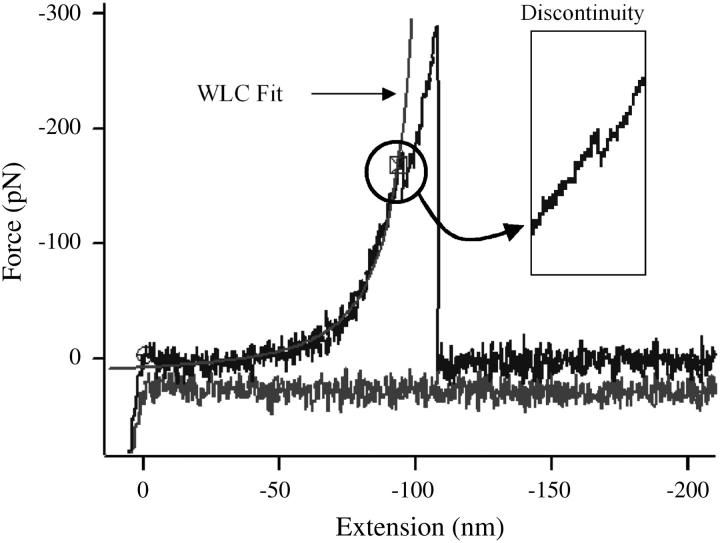
FIGURE 5.
Wormlike chain model fitting of the curve obtained for pulling on a singe molecule. The curve also presents a discontinuity along is curvature as highlighted in the inset. The model tends to fit best the lower part of the curvature of the stretching peak.
FIGURE 6.
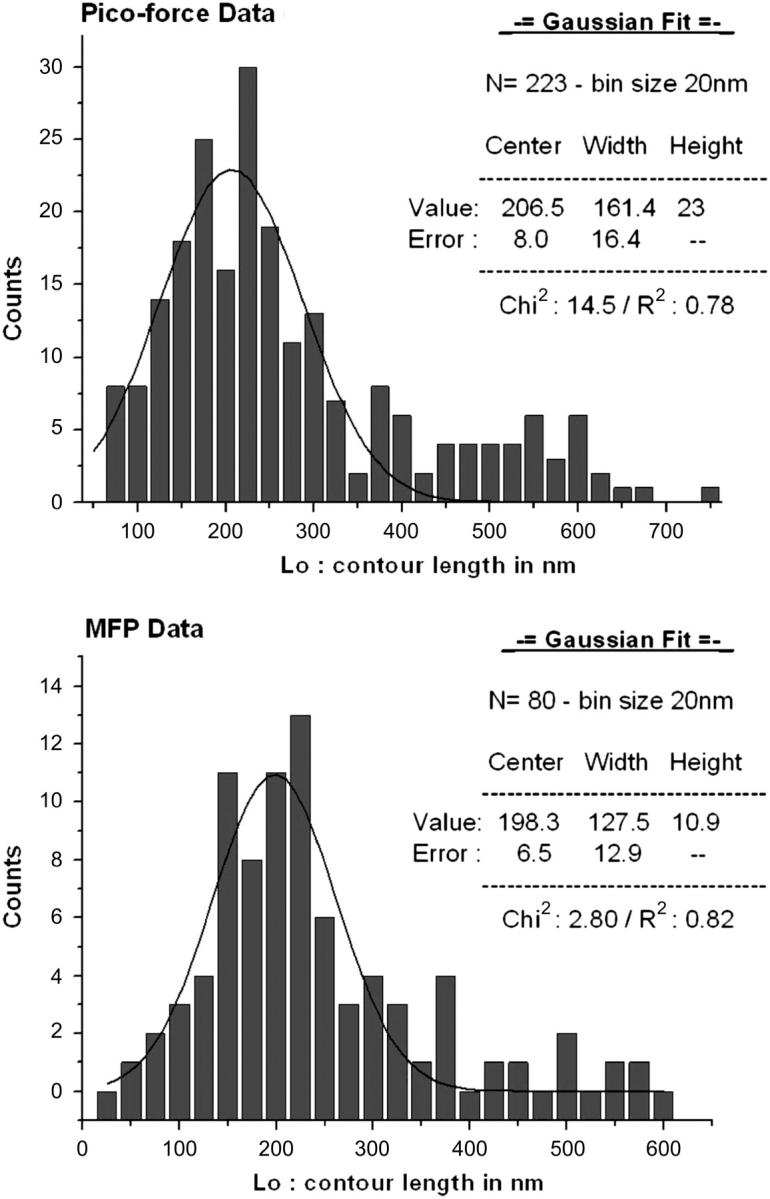
Distribution of the contour length values obtained by modeling the force-distance curves measured on two different instruments: (top) Picoforce (Veeco) and (bottom) Molecular Force Puller, MFP1 (Asylum Research). The two distributions were modeled using a Gaussian fit.
Mechanical behavior of the collagen monomer
While performing the fitting of the WLC model on the peaks, a characteristic feature occurred on some of the peaks; the inset in Fig. 5 presents one of these events (“discontinuity”). During the stretching events, a discontinuity can occur in the curvature of the peak, occurring in ∼18% of the entire data set. The WLC fitted the lower part of the curvature of the peak below the discontinuity well, whereas the upper part did not (a similar behavior is observed with peaks that do not show this discontinuity). This suggests that the curvature of the peak contains two mechanical behaviors, of which the WLC can only model one effectively. In force-distance measurements as performed on titin (Rief et al., 1997), for example, it is common not to include the last peak in the fitting of the WLC, as it is often considered as the pull-off the probe. However, in most of the force curves recorded herein, there is only one peak, compatible with collagen not being a modular protein. Thus, it is probable, as argued above, that information regarding the stretching of the monomer is included in the same peak as the pull-off peak and the presence of this discontinuity marks a transition between two mechanical regimes. The first one is situated below the discontinuity and corresponds to the portion of the curves that can be fitted by the WLC where there is true elastic stretching of the monomer. Other physical techniques will be required to examine the changes in topology that occur during this transition. Though we have no direct evidence, it is may be that it represents stretching and/or unwinding of the monomer. The second regime present in the stretching peak is more generally known as the pull-off peak. This occurs when the probe is still being retracted and therefore still applying an increasing force to the molecule, despite the molecule itself having undergone a transition. In this regime, the molecule is inelastic and the WLC model does not fit, meaning that if the binding forces in the probe-molecule-substrate system are strong enough, the internal structure of the monomer may undergo irreversible changes. Once the force exerted by the retraction of the probe exceeds that of the binding of the probe-molecule-substrate system, there is a rupture either of the probe-monomer bound or substrate-monomer bound. There have been several accounts in the literature where partial relaxation or even plateau occurring in the stretching curve of single molecule could be related to conformational changes in the system. Marszalek et al. (2002) have, for example, studied the chair-boat transitions occurring while stretching polysaccharides molecules. They found that structural rearrangement may occur when the molecule of amylose is submitted to large extension force. The molecule undergoes that release of tension by changing its native conformation (chair) to a longer conformation (boat). In the case of the collagen molecule, the nature of the phenomenon that leads to a discontinuity in the stretching peak or in the transition in the mechanical behavior is yet to be elucidated, but could be related to the winding of the monomer. As mentioned earlier, the tropocollagen molecule consists of three left-handed coiled polypeptide chains intertwined in an overall right-handed coil. As the molecule is being stretched, it is possible that the right-handed coil unwinds, leading to a small extension of the molecule length. Once this occurs, further stretching may involve a stress being applied to the hydrogen bonds that link the polypeptide chains together. A large stress may induce breakage in those bonds and therefore prevent the molecule for regaining its original conformation. A more rational explanation may come from structural variations that occur throughout the molecule. It is possible for the monomer to experience some micro-unfolding if there is an interruption in the continuity of the polypeptide chains due to the absence of prolines and hydroxyprolines at the respective X- and Y-sites of the Gly-X-Y repeat sequence. These localized structural differences could affect the local flexibility of the molecule sufficiently to influence the mechanical properties measured by AFM pulling. Perret et al. (2001) have assayed unhydroxylated recombinant collagen I homotrimer, native collagen I homotrimer and heterotrimer using dynamic light scattering. They found the absence of hydroxylated groups does influence the elasticity of the whole system, reinforcing that the basis of the inherent flexibility of the collagen molecule, is not only due to its conformation, but also to the its nature and the amino-acids contents.
SUMMARY
Herein, we study the topographic and mechanical properties of type I collagen monomers using AFM. Our focus was to understand the fundamental mechanical behavior of the monomer, such as the response of the monomer to an applied stress. This was only possible, once a controlled surface coverage of the monomers could be achieved. While adjusting the deposition parameters, several experiments were performed that led to the study of the topology of the monomer using high-resolution imaging. A repeat pattern was identified along the length of the monomer, whose pitch corresponds to the coil-pitch theoretical value. Other results also showed that molecular orientation was obtained on a mica surface depending on the concentration of monomer used. Even if this was not the prime goal of these experiments, this is a very interesting result as it may be a first step toward controlling deposition, surface patterning and fibrillogenesis, processes that would need to be regulated in the development of artificial, collagen-based materials for tissue engineering and implantable device applications (Wilson et al., 2001). It is interesting that the conditions under which these occurs are not similar to those used in published work on collagen fibrillogenesis (Christiansen et al., 2000).
The main focus of this publication revolves around understanding the mechanical behavior of the collagen monomer under applied stress. The force-distance curves obtained included those with multiple stretching peaks that were also observed in a more complex manner in rat tail collagen fibers in the recent publication by Gutsmann et al. (2004). The apparent simplicity of the force-distance curves used in our study emerged from the fact single monomers were pulled whereas others used macromolecular complexes of collagen fibers. The generic aim of this study was to further the understanding of the mechanical behavior of the basic building block, collagen, of skeletal tissues. There have been many studies involving macro- or mesoscale mechanical studies of these collageneous tissues, but none include the contribution of the monomeric mechanical response. Understanding the mechanical response of type I tropocollagen could, for example, may contribute to a more complete knowledge of the characteristic stress-strain curve of tendons. The implication of such data could lead to a better understanding of the tendon damage such as occurs in tendinitis or partial tendon rupture (Karlsson et al., 1992; Uhthoff and Sano, 1997). These experiments offer a precursor approach to a complete model of collagen biomechanics ranging from single molecules to entire tissues.
Acknowledgments
We thank the Wellcome Trust, UK, for an award of a program grant to M.A.H., secondly Drs. R. McKendry and B. Leitinger for helpful discussions and advice, and finally Dr. J. Clarke and R. Rounsevell for their expertise in single molecule force measurement and loan of equipment.
References
- Baselt, D., J. Revel, and J. Baldeschwieler. 1993. Subfibrillar structure of type I collagen observed by atomic force microscopy. Biophys. J. 65:2644–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann, C. G., S. B. Smith, C. Bustamante, and V. A. Bloomfield. 1997. Effects of ionic strength and cation valence on the elastic response of single DNA molecules. Proc. Natl. Acad. Sci. USA. 94:6185–6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, K., and B. Brodsky. 1998. Supercoiled protein motifs: the collagen triple-helix and the alpha-helical coiled coil. J. Struct. Biol. 122:17–29. [DOI] [PubMed] [Google Scholar]
- Bella, J., B. Brodsky, and H. M. Berman. 1995. Hydration structure of a collagen peptide. Structure. 3:893–906. [DOI] [PubMed] [Google Scholar]
- Bella, J., M. Eaton, B. Brodsky, and H. M. Berman. 1994. Crystal-structure and molecular-structure of a collagen-like peptide at 1.9-Angstrom resolution. Science. 266:75–81. [DOI] [PubMed] [Google Scholar]
- Bustamante, C., J. F. Marko, E. D. Siggia, and S. Smith. 1994. Entropic elasticity of lambda-phage DNA. Science. 265:1599–1600. [DOI] [PubMed] [Google Scholar]
- Chernoff, E. A. G., and D. A. Chernoff. 1992. Atomic force microscope images collagen fibres. J. Vac. Sci. Technol. A. 10:596–599. [Google Scholar]
- Christiansen, D. L., E. K. Huang, and F. H. Silver. 2000. Assembly of type I collagen: fusion of fibril subunits and the influence of fibril diameter on mechanical properties. Matrix Biol. 19:409–420. [DOI] [PubMed] [Google Scholar]
- Conti, M., Y. Bustanji, G. Falini, P. Ferruti, S. Stefoni, and B. Samori. 2001. The desorption process of macromolecules adsorbed on interfaces: the force spectroscopy approach. ChemPhysChem. 2:610–613. [DOI] [PubMed] [Google Scholar]
- Fritz, M., M. Radmacher, J. P. Cleveland, M. W. Allersma, R. J. Stewart, R. Gieselmann, P. Janmey, C. F. Schmidt, and P. K. Hansma. 1995. Imaging globular and filamentous proteins in physiological buffer solutions with tapping mode atomic-force microscopy. Langmuir. 11:3529–3535. [Google Scholar]
- Gutsmann, T., G. E. Fantner, J. H. Kindt, M. Venturoni, S. Danielsen, and P. K. Hansma. 2004. Force spectroscopy of collagen fibers to investigate their mechanical properties and structural organisation. Biophys. J. 86:3186–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsmann, T., G. E. Fantner, M. Venturoni, A. Ekani-Nkodo, J. B. Thompson, J. H. Kindt, D. E. Morse, D. K. Fygenson, and P. K. Hansma. 2003. Evidence that collagen fibrils in tendons are inhomogeneously structured in a tubelike manner. Biophys. J. 84:2593–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, W., J. Mou, J. Yang, and Z. Shao. 1995. Cryoatomic force microscopy: a new approach for biological imaging at high resolution. Biochemistry. 34:8215–8220. [DOI] [PubMed] [Google Scholar]
- Hansma, H. G., and P. K. Hansma. 1993. Potential applications of atomic force microscopy of DNA to the human genome project. Proc. SPIE Int. Opt. Eng. 1891:66–70. [Google Scholar]
- Hinterdorfer, P., H. J. Gruber, F. Kienberger, G. Kada, C. Riener, C. Borken, and H. Schindler. 2002. Surface attachment of ligands and receptors for molecular recognition force microscopy. Colloids Surf. B Biointerfaces. 23:115–123. [Google Scholar]
- Hutter, J. L., and J. Bechhoefer. 1993. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 64:3342. [Google Scholar]
- Jiang, F. Z., H. Horber, J. Howard, and D. J. Muller. 2004. Assembly of collagen into microribbons: effects of pH and electrolytes. J. Struct. Biol. 148:268–278. [DOI] [PubMed] [Google Scholar]
- Karlsson, J., P. Kalebo, L. A. Goksor, R. Thomee, and L. Sward. 1992. Partial rupture of the patellar ligament. Am. J. Sports Med. 20:390–395. [DOI] [PubMed] [Google Scholar]
- Marszalek, P. E., H. Li, A. F. Oberhauser, and J. M. Fernandez. 2002. Chair-boat transitions in single polysaccharide molecule observed with force-ramp AFM. Proc. Natl. Acad. Sci. USA. 99:4278–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertig, M., U. Thiele, J. Bradt, G. Leibiger, W. Pompe, and H. Wendrock. 1997. Scanning force microscopy and geometric analysis of two-dimensional collagen network formation. Surf. Interfac. Anal. 25:514–521. [Google Scholar]
- Morris, V. J., A. R. Kirby, and A. P. Gunning. 1999. Basic Principles. Atomic Force Microscopy for Biologists. Imperial College Press, London.
- Olsen, B. R. 1991. Collagen biosynthesis. In Cell Biology of the Extracellular Matrix, 2nd ed. E. D. Hay, editor. Plenum Press, New York. 177–220.
- Orgel, J. P., T. J. Wess, and A. Miller. 2000. The in situ conformation and axial location of the intermolecular cross-linked non-helical telopeptides of type I collagen. Struct. Fold. Des. 8:137–142. [DOI] [PubMed] [Google Scholar]
- Perret, S., S. Merle, J. Nettleton, and J. Howard. 2001. Unhydroxylated triple-helical collagen I produced in transgenic plants provides new clues on the role of hydroxyproline in collagen folding and fibril formation. J. Biol. Chem. 276:43693–43698. [DOI] [PubMed] [Google Scholar]
- Ramachandra, G. N., and G. Karthan. 1955. Structure of collagen. Nature. 176:593–595. [DOI] [PubMed] [Google Scholar]
- Revenko, I., F. Sommer, D. T. Minh, R. Garrone, and J. M. Franc. 1994. Atomic force microscopy study of the collagen fibre structure. Biocell. 80:67–69. [DOI] [PubMed] [Google Scholar]
- Rief, M., M. Gautel, F. Oesterhelt, J. M. Fernandez, and H. E. Gaub. 1997. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 276:1109–1112. [DOI] [PubMed] [Google Scholar]
- Rounsevell, R., J. R. Forman, and J. Clarke. 2004. Atomic force microscopy: mechanical unfolding of proteins. Methods. 34:100–111. [DOI] [PubMed] [Google Scholar]
- Schabert, F. A., and J. P. Rabe. 1996. Vertical dimension of hydrated biological samples in tapping mode scanning force microscopy. Biophys. J. 70:1514–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, F. D., C. E. Hall, and M. A. Jakns. 1942. Electron microscopy investigations of the structure of collagens. J. Cell. Comp. Physiol. 20:11–33. [Google Scholar]
- Sun, Y., Z. Luo, A. Fertala, and K. An. 2002. Direct quantification of the flexibility of type I collagen monomer. Biochem. Biophys. Res. Commun. 295:382–386. [DOI] [PubMed] [Google Scholar]
- Tskhovrebova, L., J. Trinick, J. A. Sleep, and R. M. Simmons. 1997. Elasticity and unfolding of single molecules of the giant muscle protein titin. Nature. 387:308–312. [DOI] [PubMed] [Google Scholar]
- Uhthoff, H. K., and H. Sano. 1997. Pathology of failure of the rotator cuff tendon. Orthop. Clin. North Am. 28:31–41. [DOI] [PubMed] [Google Scholar]
- Walters, D. A., J. P. Cleveland, N. H. Thomson, P. K. Hansma, M. A. Wendman, G. Gurley, and V. Elings. 1996. Short cantilevers for atomic force microscopy. Rev. Sci. Instrum. 67:3583–3590. [Google Scholar]
- Wang, M. D., H. Yin, R. Landick, J. Gelles, and S. M. Block. 1997. Stretching DNA with optical tweezers. Biophys. J. 72:1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, D. L., R. Martin, S. Hong, M. Cronin-Golomb, C. A. Mirkin, and D. L. Kaplan. 2001. Surface organization and nanopatterning of collagen by dip-pen nanolithography. Proc. Natl. Acad. Sci. USA. 98:13660–13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., S. J. Sheng, and Z. Shao. 1996. Imaging biological structures with the cryoatomic force microscope. Biophys. J. 71:2168–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]