Abstract
Raman spectra of cells and nuclei from cultures in the plateau (nonproliferating) and exponential (proliferating) phases of growth were measured and show that Raman spectroscopy can monitor changes due to cell proliferation. A simple fitting routine was developed using a basis set (lipid, protein, DNA, RNA) to estimate the relative amounts of biochemical components in cells and nuclei. Using relative amounts and ratios of biochemical components, reproducible differences can be detected and quantified that are not readily apparent by visual analysis of vibrational bands in the spectra. These differences, due to cell proliferation, can be assigned to specific biochemical changes. They include a decrease in the relative lipid and increases in the relative protein and RNA for both nontumorigenic exponential cells and nuclei, and an increase in the relative RNA for tumorigenic exponential cells. The lipid/RNA ratio decreases for nontumorigenic exponential cells and nuclei and tumorigenic exponential cells. The protein/lipid ratio increases for both tumorigenic and nontumorigenic exponential cells and nuclei. Finally, the lipid/DNA ratio decreases for tumorigenic exponential nuclei. This knowledge will be important for Raman detection of rapidly dividing populations of cancer cells in vivo.
INTRODUCTION
There is much interest in using vibrational spectroscopy as a diagnostic tool. It is a technique that promises to allow rapid in vivo characterization of tissue and bodily fluids in a nondestructive and less invasive way compared to methods now in general use. Raman spectroscopy is one method currently being tested as a diagnostic tool (Wolthuis et al., 1999; Naumann, 2001; Petrich, 2001; Choo-Smith et al., 2002). Compared to infrared absorption, Raman has the advantage of having only small and easily subtracted water bands. In biological samples Raman spectra often exhibit a number of rather sharp bands, whereas infrared spectra of cells and tissue often show broader spectral features (Naumann, 2001). This empirical observation is particularly important in analyzing complex biochemical systems. That is because, whereas infrared spectroscopy is able to yield information about cellular components (e.g., proteins, lipids, nucleic acids) (Mourant et al., 2003a), Raman spectroscopy gives this information as well as much more information about some of the specific molecules in these groups of components (e.g., phenylalanine, tyrosine, and adenine) that is not available from infrared spectra.
Differences between Raman spectra of normal and malignant tissues, both in vivo and in vitro, are being investigated as a method for the early detection of cancer. For example, in vitro studies of neoplasia in Barrett's esophagus have used principal component analysis (PCA) and linear discriminant analysis (LDA) for pathological classification of Raman spectra (Kendall et al., 2003). Barrett's esophagus has also been studied in a rat model, using ex vivo samples, in which a multivariate classification model was able to discriminate between spectra of Barrett's and normal epithelium (Boere et al., 2003). Raman spectroscopy has been used ex vivo and in vivo, along with PCA and LDA, to differentiate adenomatous from hyperplastic polyps of the colon (Molckovsky et al., 2003). PCA and LDA have been used to classify prostatic adenocarcinoma using spectra taken in vitro (Crow et al., 2003). Spectral band analysis of ex vivo samples has been used to show differences between normal and cancerous tissue of the nasopharynx (Lau et al., 2003). Epithelial precancers and cancers of six different tissues have been classified from spectra taken in vitro, and it was suggested that there is a biochemical continuum of progression from the normal to the diseased state (Stone et al., 2002). Normal, benign, and malignant skin samples have been studied in vitro, and the spectra have been classified using cluster analysis (Fendel and Schrader, 1998), PCA (Nunes et al., 2003), and PCA combined with cluster analysis (Nijssen et al., 2002). Cervical precancers have been studied in vitro and the spectra classified using PCA for data reduction and Fisher discriminant analysis for classification (Mahadevan-Jansen et al., 1998). A pilot study obtaining in vivo cervical tissue spectra has been performed and an algorithm was developed for separating high-grade dysplasia from all other diagnoses using band intensity ratios (Utzinger et al., 2001). Raman spectra of oral tissues taken ex vivo have been classified into normal and malignant sets using scores from a PCA analysis (Venkatakrishna et al., 2001). Normal and diseased breast tissue has been studied in vitro, and the spectra analyzed by fitting them with a linear combination of nine cell and chemical components (Shafer-Peltier et al., 2002). In a study using cell cultures, factor analysis of Raman spectra was used to differentiate tumorigenic and nontumorigenic cell lines (Omberg et al., 2002).
These studies show that differences are observed between normal and abnormal states, but there is usually only a qualitative understanding of the underlying biochemical changes. In addition, the proliferative status of the cells may affect the observations. The fraction of actively dividing cells in tumors is generally higher than in the tissue of origin. We report here that there are differences between populations of actively dividing and quiescent cells that can be detected and quantitatively described using Raman scattering. Cell cultures in two proliferative states, “plateau” and “exponential”, were used in this study. Plateau cells are defined as a monolayer culture that has reached a maximal cell number in the culture vessel. These cultures have a very low proliferative index, with most of the cells in the G1/G0 phase of the cell cycle. Exponential cells are defined as a monolayer culture where the cell number is increasing exponentially with time. These cultures are essentially 100% proliferating with most of their cells in the S and G2 phases of the cell cycle (Tannock and Hill, 2004). This simple model approximates the proliferative status of most solid tumors (proliferating) and the normal tissues surrounding them (nonproliferating). In addition, within tumors there are both temporal and microregional variations in the growth rate of cells (Sutherland et al., 1996; Tannock, 2001). Thus, the ability to determine the location of cell populations with certain proliferative state characteristics may prove useful not only in initial diagnosis, but also for monitoring the growth of a tumor and its response to treatment.
Four vibrational spectroscopy studies have considered cells at different points in the cell cycle. A synchrotron radiation based Fourier transform infrared spectroscopy study (Holman et al., 2000) found it was possible to detect changes in spectral regions corresponding to proteins, DNA, and RNA at different points in the cell cycle. A 1999 infrared spectroscopy study (Boydston-White et al., 1999) of myeloid leukemia cells reported changes in DNA content and possibly structure at different points of the cell cycle. One Raman spectroscopy study (Schuster et al., 2000a) presented spectra of single cells from the solventogenic organism Clostridium acetobutylicum at different points in its cell cycle. At different times during solventogenic fermentation, changes were detected in some Raman bands assigned to nucleic acids and proteins. In a second Raman study (Huang et al., 2003), a confocal Raman microspectrometer was focused at the center of a living yeast cell just before mitosis, and spectra were recorded at this position through the M, G1, S, and into the early G2 phases of the cell cycle. Raman spectral changes, with the aid of fluorescence microscope pictures, were attributed to separation of daughter nuclei, the presence of mitochondria, and the formation of the primary septum between two daughter cells. One infrared study (Boydston-White et al., 1999), and the first Raman study above, used cells dried onto solid supports. We have recently shown (Mourant et al., 2003b) that infrared and Raman spectra of dried cells have distorted features compared to spectra of cells that have not been dried. Raman spectra reported here are of viable cells suspended in buffer, more closely mimicking conditions in vivo.
Spectra of two cell lines derived from rat embryo fibroblast cells are presented. M1 cells are an immortalized, nontumorigenic derivative of normal cells made by stable transfection with a myc oncogene. MR1 cells are a tumorigenic line derived from M1 cells by stable transfection with a mutated ras oncogene (Kunz-Schughart et al., 1995). Raman spectra were analyzed for both intact cells and isolated nuclei from each cell line under exponential and plateau culture states. Specific vibrational bands are significantly altered when comparing proliferating and quiescent cells and nuclei. Changes in specific vibrational bands, however, do not give a complete picture of the differences between plateau and exponential proliferative states. Spectral fits, using a basis set of component spectra, give a more quantitative understanding of how the relative amounts of cellular components vary due to changes in the proliferative state.
MATERIALS AND METHODS
Cell culture and characterization
M1 and MR1 cells were propagated in monolayer culture using standard mammalian cell culture techniques as described previously (Kunz-Schughart et al., 1995; Mourant et al., 2000). Briefly, cells were grown as monolayers in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Grand Island, NY) supplemented with 5% (v/v) of fetal calf serum (Hyclone, Logan, UT), 4.5 g/L of D-glucose, 100 IU/ml of penicillin (Invitrogen), and 100 μg/mL of streptomycin (Invitrogen), hereafter referred to as complete DMEM. To obtain cell suspensions for Raman measurements, cells were harvested from monolayer cultures by treatment with 0.25% of trypsin in a phosphate-buffer (pH 7.4) containing 1 mM of EDTA and 25 mM of HEPES, followed by the addition of complete DMEM. Cells were passed once through an 18-gauge needle, centrifuged to obtain a pellet, resuspended in phosphate-buffered saline (PBS; Invitrogen), centrifuged again to remove any residual medium, after which PBS was added to obtain a final concentration of 1.01 × 108 to 3.2 × 108 cells/mL. Cell concentrations were determined as described previously (Freyer, 1998; Mourant et al., 2003a), using an electronic particle counter equipped with a pulse-height analyzer (Coulter Electronics, Hialeah, FL) for measuring the cell volume distribution and mean cell size. All cell counts were corrected for the presence of acellular debris using the cell volume distributions.
Nuclei isolation
Nuclei were isolated using the following solutions: 1), hypotonic lysis buffer (HLB) (10 mM Tris pH 7.9, 5 mM KCl, 1.5 mM MgCl2); 2), first sucrose cushion—800 mM sucrose in HLB; 3), first resuspension buffer (250 mM sucrose, 10 mM Tris, pH 7.9, 3.3 mM MgCl2); 4), second resuspension buffer (250 mM sucrose, 10 mM MgCl2); and 5), second sucrose cushion—350 mM sucrose, 0.5 mM MgCl2. Whole cells (0.7–1.2 × 108 cells) were incubated in HLB (10–15 ml) on ice for 10 min. Using a 22-gauge needle, cells were syringed six times up and six times down, to disrupt the outer cell membrane, then layered on top of the first sucrose cushion (5 ml) in a centrifuge tube and spun at 4°C and 5000 rpm for 10 min (Heraeus highconic rotor No. 3046, Sorvall, Newtown, CT); the supernatant was suctioned off, and the pellet resuspended in 5 ml of the first resuspension buffer and then spun at 4°C and 5000 rpm for 5 min. The supernatant was suctioned off and the pellet resuspended in 5 ml of the second resuspension buffer. This was layered on top of the second sucrose cushion (5 ml) in a centrifuge tube and spun at 4°C and 5000 rpm for 10 min. The supernatant was suctioned off, and the pellet was resuspended in 5 ml of cold PBS and spun at 4°C and 5000 rpm for 10 min. The PBS wash was repeated, the supernatant decanted, and the nuclei pellet resuspended in 0.5 ml of cold PBS. Before Raman measurements, a 10-μl nuclei aliquot was added to 1 ml of cold PBS and counted using a hemacytometer. Cytospin slides (Shandon, Pittsburgh, PA) were prepared and stained with the DNA fluorophore Hoechst 33342 (Calbiochem, San Diego, CA) to verify the integrity of the nuclei and the absence of intact cells. One hundred intact cells and 100 isolated nuclei were analyzed from each cell culture experiment in which nuclei were isolated, to monitor the integrity and the mean nuclear size. Unless otherwise noted, all chemicals were from Sigma (St. Louis, MO).
Proliferative status
Flow cytometry was used to characterize the proliferative status of cells (Krishan, 1975; Mourant et al., 2003a). Briefly, cell suspensions (∼1 × 106 cells) were fixed in 70% ethanol and stored at 4°C. Before flow cytometry analysis, the cells were centrifuged for 10 min at 1500 rpm and 4°C, and the fixation solution was removed. The cells were stained using 1 ml of propidium iodide (50 μg/ml) in PBS containing CaCl2 and MgCl2 (Invitrogen), and RNase (100 μg/ml) was added to remove cellular RNA and ensure a fluorescence signal proportional to DNA content. The samples were allowed to sit overnight at 4°C before flow measurements to ensure complete RNA digestion. DNA content histograms were then collected using a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ). DNA content histograms were deconvolved using the WinList and ModFit LT software packages (Verity Software House, Topsham, ME) to estimate fractions of cells in the G1/G0, S, and G2 phases of the cell cycle. From the flow cytometry data, we verified which cell cultures were in the “plateau” proliferative state and which were in the “exponential” proliferative state (see Table 1). “Plateau” cells had 80% or more of their cells in the G1/G0 phase of the cell cycle. “Exponential” cells had 50% or less of their cells in the G1 phase of the cell cycle.
TABLE 1.
Percentage of cells in the G1/G0, S, and G2 portions of the cell cycle
| M1P (n = 7) | M1E (n = 8) | MR1P (n = 8) | MR1E (n = 7) | |
|---|---|---|---|---|
| G1/G0 | 80.0 ± 6.8 | 43.9 ± 3.6 | 86.6 ± 4.1 | 50.1 ± 8.0 |
| S | 12.0 ± 6.0 | 41.4 ± 2.4 | 9.0 ± 3.8 | 35.7 ± 6.0 |
| G2 | 7.9 ± 2.9 | 14.6 ± 3.2 | 4.3 ± 2.1 | 14.0 ± 2.9 |
The number of experiments is shown in parentheses. M1P, M1 plateau; M1E, M1 exponential; MR1P, MR1 plateau; and MR1E, MR1 exponential.
Collection of the Raman spectra
Raman spectra were measured using 785 nm light for sample excitation. The system includes an Invictus 785 nm diode laser coupled by an optical fiber to an excitation/collection probe (Kaiser Optical Systems, Ann Arbor, MI). Collection is by direct backscattering into the probe. The excitation light was focused and the scattered light collimated using an aspheric lens, f/0.7, focal length 10.8 mm (Edmund Industrial Optics, Barrington, NJ) on the end of the probe. The Raman scattered light was coupled by an optical fiber into a holographic imaging spectrograph (HoloSpec f/1.8, Kaiser Optical Systems). The dispersed light was measured using a liquid nitrogen-cooled, deep-depletion, back-illuminated charge-coupled device detector connected to an ST-133 controller (Princeton Instruments, Trenton, NJ). The spectrograph was equipped with a holographic grating for either the low or the high wavenumber Stokes shifted regions. These gratings had an approximate spectral coverage of 100–1900 and 1780–3250 cm−1 for the low and the high wavenumber spectral regions, respectively. Data were collected for each experiment over both spectral regions. The spectral resolutions, defined as twice the spectral bandpass (full width at half maximum), were ∼6.2 and 5.0 cm−1 for the low and the high wavenumber regions, respectively. The spatial cosmic ray correction of the WinSpec/32 program (Princeton Instruments) was used at 100% during data collection. This correction had a slight smoothing effect on the data, equivalent to about a five-point boxcar smooth. For a given experiment, collection times were normally 300 s/spectrum with a laser power at the sample of ∼200 mW. The diameter of the unfocused laser is 4 mm, so the minimum spot size of the focused beam is 55 μm. The samples were contained in 5-mm diameter suprasil NMR tubes (Wilmad/Lab Glass, Buena, NJ). Samples were chilled in an ice water bath during data collection. For each experiment, two spectra were collected of the empty NMR tube, two spectra of the NMR tube containing the buffer, and 10 spectra of the NMR tube containing the cell culture or isolated nuclei sample. For the DNA, RNA, and lipid basis set components six spectra of the NMR tube containing the sample were collected. For the isolated protein basis set component 10 spectra of the NMR tube containing the sample were collected. The cell spectral data reported here are the averages of seven or eight separate cell culture experiments for each of M1 and MR1 plateau and exponential proliferative-state cells. The nuclei spectral data reported here are the averages of three separate nuclei isolations from each of M1 and MR1, plateau and exponential proliferative-state cells.
Analysis of the Raman spectra
Spectra used to subtract background signal due to the optics and NMR tube, and due to the PBS buffer, were smoothed using a seven-point boxcar (sliding average) smooth. The cell culture, isolated nuclei, and basis set component spectra were not smoothed. A standard tungsten source (Optronic Laboratories, Orlando, FL) was used to measure the instrumental response. This response was divided by the spectral irradiance of the tungsten lamp and then smoothed three times using a 15-point boxcar smooth to give the final spectrum used in the correction. The data spectra were then divided by the instrumental response correction spectrum.
For the low wavenumber region of the basis set components, the spectrum of the optics and sample holder contribution was subtracted from the spectra, until the broad band centered near 435 cm−1, due to the NMR tube, was no longer evident above the background fluorescence. The buffer spectrum was then subtracted from the basis-set spectra, until the broad band centered near 180 cm−1 was no longer evident above the background. The background fluorescence was subtracted using a cubic spline (Igor Pro, WaveMetrics, Lake Oswego, OR). Using a cubic spline eliminated most regions of negative Raman intensity that were present if a fourth- or fifth-order least-squares polynomial was used to subtract background fluorescence. This makes the subsequent component fits easier. The parameters for the cubic spline were chosen by trial and error. The number of nodes was 14. Higher and lower numbers, such as 30 and 0, caused obvious distortions to the fits. The end nodes were turned off, as estimating them by extrapolation sometimes caused distortion in the fits of slightly noisier spectra. The match of the fit was to the second derivative. There were only minor differences if the match was to the first derivative. The spline was fit to spectral regions near 283, 482, 714, 877, 993, 1165, 1405, 1618, 1712, 1806, 1830, and 1857 cm−1 for DNA; near 285, 359, 480, 649, 749, 843, 990, 1155, 1279, 1435, 1607, 1653, 1744, and 1783 cm−1 for RNA; near 200, 476, 675, 1187, 1582, and 1685 cm−1 for lipid; and near 391, 686, 792, 1021, 1143, 1496, 1746, and 1807 cm−1 for protein. These regions were chosen in places where there appeared to be no Raman scattering from the biochemical chromophore.
For the low wavenumber region of the cell culture samples and the isolated nuclei, the spectrum of the optics and sample holder contribution was subtracted from the data spectra. The background sample fluorescence was then subtracted using a cubic spline fit. For most low wavenumber spectra the spline was fit to spectral regions near, but not limited to, 325–342, 580, 614, 644, 696, 842, 911, 1019, 1142, and 1780–1900 cm−1. The buffer contribution to the spectra was estimated using the Levenberg-Marquardt algorithm to minimize the difference between the data and the fit. Basis-set spectra of RNA, DNA, lipid, and protein (all described below), and PBS buffer were used for the fit. The criterion for minimizing the fits here, and in the section below, was to minimize χ2. No constraints were placed on the values of the fit coefficients. The estimated buffer contribution was subtracted from the data spectra. A second cubic spline was used to rezero the baseline of each spectrum, using the same regions as the first cubic spline and additionally the region near 1518 cm−1. The low wavenumber spectra were normalized to the CH2 deformation band near 1445 cm−1, using the integrated intensity from 1437 to 1453 cm−1. Average spectra and standard deviations were then calculated.
For the high-wavenumber region, the buffer contribution was subtracted from the spectra. This subtraction was performed until there was no visible buffer contribution in the region between 3100 and 3200 cm−1. Background sample fluorescence was removed using a curve generated by a cubic spline fit to selected regions of the data spectra near, but not limited to, 2027, 2179, 2424, 2605, 2661, 2780, 3106, and 3150 cm−1. The high-wavenumber spectra were normalized to the CH2 asymmetric stretch near 2933 cm−1, using the integrated intensity from 2929 to 2940 cm−1.
Spectral fitting
To determine the relative amounts of the different components in the cells and the isolated nuclei, experimental spectra with optics and PBS contributions subtracted were fit to basis-set spectra of isolated cellular components using the Levenberg-Marquardt algorithm. The basis set of spectra used were: calf liver RNA, Type IV, highly polymerized calf thymus DNA, Type I, a 4:1 egg yolk phosphatidylcholine/phosphatidylethanolamine mixture, and a straight line. For experiments to characterize mixtures of components liver total lipid extract (bovine) or egg phosphatidylcholine (both from Avanti Polar Lipids, Alabaster, AL) were used for the lipid component. A phosphatidylcholine/phosphatidylethanolamine mixture was not used, as we were unable to obtain satisfactory egg yolk phosphatidylethanolamine at the time those experiments were done. The protein was isolated from M1 and MR1 cells as described previously (Mourant et al., 2003b). The protein spectrum used in the fits here was from MR1 plateau cells. The protein was isolated from the fibroblast cells, rather than using a mixture of commercially available proteins because we believe that this will more closely mimic the protein contribution of the cells. For instance, the spectrum of bovine serum albumin (BSA) is quite different from that of our isolated protein. No significant differences were noted in the spectra of different protein isolations from the two related rat fibroblast cell lines used here. So in the future, it may be possible that a protein basis spectrum could be measured for a particular class of cells and that spectrum then used for similar cells. BSA that is globulin- and fatty acid-free was used as the protein component in experiments to characterize mixtures of components. The 4:1 ratio for lipids was chosen because the phosphatidylcholine/phosphatidylethanolamine ratio in fibroblasts has been reported to be 3:1–4:1 (Zhang et al., 1998). Component spectra were scaled to represent equal concentrations at 40 mg/ml. The spectral region between 575 and 1800 cm−1 was fit to the basis set. Fit coefficients for each spectrum were normalized to sum to 1. It has been shown that the Raman spectral intensity scales linearly with the amount of a given component in a mixture, and that a linear superposition of spectra of individual components can be used to determine the composition of a biological sample (Manoharan et al., 1992; Brennan et al., 1997; Salenius et al., 1998; Römer et al., 1998; Boustany et al., 1999). Thus, the normalized fit coefficients for each individual component give an estimate of the relative amount of that component in the cells or the nuclei.
RESULTS
Cell proliferative state and nuclei characterization
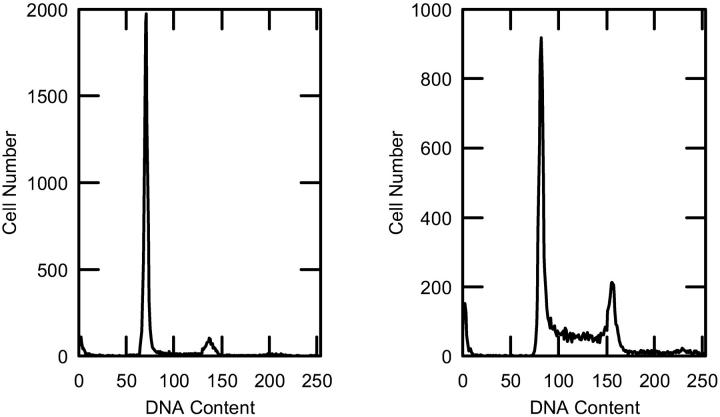
The proliferative state of the cell cultures was monitored by measuring DNA content using flow cytometry. Typical results of M1 cell cultures in the plateau and in the exponential proliferative states are shown in Fig. 1. Average cell cycle distributions of the percentages of cells in the G1/G0, S, and G2 parts of the cell cycle are shown in Table 1. Plateau cells had 80% or more of their cells in the G1/G0 phase of the cell cycle. Exponential cells had 50% or less of their cells in the G1 phase of the cell cycle. A second parameter used to monitor the proliferative state is the density of cells (cells/cm2) on the culture plates at the time of harvesting. Since plateau cells have reached confluence, they are expected to have a higher value of cells/cm2 on the plates than exponential cells. For M1 cells the values were 170.5 × 103 ± 31.2 × 103 (n = 7) and 56.4 × 103 ± 6.6 × 103 cells/cm2 (n = 8) for plateau and exponential cells, respectively (p > 99.5%). For MR1 cells the values were 433.8 × 103 ± 42.4 × 103 (n = 8) and 245.7 × 103 ± 69.2 × 103 cells/cm2 (n = 7) for plateau and exponential cells, respectively (p > 99.5%). A third parameter that can be used to monitor the proliferative state is the cell volume, because cell size increases during the cell cycle and is expected to be largest in the G2 phase just before mitosis. For M1 plateau and exponential proliferative-state cells the mean volumes and standard deviations were 1.78 × 103 ± 0.23 × 103 μm 3 (n = 6) and 2.11 × 103 ± 0.18 × 103 μm3 (n = 8), respectively (p > 99.5%). For MR1 plateau and exponential proliferative-state cells these numbers were 0.98 × 103 ± 0.16 × 103 μm3 (n = 6) and 1.53 × 103 ± 0.20 × 103 μm3 (n = 6), respectively (p > 99.5%).
FIGURE 1.
Examples of the flow cytometry analysis for M1 cells are shown. For the plateau proliferative state (left), 85% of the cells were in G1/G0, 11% were in S, and 4% were in G2. For exponential-state cells (right), 43% of the cells were in G1, 42% were in S, and 15% were in G2.
Analysis of cytospin preparations used to monitor the integrity and size of isolated nuclei are shown in Table 2. For both M1 and MR1 plateau and exponential cells the mean diameters and standard deviations of nuclei inside cells and of isolated nuclei are listed. The isolated and intact nuclei are the same size within experimental error. Also, micrographs taken of the isolated nuclei for each experiment showed no signs of nuclear disruption. A comparison between plateau and exponential proliferative-state nuclei showed that they differ in size at a confidence level of >75.0% for M1 and MR1 nuclei inside cells, do not differ for M1 isolated nuclei, and differ at a confidence level >99.5% for MR1 isolated nuclei. The apparent slight increase in nuclear size is expected, as the exponential proliferative-state cells have a higher proportion of cells in the S and G2 portions of the cell cycle (see Table 1) and will have more DNA than plateau cells.
TABLE 2.
Nuclei sizes from cytospin preparations
| Intact nuclei (μm) | Isolated nuclei (μm) | ||
|---|---|---|---|
| M1 nuclei | Plateau | 10.62 ± 0.43 (3) | 9.87 ± 0.76 (3) |
| Exponential | 11.20 ± 0.58 (3) | 10.30 ± 0.68 (3) | |
| MR1 nuclei | Plateau | 9.02 ± 0.42 (4) | 9.10 ± 0.40 (4) |
| Exponential | 9.46 ± 0.55 (3) | 10.41 ± 0.63 (3) |
Measurements are nuclei diameters. The number of nuclei preparations is given in parentheses after the measurements.
Low-wavenumber data
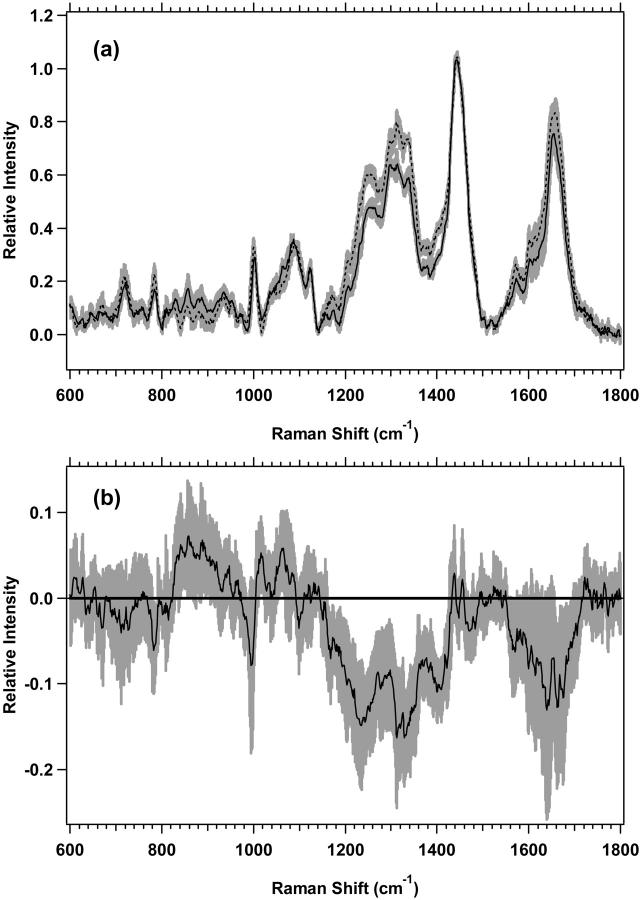
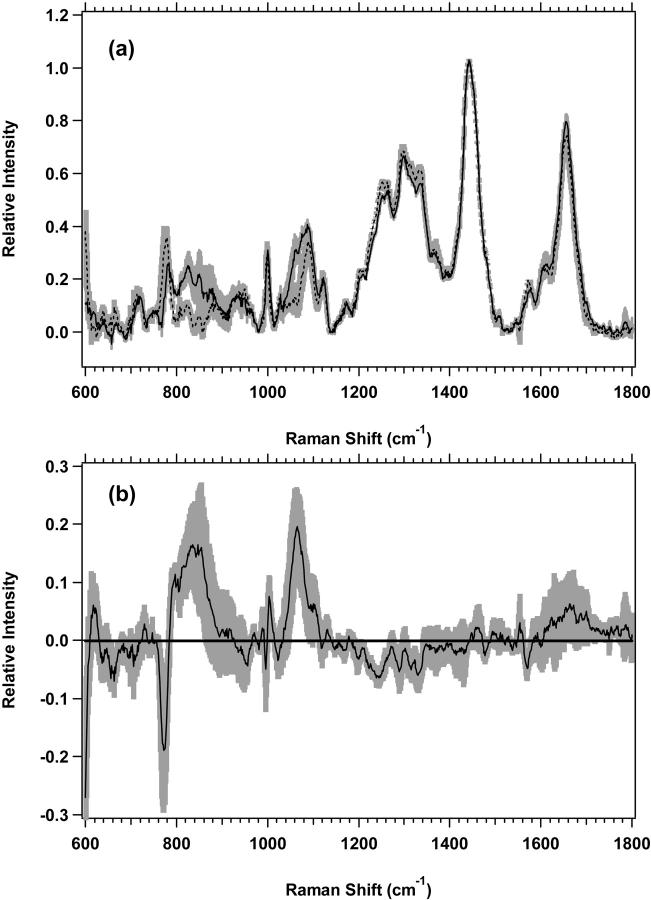
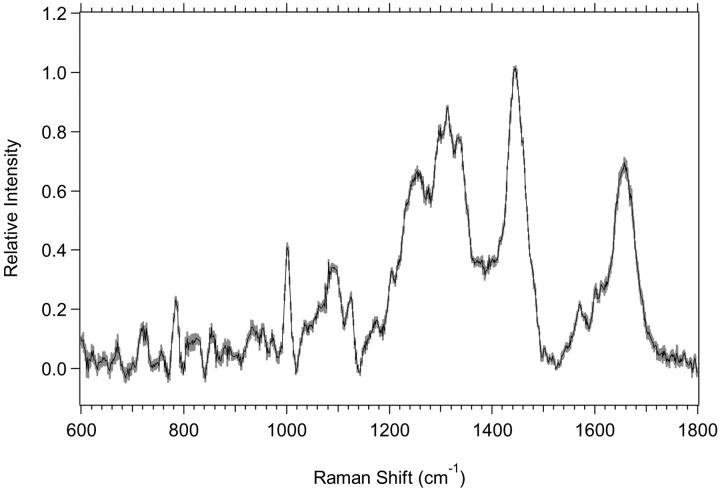
Shown in Fig. 2 a are the average spectra of the low-wavenumber region from 600 to 1800 cm−1 for M1 plateau and exponential cell cultures with the experimental standard deviations overlying the averages. There are spectral regions where the standard deviations do not overlap, and the differences are thus significant and reproducible. The changes are more clearly observed in Fig. 2 b, which shows the difference spectrum of the averages for M1 plateau and exponential cell cultures. The vibrational regions that change, have nonoverlapping standard deviations, and are shown to be different by Student's t-test at >99.5% confidence are listed in Table 3. Assignments of vibrations that change have been made based on assignments by others (Barry et al., 1992; Gniadecka et al., 1997a; Fendel and Schrader, 1998; Schuster et al., 2000b; Naumann, 2001; Venkatakrishna et al., 2001). It is important to note that although some specific assignments can be made, many of the changes are in spectral regions with significant overlap between protein, lipid, and nucleic acid peaks (see Fig. 11).
FIGURE 2.
(a) Standard deviations (shaded area) overlying the averages are shown from 600 to 1800 cm−1 for the M1 plateau (solid line) (n = 7) and exponential (dotted line) (n = 8) cells. The spectra have been normalized to the integrated intensity from 1437 to 1453 cm−1. (b) Difference spectrum for the averages of M1 plateau minus M1 exponential cells, with its standard deviation, is shown.
TABLE 3.
Spectral regions with nonoverlapping standard deviations for M1 cells
| Region (cm−1) | Assignment | Change* | t-test† |
|---|---|---|---|
| 781–790 | Cytosine, uracil | Negative | 99.5 |
| 842–855 | Tyrosine, lipid | Positive | 99.5 |
| 887–905 | CH2 rock (lipid), structural protein | Positive | 99.5 |
| 1055–1069 | C-C stretch (lipid) | Positive | 99.5 |
| 1162–1184 | Aromatic amino acids, C-C stretch | Negative | 99.5 |
| 1185–1211 | Aromatic amino acids | Negative | 99.5 |
| 1212–1290 | Amide III, nucleic acid O-P-O | Negative | 99.5 |
| 1291–1324 | CH2 twist (protein, lipid), nucleic acids | Negative | 99.5 |
| 1325–1424 | CH2 twist and bend, CH3 deformation (protein, lipid), nucleic acids | Negative | 99.5 |
| 1562–1573 | Guanine, adenine | Negative | 99.5 |
| 1656–1690 | Amide I | Negative | 99.5 |
The “change” is defined as M1 plateau minus M1 exponential cells.
The Student's t-test results give the percent of confidence that the means of the spectral regions are different.
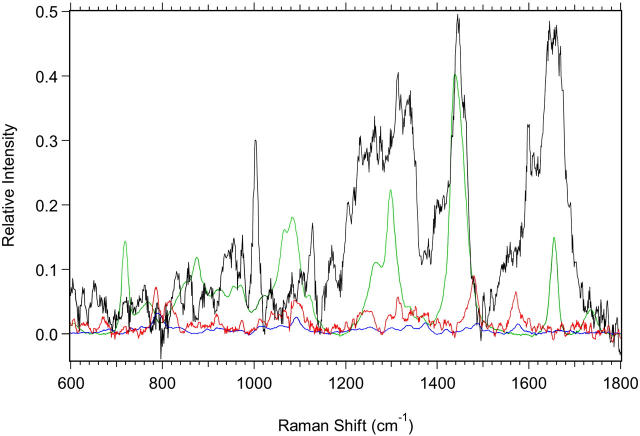
FIGURE 11.
Spectra of the biochemical components used in the fits are shown. The components are scaled to reflect the relative amounts used in the fit of M1 cells shown in Fig. 10. Black, protein; green, lipid; red, RNA; and blue, DNA.
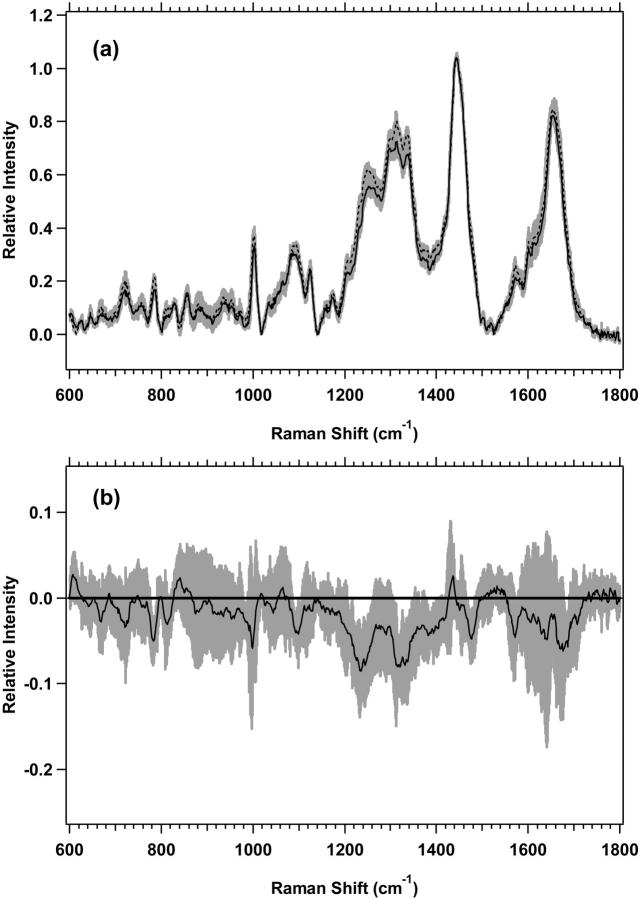
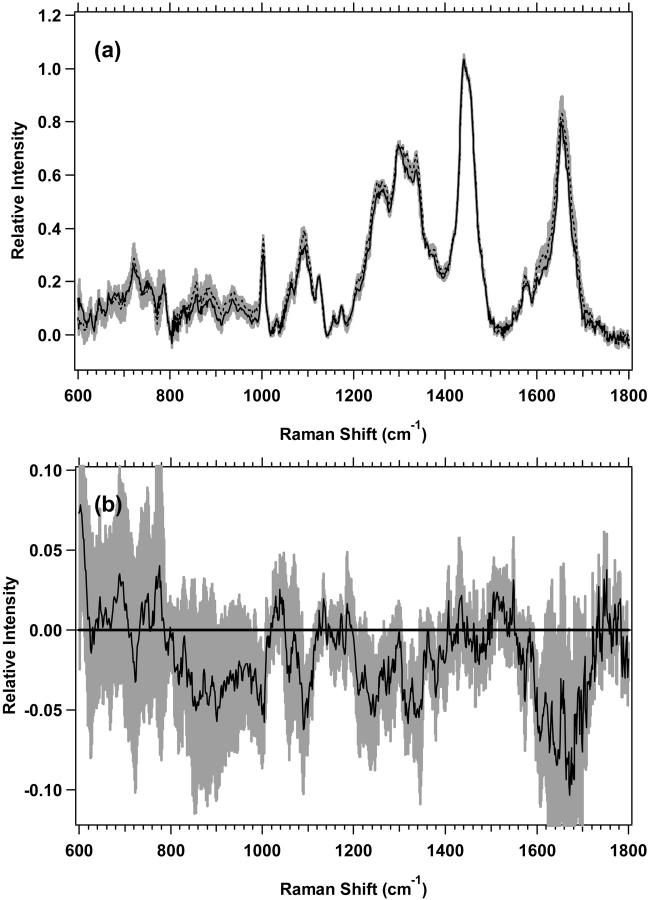
The low-wavenumber region spectra for MR1 plateau and exponential proliferative-state cells are shown in Fig. 3 a, with the difference spectrum of the averages shown in Fig. 3 b. The changes in Fig. 3 b, with nonoverlapping standard deviations, which are shown to be different by Student's t-test at >99.0% confidence, are listed in Table 4. The magnitude of the changes is smaller than for M1 cells, and again, there are many overlaps in the changes of protein, lipid, and nucleic acids. The difference spectra of the averages for both M1 and MR1 cells (Figs. 2 b and 3 b) have other vibrational bands that appear to change, but they have overlapping standard deviations between the plateau and exponential states and have lower confidence levels of difference by Student's t-test.
FIGURE 3.
(a) Standard deviations (shaded area) overlying the averages are shown from 600 to 1800 cm−1 for MR1 plateau (solid line) (n = 8) and exponential (dotted line) (n = 7) cells. The spectra have been normalized to the integrated intensity from 1437 to 1453 cm−1. (b) Difference spectrum for the averages of MR1 plateau minus MR1 exponential cells, with its standard deviation, is shown.
TABLE 4.
Spectral regions with nonoverlapping standard deviations for MR1 cells
| Region (cm−1) | Assignment | Change* | t-test† |
|---|---|---|---|
| 776–790 | Cytosine, uracil | Negative | 99.5 |
| 980–988, 1000–1003 | Phenylalanine | Negative | 99.5 |
| 1087–1105 | C-C, nucleic acid O-P-O | Negative | 99.0 |
| 1205–1219 | Aromatic amino acids | Negative | 99.5 |
| 1220–1260 | Amide III, nucleic acid O-P-O | Negative | 99.0 |
| 1308–1347 | CH2 twist and bend (protein, lipid), nucleic acids | Negative | 99.5 |
| 1376–1398 | CH3 deformation | Negative | 99.5 |
| 1466–1482, 1572–1577 | Guanine, adenine | Negative | 99.5 |
| 1679–1690 | Amide I | Negative | 99.5 |
The “change” is defined as MR1 plateau minus MR1 exponential cells.
The Student's t-test results give the percent of confidence that the means of the spectral regions are different.
High-wavenumber data
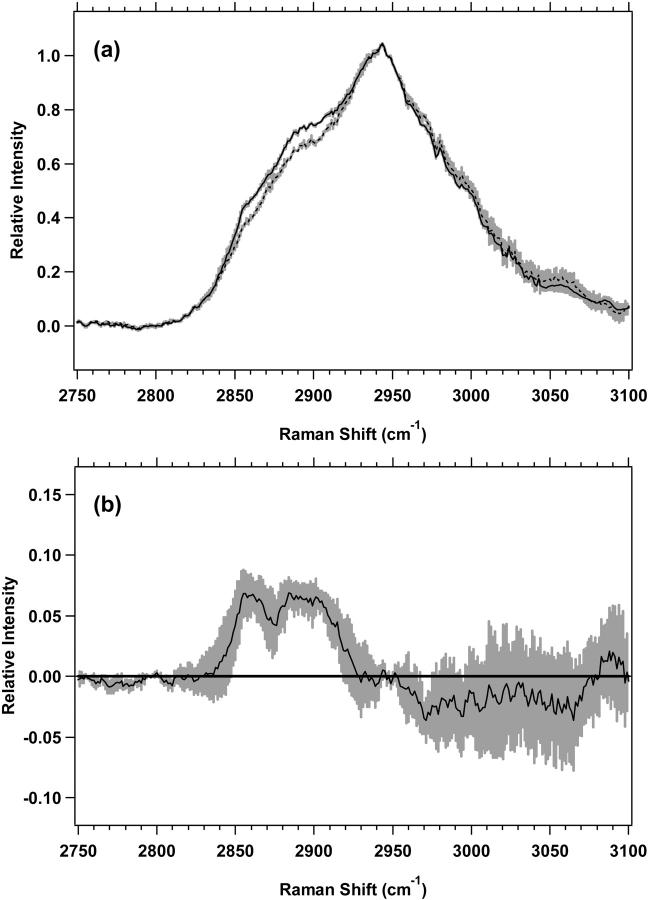
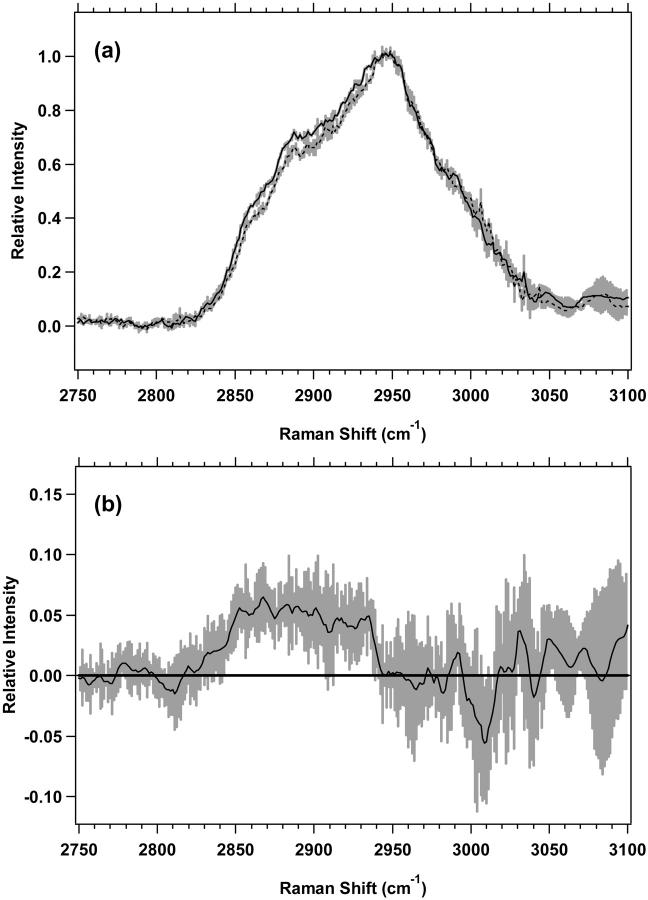
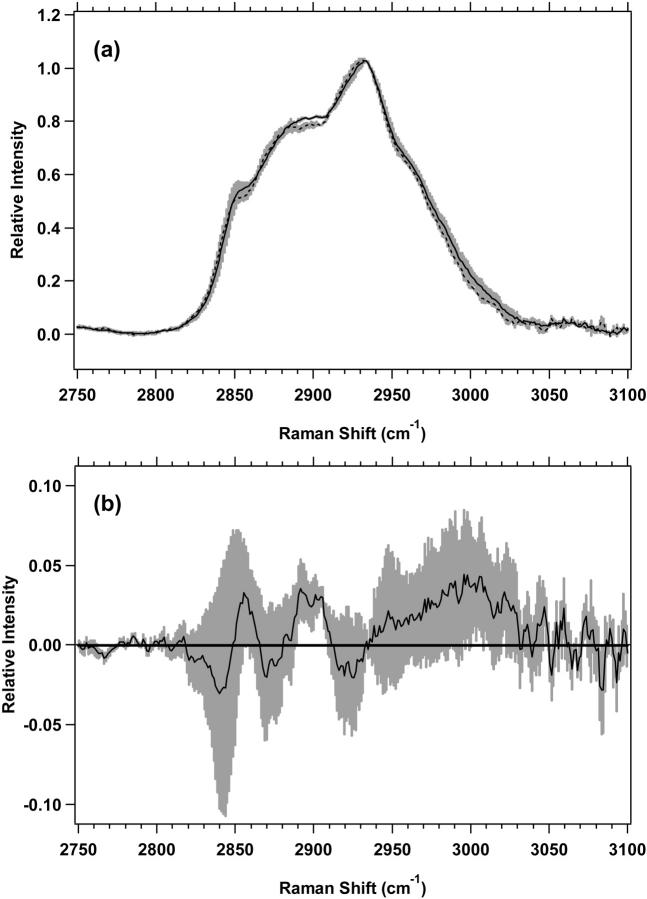
The high-wavenumber spectra of the plateau and exponential cell cultures for M1 and MR1 cells are shown in Figs. 4 a and 5 a, respectively. The difference spectra, in Figs. 4 b and 5 b further emphasize the changes between plateau and exponential cell cultures. The differences are similar for both M1 and MR1 cells. For M1 cells (Fig. 4 b), there are two distinct positive components present with nonoverlapping standard deviations. First, there is a band between 2850 and 2875 cm−1 assigned to the CH2 symmetric stretch of lipids. The second, broader band, between 2876 and 2919 cm−1, is assigned to the CH2 asymmetric stretch near 2883 cm−1 and the CH stretch near 2900 cm−1, both of which contain contributions from lipids and proteins. For MR1 cells, (Fig. 5 b), the areas of nonoverlapping standard deviations are split between two regions, 2842–2906 and 2913–2938 cm−1. The vibrational assignments are similar to M1 cells. The region 2842–2875 cm−1 is largely the CH3 symmetric stretch of lipids, the region 2876–2906 is mostly the CH2 asymmetric stretch and the CH stretch of lipids and proteins, and the region 2913–2938 cm−1 is assigned to the CH stretch of lipids and proteins. These assignments correspond to those used previously (Barry et al., 1992; Lawson et al., 1998; Venkatakrishna et al., 2001).
FIGURE 4.
(a) Standard deviations (shaded area) overlying the averages are shown from 2750 to 3100 cm−1 for M1 plateau (solid line) (n = 7) and exponential (dotted line) (n = 7) cells. The spectra have been normalized to the integrated intensity from 2929 to 2940 cm−1. (b) Difference spectrum for M1 plateau minus M1 exponential cells, with its standard deviation, is shown.
FIGURE 5.
(a) Standard deviations (shaded area) overlying the averages are shown from 2750 to 3100 cm−1 for MR1 plateau (solid line) (n = 7) and exponential (dotted line) (n = 7) cells. The spectra have been normalized to the integrated intensity from 2929 to 2940 cm−1. (b) Difference spectrum for MR1 plateau minus MR1 exponential cells, with its standard deviation, is shown.
Isolated nuclei data
When looking for spectral changes due to differences in the proliferative state of the cell cultures, it is informative to determine if the detected changes also occur in cell nuclei or are limited to the cytoplasmic region. Average spectra of the low-wavenumber region for both plateau and exponential M1 and MR1 nuclei are shown in Figs. 6 a and 7 a, respectively. The difference spectra of the averages are shown in Figs. 6 b and 7 b, respectively. Vibrational regions, in which the difference has nonoverlapping standard deviations, and which are shown to be different by Student's t-test at >95% confidence, are listed in Table 5. The data suggest that between plateau and exponential nuclei there are changes in protein, nucleic acids, and lipids for both M1 and MR1 nuclei. In all but a couple of instances, such as for phenylalanine, the changes are in regions of overlapping vibrational bands. Average spectra of the high-wavenumber regions of M1 and MR1 cells, for both plateau and exponential nuclei, are shown in Figs. 8 a and 9 a, respectively. The difference spectra are shown in Figs. 8 b and 9 b. Both of these figures show changes in the high-wavenumber region between nuclei of the two proliferative states. For M1 plateau nuclei there is an increase in the average intensity from 2889 to 2908 cm−1 that can be assigned to the CH2 asymmetric stretch of lipids and proteins. For MR1 nuclei there are small increases in mean intensity for plateau nuclei from 2817 to 2849 cm−1 assigned to the CH2 symmetric stretch of lipids, and from 2853 to 2881 cm−1 assigned to the CH2 symmetric stretch of lipids and the CH2 asymmetric stretch of lipids and proteins.
FIGURE 6.
(a) Standard deviations (shaded area) overlying the averages are shown from 600 to 1800 cm−1 for M1 plateau (solid line) (n = 3) and exponential (dotted line) (n = 3) nuclei. The spectra have been normalized to the integrated intensity from 1437 to 1453 cm−1. (b) Difference spectrum for M1 plateau minus M1 exponential nuclei, with its standard deviation, is shown.
FIGURE 7.
(a) Standard deviations (shaded area) overlying the averages are shown from 600 to 1800 cm−1 for MR1 plateau (solid line) (n = 3) and exponential (dotted line) (n = 3) nuclei. The spectra have been normalized to the integrated intensity from 1437 to 1453 cm−1. (b) Difference spectrum for MR1 plateau minus MR1 exponential nuclei, with its standard deviation, is shown.
TABLE 5.
Spectral regions with nonoverlapping standard deviations for M1 and MR1 nuclei
| Region (cm−1) | Assignment | Change* | t-test† |
|---|---|---|---|
| M1 Nuclei | |||
| 653–673 | Guanine | Negative | 99.0 |
| 763–779 | Cytosine, uracil | Negative | 97.5 |
| 789–861 | Nucleic acids, protein | Positive | 99.5 |
| 1000–1009 | Phenylalanine | Positive | 99.0 |
| 1049–1088 | Lipid C-C stretch, nucleic acid O-P-O | Positive | 99.0 |
| 1212–1266 | Amide III, nucleic acid O-P-O | Negative | 99.5 |
| 1278–1293, 1308–1338 | CH2 twist and bend (protein, lipid), nucleic acids | Negative | 95, 99.5 |
| 1565–1573 | Guanine, adenine | Negative | 99.5 |
| MR1 Nuclei | |||
| 986–1006 | Phenylalanine | Negative | 99.0 |
| 1089–1111 | Nucleic acid O-P-O | Negative | 95.0 |
| 1229–1264 | Amide III, nucleic acid O-P-O | Negative | 95.0 |
| 1305–1342 | CH2 twist and bend (protein, lipid), nucleic acids | Negative | 99.5 |
| 1594–1617 | Aromatic amino acids | Negative | 95.0 |
| 1673–1681 | Amide I | Negative | 95.0 |
The “change” is defined as M1/MR1 plateau minus M1/MR1 exponential nuclei.
The Student's t-test results give the percent of confidence that the means of the spectral regions are different.
FIGURE 8.
(a) Standard deviations (shaded area) overlying the averages are shown from 2750 to 3100 cm−1 M1 plateau (solid line) and exponential (dotted line) nuclei. The spectra have been normalized to the integrated intensity from 2929 to 2940 cm−1. (b) Difference spectrum for M1 plateau minus M1 exponential nuclei, with its standard deviation, is shown.
FIGURE 9.
(a) Standard deviations (shaded area) overlying the averages are shown from 2750 to 3100 cm−1 MR1 plateau (solid line) and exponential (dotted line) nuclei. The spectra have been normalized to the integrated intensity from 2929 to 2940 cm−1. (b) Difference spectrum for MR1 plateau minus MR1 exponential nuclei, with its standard deviation, is shown.
Spectral fitting
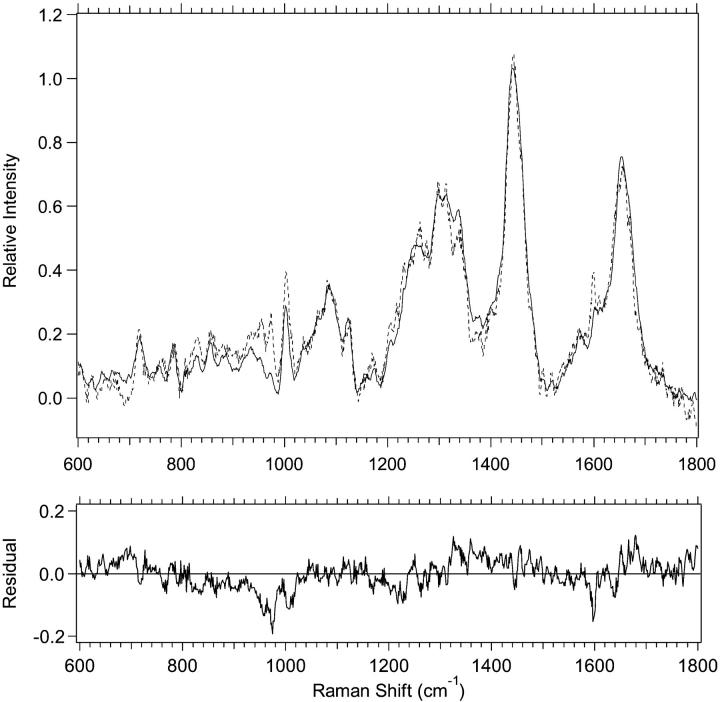
With a basis set of component spectra, it is possible, by minimizing χ2, to fit the cell and nuclei spectra and thereby estimate the relative amounts of lipid, protein, RNA, and DNA. Fig. 10 shows an example of a fit for plateau M1 cells along with the residual of the fit. The quality of this fit is typical of most of our fits. With the method used here we sometimes have difficulty fitting portions of the region between ∼820 and 1000 cm−1. This may be partially due to imperfect subtraction of contributions from the sample cell and optical components that are present in this region. Also, although there are contributions from all cell components in this region, there are many contributions from protein components. Due to the method of protein isolation, our basis spectrum may not have some proteins that contribute to cell spectra in this region. If these “missing” proteins have a high proportion of tyrosine, proline, hydroxyproline, and phenylalanine, it would affect the quality of our fits. Table 6 lists averages and standard deviations for relative amounts of the biochemical components obtained by fits of the low-frequency region. Spectra of the basis-set components scaled to represent their contributions to the fit in Fig. 10 are shown in Fig. 11. The relative amounts of lipid, protein, and RNA are found to have differences outside of the errors between the plateau and exponential proliferative states for M1 cells. Although the trends are the same for MR1 cells, only RNA shows changes outside of experimental error. There are also significant changes in the relative amounts of lipid, protein, and RNA for M1 nuclei between the plateau and exponential proliferative states. For MR1 nuclei, none of the changes are outside of experimental error, although there are suggestive trends for lipid, protein, and DNA. Another way to look at differences between plateau and exponential cells and nuclei is to consider changes in the ratios of the various components, listed in Table 7, along with their errors. There are changes outside the errors between the plateau and exponential proliferative states in the protein/lipid and lipid/RNA ratios for both M1 and MR1 cells. For M1 nuclei, the protein/lipid and lipid/RNA ratios show changes. Finally, for MR1 nuclei the protein/lipid and lipid/DNA ratios show changes outside the errors. Other trends for the ratios between plateau and exponential proliferative-state cells and nuclei can be noted but they are not outside the errors.
FIGURE 10.
Example of a spectral fit and its residual are shown. The experimental data from 600 to 1800 cm−1 for M1 plateau cells (solid line) and the spectral fit using biochemical components (dotted line) are shown.
TABLE 6.
Relative amounts of biochemical components from spectral fitting
| Cells | Lipid | Protein | DNA | RNA |
|---|---|---|---|---|
| M1P* | .425 ± .050† | .483 ± .034 | .018 ± .012 | .073 ± .013 |
| M1E | .253 ± .031 | .595 ± .028 | .040 ± .022 | .110 ± .018 |
| MR1P | .314 ± .042 | .577 ± .033 | .043 ± .021 | .066 ± .018 |
| MR1E | .244 ± .032 | .593 ± .028 | .042 ± .023 | .112 ± .022 |
| Nuclei | ||||
| M1P | .391 ± .016 | .425 ± .023 | .149 ± .033 | .035 ± .007 |
| M1E | .344 ± .004 | .460 ± .006 | .139 ± .013 | .057 ± .014 |
| MR1P | .409 ± .022 | .482 ± .006 | .068 ± .011 | .042 ± .014 |
| MR1E | .371 ± .019 | .513 ± .027 | .082 ± .005 | .034 ± .032 |
Abbreviations as in Table 1.
The values have been normalized for the different proliferative states of the two cell lines. Bold type is used where the difference between the plateau and exponential proliferative state is outside the error bars.
TABLE 7.
Ratios of biochemical components from spectral fitting
| Cells | Lipid/DNA | Lipid/RNA | Protein/Lipid | Protein/DNA | Protein/RNA | RNA/DNA |
|---|---|---|---|---|---|---|
| M1P* | 23.611 ± 15.984 | 5.822 ± 1.243† | 1.136 ± 0.156 | 26.833 ± 18.438 | 6.616 ± 1.267 | 4.056 ± 2.799 |
| M1E | 6.325 ± 3.564 | 2.300 ± 0.470 | 2.352 ± 0.309 | 14.875 ± 8.211 | 5.409 ± 0.950 | 2.750 ± 1.578 |
| MR1P | 7.302 ± 3.697 | 4.758 ± 1.445 | 1.838 ± 0.267 | 13.419 ± 6.840 | 8.742 ± 2.436 | 1.535 ± 0.859 |
| MR1E | 5.809 ± 3.271 | 2.179 ± 0.515 | 2.430 ± 0.325 | 14.119 ± 7.762 | 5.295 ± 1.070 | 2.667 ± 1.552 |
| Nuclei | ||||||
| M1P | 2.624 ± 0.591 | 11.171 ± 2.280 | 1.087 ± 0.074 | 2.852 ± 0.650 | 12.143 ± 2.516 | 0.235 ± 0.070 |
| M1E | 2.475 ± 0.233 | 6.035 ± 1.484 | 1.337 ± 0.023 | 3.309 ± 0.312 | 8.070 ± 1.985 | 0.410 ± 0.108 |
| MR1P | 6.015 ± 1.025 | 9.738 ± 3.288 | 1.178 ± 0.067 | 7.088 ± 1.150 | 11.476 ± 3.828 | 0.618 ± 0.229 |
| MR1E | 4.524 ± 0.360 | 10.912 ± 10.285 | 1.383 ± 0.102 | 6.256 ± 0.504 | 15.088 ± 14.234 | 0.415 ± 0.391 |
Abbreviations as in Table 1.
Bold type is used where the difference between the plateau and exponential proliferative state is outside the error bars.
As a check of our method, mixtures of components were made, their spectra collected, and their relative compositions determined using the spectral fitting method described here. In one mixture the ratios of BSA/DNA/RNA/lipid were 2:1:1:0, the measured ratios from the fits 2:0.91:1.18:0. A second mixture had BSA/DNA/RNA/egg phosphatidylcholine ratios of 4:4:4:1, with ratios from the fit of 4:3.95:3.91:3.64. This second solution was visually observed to be highly scattering of light. Finally, a third mixture had BSA/nucleic acid/liver total lipid extract ratios of 6:1:2, with those from the fit of 6:1.08:2.22. This mixture was visually less scattering of light than the mixture containing egg phosphatidylcholine. The third mixture also had higher background fluorescence than the first two mixtures and so lower signal to noise. This caused the nucleic acid portion of the fit to be weighted almost entirely toward DNA, but the total nucleic acid component of the fit was near the expected value.
One other point deserves comment. There are differences, apparent in Tables 6 and 7, between the nontumorigenic (M1) and the tumorigenic (MR1) cell lines. We will discuss these differences and their meaning along with additional data from other nontumorigenic and tumorigenic cell lines in a future publication (Mourant et al., 2005).
Single experiment results
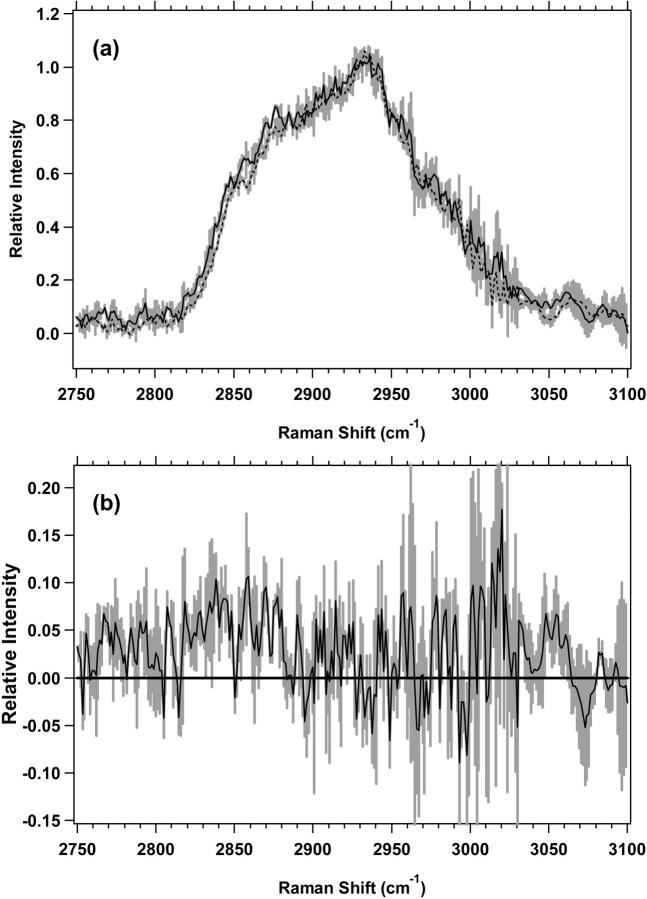
In Figs. 2 a and 3 a, low-wavenumber average spectra for seven or eight experiments are shown along with their overlaid standard deviations. These standard deviations for day-to-day variation between different cell preparations are small but not insignificant. When a single experiment is considered, the results are somewhat different. Fig. 12 shows the average, with the standard deviation overlaid, of 10 consecutive 5-min scans of a single MR1 exponential proliferative-state cell preparation. It is apparent that the variation between scans of a single cell preparation is much smaller than the variation between the average spectra of different cell preparations, indicating that most of the variation seen in our Raman data is due to variation in the cell preparations. This high degree of scan-to-scan reproducibility will likely be important during future in vivo experiments when adjacent sections of normal and abnormal tissue are sampled with a Raman probe.
FIGURE 12.
Standard deviation (shaded area) overlying the average (black) for a single 10-scan experiment of MR1 exponential cells is shown.
DISCUSSION
Changes in the cells
Changes in the Raman spectra that can be attributed to differences in the proliferative status of the cells are clearly evident in Figs. 2–5. Many of the changes were reproducible in all experiments. Consider first the differences between the plateau and exponential M1 cells. Fig. 2 b and Table 3 show that there are negative changes in the difference for plateau minus exponential spectra due to changes in vibrational regions assigned to nucleic acids and proteins. This indicates that the relative amounts of nucleic acids and proteins increase in the proliferating cells. High-wavenumber spectra in Fig. 4, a and b, suggest that the relative amount of lipid is higher in plateau cells and that there is likely a change in the amount of protein. These conclusions are supported by the results in Table 6, which show increases in the spectral contribution of protein and RNA, and a decrease in the spectral contribution of lipid between the plateau and the exponential M1 cells. Additional support is given in Table 7 by a decrease in the lipid/RNA ratio and an increase in the protein/lipid ratio between plateau and exponential M1 cells.
The difference spectrum for MR1 cells, in Fig. 3 b, shows that the changes between the plateau and exponential proliferative state are similar to those found for the M1 cell line. However, the spectral changes appear smaller than for M1 cells. Many bands showing changes, as listed in Table 4, are partially due to changes in the nucleic acids. This indicates that the relative amount of nucleic acids is higher in the exponential cells. The results in Tables 6 and 7 show that this is true, as the amount of RNA increases and the RNA/lipid ratio increases for MR1 exponential cells. Increased RNA content in proliferating cells has been demonstrated previously using flow cytometry (Frankfurt, 1990). The high-wavenumber MR1 spectra, shown in Fig. 5, a and b, suggest that the relative amount of lipid is lower in the exponential cells. The averages of the component fits in Table 6 suggest this is true, although the change is not outside the errors. However, a decrease in the lipid/RNA ratio and an increase in the protein/lipid ratio show that there is likely a decrease in the relative amount of lipid between plateau and exponential cells. The fact that the trends of the averages in Table 6 for lipid and protein are not outside of error agrees with the empirical observation that the spectral changes are smaller for MR1 cells than for M1 cells between the plateau and exponential proliferative states.
Changes in the nuclei
The data for M1 nuclei in Fig. 6 b and Table 5 suggest that there is an increase in nucleic acids and a decrease in lipid between plateau and exponential nuclei. In Fig. 8 b, the increase of intensity between 2889 and 2908 cm−1 also suggests a decrease in lipid. It is unclear what happens to the protein, since the positive change in the phenylalanine at 1003–1009 cm−1 and in the high-wavenumber region in Fig. 8 b suggest a decrease, and the negative change in the 1216–1252 cm−1 region containing amide III intensity suggests an increase. The spectral fitting results in Table 6 show that there is indeed a decrease in the relative amount of lipid and an increase in the relative amount of RNA for M1 exponential nuclei. The fits also show an increase in the relative amount of protein for M1 exponential nuclei. Ratios of components in Table 7 give further evidence of these changes since the lipid/RNA ratio decreases and the protein/lipid ratio increases between M1 plateau and exponential nuclei.
For MR1 nuclei, Fig. 7 b and Table 5 suggest that the relative amount of both protein and nucleic acids increases for exponential nuclei. The spectral region containing lipid intensity at 1305–1342 cm−1 suggests an increase, and changes in the high-wavenumber region of Fig. 9 b at 2817–2849 cm−1 suggests a decrease, in the amount of lipid. For the spectral fits in Table 6, although the standard deviations are very slightly overlapping, the averages show a decrease in the amount of lipid and an increase in the amounts of protein and DNA between MR1 plateau and exponential nuclei. In Table 7 a decrease in the lipid/DNA ratio and an increase in the protein/lipid ratio agrees with these suggested changes between MR1 plateau and exponential nuclei.
Raman spectroscopy is also able to detect differences in the biochemical composition between whole cells and nuclei. M1 plateau and exponential nuclei show a decrease in the relative amount of protein and RNA, and an increase in the relative amount of DNA when compared to whole cells (Table 6). M1 exponential nuclei also show an increase in relative lipid content when compared to whole cells. For MR1 plateau and exponential nuclei there is an increase in relative lipid and a decrease in relative protein compared to whole cells. MR1 exponential nuclei also have increased relative DNA and decreased relative RNA compared to whole cells. Many changes in the ratios of cellular components (Table 7) between whole cells and nuclei are also observed. When compared to whole cells, M1 plateau and exponential nuclei show decreased lipid/DNA, protein/DNA, and RNA/DNA ratios, and an increased lipid/RNA ratio. M1 plateau nuclei also show an increased protein/RNA ratio and M1 exponential nuclei show a decreased protein/lipid ratio. MR1 plateau nuclei show a decreased protein/lipid ratio and an increased lipid/RNA ratio, and MR1 exponential nuclei show decreased protein/lipid and RNA/DNA ratios when compared to whole cells.
Biochemical component analysis
The results of the biochemical component analysis, in Tables 6 and 7, agree in general with a qualitative analysis of the spectra, but there are some important advantages. Visual inspection of the spectra and of the difference spectra shows that there are clearly changes and that they can often be assigned to specific vibrational regions. However, the complexity of the spectra and the large number of overlapping bands in these regions sometimes makes this a process of “guessing”, in which there is likely to be error. That there are many overlapping bands is shown by looking at the spectral contributions of the individual components to the overall Raman signal, which are shown in Fig. 11. The spectra in Fig. 11 also make clear the differences in the contributions of the components to the spectral intensity of the cells. For instance, the small contribution of DNA to the overall intensity and the similarity of the RNA and DNA spectra make it more likely that any spectral changes attributed to nucleic acids are due to changes in relative RNA content. When visual inspection is done, the complexity of the spectra obscures important changes that occur. For example, although the low wavenumber region contains many lipid bands, visual inspection gives very little indication that there is a difference in the relative amount of lipid between the plateau and exponential proliferative states. Although the high-wavenumber region may suggest that there is a lipid change, there are overlapping protein contributions. The biochemical component analysis clearly shows the lipid change. A second example involving the nucleic acids clearly shows that many of the changes reported in Table 6 would be difficult to assign simply from an analysis of spectral band differences. Visual inspection of the spectra (Figs. 2, 3, and 6) suggests that the nucleic acids, particularly for M1 cells and nuclei and MR1 cells, change between the plateau and exponential proliferative states, but are the relative changes greater in the DNA or in the RNA? Component analysis is able to show that the changes are greater for RNA. Thus, the real advantage of this analysis is that it provides a quantitative means of determining changes in the relative amounts of cellular components and in their ratios. It therefore provides an easy and noninvasive way of elucidating the biochemical differences among different spectra from cells or tissue that may only have subtle biochemical differences. This is an advantage over methods such as PCA analysis (Haka et al., 2002), PCA combined with Fisher discriminant analysis (Mahadevan-Jansen et al., 1998), or PCA combined with LDA or cluster analysis (Deinum et al., 1999; Venkatakrishna et al., 2001), which can determine that there are components of the spectra changing but cannot quantify, and sometimes cannot identify, the biochemical changes.
Many of the changes noted in Table 6 correspond to expected biochemical alterations. Exponential cells are expected to have a higher relative protein, RNA, and DNA content than plateau cells. We observe the differences for protein and RNA, but not for DNA, likely because the relative amount of DNA in the cell lines used here is small compared to the other cellular components (see Fig. 11). Isolated nuclei are expected to have a higher relative DNA content and lower relative RNA content than whole cells since a large fraction of cellular RNA is in the ribosomes. These changes are observed, except for MR1 plateau cells, where the error bars of the relative amounts overlap. We are currently measuring the DNA, RNA, and protein content of the cells using both flow cytometric and biochemical techniques for comparison with the Raman data.
Spectral fits of component mixtures
When our method of biochemical component analysis is used to analyze artificial mixtures of the components, the results compare favorably to those expected. In the three mixtures tested the amounts of protein and total nucleic acids in the spectral fits were near expected values. When there were higher amounts of DNA and RNA, and lower background fluorescence, their individual amounts in the analysis were as expected. At lower signal to noise, for the artificial mixtures it was found that the fits tended to be weighted more toward DNA than RNA. This is likely related to the high similarity between the basis spectra of RNA and DNA. So at low signal to noise, spectral differences that could distinguish between them, such as near 800 cm−1, may be less prominent. The amounts of lipid in the fits were near expected values when liver total lipid extract was used, but were consistently too high when egg phosphatidylcholine was used. How lipid content is overestimated can be understood as follows. We have experimental evidence that for our experimental setup the Raman signal from a turbid sample is reduced relative to the Raman signal of a clear sample that contains the same amount of biochemical components. For example, assume there are two components, and component 1 is clear whereas component 2 is turbid. Both components are measured at a concentration of 10 mg/ml; then a sample is made that contains 5 mg/ml of each sample. The sample will be turbid because the second component is turbid. Consequently, the fit coefficient for the basis spectra for component 2 will be ∼0.5 because the turbidity is similar to that of component 2. However, the fit coefficient for component 1 will be <0.5 because the sample is more turbid than that of component 1 by itself. Thus, the estimated relative amount of component 1 will be less than that of component 2. Given this analysis, it is possible Raman spectroscopy may in certain situations slightly overestimate the relative amount of lipid in cells. Further evidence for this conclusion comes from simultaneous Raman and infrared spectroscopy studies of fibroblast cell preparations in which infrared spectroscopy shows slightly lower amounts of lipid in the cells than does Raman spectroscopy (Mourant et al., 2004). There is not a problem with turbidity in the infrared experiments because the measurement path is only 50 μm.
Prospects for using Raman spectroscopy as a diagnostic tool
We have shown that there is an advantage in using biochemical component analysis, instead of simply analyzing spectral bands, to detect cellular markers that change between plateau and exponential cells. This is because the individual spectral bands of the cells may overlap with other bands in the spectrum. In vivo, because of the intercellular matrix, circulating blood, or bone (Bakker Schut et al. 2000), the Raman spectra will be even more complex and have more overlapping bands than we observe here (although in vivo the spectral contribution from water will be smaller than we observe in our cell suspensions). It is in this situation that biochemical component analysis may quickly be able to characterize any differences in the relative amounts of components present between two areas of the tissue.
For each of the cell cultures grown for this study we have measured the average volume of the cells and the number of cells/ml in the dispersions. A minimum excitation volume is approximated as a cylinder with a diameter of 55 μm, equal to the minimum laser spot size, and with a minimum length of 60 μm. The minimum spot size was determined by taking into account the transverse spherical aberration. The minimum length was made the same as the longitudinal spherical aberration for our setup, which is the distance along the optical axis between where the marginal rays and the paraxial rays from our detection lens are focused. Experimentally, the laser at the focus has a measured diameter of 55–60 μm and there is detectable Raman signal along ∼1 mm of the optical axis, with the maximum signal at the focus. Using the minimum spot size and minimum length, a minimum excitation volume (in mm3) can be determined and multiplied by the number of cells/mm3, to give the number of cells in the excitation volume. In 26 experiments the lower bound of this quantity varied between 15 and 47 cells in the excitation volume. Using the experimental parameters, the upper bound would be between 640 and 1960 cells making some contribution to the Raman signal. The number of cells in the excitation volume can be multiplied by the volume/cell and divided by the excitation volume (both in μm3), and multiplied by 100 to give a percent of the excitation volume filled by cells. Again, for 26 experiments, 16–49% of the excitation volume was filled with cells. The high value is near the lower end of comparable values for normal tissues and tumors. So for comparable signal to noise, we can likely reduce in vivo spectral acquisition times below those used here. Improvements in probe design (Motz et al., 2004) and smoothing of spectra would likely further reduce the necessary acquisition time.
Recently, the Feld group reported a component analysis study of normal and abnormal breast tissue (Shafer-Peltier et al. 2002). Their approach differs from ours in that the analysis does not consider the biochemical composition of the cells independently of the extracellular matrix. The cell nucleus and cell cytoplasm are considered to be unvarying components of a seven-component basis set. In their studies, this may be an appropriate approximation, as their spectral resolution appears lower than in the spectra reported here, and because there are apparently very large tissue morphology changes in the transformation from normal to cancerous breast tissue. However, we have shown here that Raman spectroscopy is able to detect small changes within cells and their nuclei that may become important in a diagnostic situation where these large morphological changes do not exist.
Our group has recently reported a Fourier transform infrared study of M1 and MR1 cells that considered the biochemical differences between plateau and exponential proliferative-state cells (Mourant et al., 2003a). In that study, the amount of biochemical components and their ratios to each other were determined. Those results compare quite favorably with the data reported here. The relative amounts of different biochemical components show trends the same as those reported in Table 6 of this study. In both studies, the relative amount of lipid decreases in the exponential state, the relative amount of RNA increases in the exponential state, the relative amount of protein is lowest for M1 plateau cells, and the relative amount of DNA changes very little between the plateau and exponential states. Also, both studies show changes outside the error bars in the relative amounts of lipid that decrease and amounts of protein that increase between the plateau and exponential proliferative state for M1 cells. These observations further support the use of vibrational spectroscopy as a viable method to monitor biochemical changes in cells and tissue.
One problem with using Raman spectroscopy as a quantitative tool in biochemical analysis has been the somewhat subjective methods used to subtract background fluorescence from the spectra. These methods, whether instrumentational or mathematical, normally require some subjective user intervention. There has been one recent report of an automated method for fluorescence subtraction based on modification of least-squares polynomial curve fitting (Lieber and Mahadevan-Jansen, 2003). We are currently developing a more automated, less subjective version of the biochemical component analysis method described here (Mourant et al., 2004). It involves including in the basis set of components used in the fits spectra of the sample buffer, empty sample cell, and a fourth- or fifth-order polynomial to account for background fluorescence. In this way there is no user intervention in determining the amounts of background spectra to subtract from the cell spectra. The method, which includes glycogen as a component and uses both low- and high-wavenumber regions, predicts amounts of protein, lipid, and total nucleic acids to within error of what is reported here, except for protein and lipid for M1 and MR1 exponential cells and lipid for M1 plateau cells. The protein values are higher, and the lipid values are lower using the automated method. In contrast to the method here, the automated method currently does not give the expected biochemical result that there is more RNA than DNA in the cells.
Some of the changes we detect by a fitting to the biochemical components are very subtle. The ability to detect small changes and also their biochemical origins will be very useful in developing Raman spectroscopy for the early detection of cancer. Some earlier studies have looked at changes in vibrational band ratios (Frank and McCreery, 1995; Yazdi et al., 1999; Utzinger et al., 2001) or changes in specific vibrational bands (Mizuno et al., 1994; Hawi et al., 1996; Gniadecka et al., 1997a,b) to classify normal and diseased tissue. In many cases, the spectral changes were larger than we observed here for the cell proliferative state analysis of the M1 and MR1 cell lines. Although this means that it is easy to detect abnormalities, there is also a possibility that analysis of specific bands, or of band ratios, will be skewed by underlying bands in the complex cell or tissue spectra. For example, as shown in Fig. 11, DNA is a very minor contribution to Raman spectra of the cells and consequently assignments of nucleic acid spectral changes to DNA should be made with caution. It has been suggested that PCA combined with K-means clustering analysis is able to detect small changes between normal and diseased epidermal tissue (Nijssen et al., 2002). This ability may prove useful in early cancer diagnosis; however, biochemical component analysis will still be necessary to determine and quantify the underlying biochemical changes. The identification of the biochemical changes and their amounts may also be useful in following the response of a tumor to therapeutic treatment, which would be expected to alter the proliferative state of the cells.
CONCLUSIONS
The results presented here show that Raman spectroscopy can be used to detect biochemical changes that occur as a result of changes in proliferation. Differences can be defined from specific vibrational regions that change, or from a fit of biochemical components to the spectra. However, the component fits can characterize changes that are often obscured by spectral complexity when looking only at specific vibrational regions. When comparing plateau and exponential cells, the component fits show that the relative amounts of protein and RNA increase and that the relative amount of lipid decreases in nontumorigenic exponential cells. For tumorigenic cells, the relative amount of RNA increases in exponential cells. For nontumorigenic exponential nuclei, the fits also show that the relative amount of lipid decreases, and that the relative amounts of protein and RNA increase. The ratio of lipid/RNA decreases and the ratio of protein/lipid increases for both tumorigenic and nontumorigenic exponential cells. Lipid/RNA decreases and protein/lipid increases for nontumorigenic exponential nuclei. Lipid/DNA decreases and protein/lipid increases for tumorigenic exponential nuclei. These changes can potentially be used as markers for the proliferative status of cells.
Acknowledgments
We thank Ms. Pratima Kunapareddy for assistance in collecting spectra of biochemical component mixtures. We are also grateful for assistance from the National Flow Cytometry Resource, Los Alamos National Laboratory, Los Alamos, New Mexico.
This work was funded by grant Nos. CA089255 and RR01315 from the National Institutes of Health.
References
- Bakker Schut, T. C., M. J. H. Witjes, H. J. C. M. Sterenborg, O. C. Speelman, J. L. N. Roodenburg, E. T. Marple, H. A. Bruining, and G. J. Puppels. 2000. In vivo detection of dysplastic tissue by Raman spectroscopy. Anal. Chem. 72:6010–6018. [DOI] [PubMed] [Google Scholar]
- Barry, B. W., H. G. M. Edwards, and A. C. Williams. 1992. Fourier transform Raman and infrared vibrational study of human skin: assignment of spectral bands. J. Raman Spectrosc 23:641–645. [Google Scholar]
- Boere, I. A., T. C. Bakker Schut, J. van den Boogert, R. W. F. de Bruin, and G. J. Puppels. 2003. Use of fibre optic probes for detection of Barrett's epithelium in the rat oesophagus by Raman spectroscopy. Vib. Spectrosc. 32:47–55. [Google Scholar]
- Boustany, N. N., J. M. Crawford, R. Manoharan, R. R. Dasari, and M. S. Feld. 1999. Analysis of nucleotides and aromatic amino acids in normal and neoplastic colon mucosa by ultraviolet resonance Raman spectroscopy. Lab. Invest. 79:1201–1214. [PubMed] [Google Scholar]
- Boydston-White, S., T. Gopen, S. Houser, J. Bargonetti, and M. Diem. 1999. Infrared spectroscopy of human tissue. V. Infrared spectroscopic studies of myeloid leukemia (ML-1) cells at different phases of the cell cycle. Biospectroscopy. 5:219–227. [DOI] [PubMed] [Google Scholar]
- Brennan III, J. F., T. J. Römer, R. S. Lees, A. M. Tercyak, J. R. Kramer, Jr., and M. S. Feld. 1997. Determination of human coronary artery composition by Raman spectroscopy. Circulation. 96:99–105. [DOI] [PubMed] [Google Scholar]
- Choo-Smith, L.-P., H. G. M. Edwards, H. P. Endtz, J. M. Kros, F. Heule, H. Barr, J. S. Robinson, Jr., H. A. Bruining, and G. J. Puppels. 2002. Medical applications of Raman spectroscopy: from proof of principle to clinical implementation. Biopolymers. 67:1–9. [DOI] [PubMed] [Google Scholar]
- Crow, P., N. Stone, C. A. Kendall, J. S. Uff, J. A. M. Farmer, H. Barr, and M. P. J. Wright. 2003. The use of Raman spectroscopy to identify and grade prostatic adenocarcinoma in vitro. Br. J. Cancer. 89:106–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinum, G., D. Rodriguez, T. J. Römer, M. Fitzmaurice, J. R. Kramer, and M. S. Feld. 1999. Histological classification of Raman spectra of human coronary artery atherosclerosis using principal component analysis. Appl. Spectrosc. 53:938–942. [Google Scholar]
- Fendel, S., and B. Schrader. 1998. Investigation of skin and skin lesions by NIR-FT-Raman spectroscopy. Photochem. Photobiol. 66:418–423. [Google Scholar]
- Frank, C. J., and R. L. McCreery. 1995. Raman spectroscopy of normal and diseased human breast tissues. Anal. Chem. 67:777–783. [DOI] [PubMed] [Google Scholar]
- Frankfurt, O. S. 1990. Flow cytometric analysis of double-stranded RNA content distributions. In Methods in Cell Biology, Vol. 33. Z. Darzynkiewicz and H. A. Crissman, editors. Academic Press, San Diego. 299–304. [DOI] [PubMed]
- Freyer, J. P. 1998. Decreased mitrochondrial function in quiescent cells isolated from multicellular tumor spheroids. J. Cell. Physiol. 176:138–149. [DOI] [PubMed] [Google Scholar]
- Gniadecka, M., H. C. Wulf, N. N. Mortensen, O. Faurskov Nielsen, and D. H. Christensen. 1997b. Diagnosis of basal cell carcinoma by Raman spectroscopy. J. Raman Spectrosc. 28:125–129. [Google Scholar]
- Gniadecka, M., H. C. Wulf, O. F. Nielsen, D. H. Christensen, and J. Hercogova. 1997a. Distinctive molecular abnormalities in benign and malignant skin lesions: studies by Raman spectroscopy. Photochem. Photobiol. 66:418–423. [DOI] [PubMed] [Google Scholar]
- Haka, A. S., K. E. Shafer-Peltier, M. Fitzmaurice, J. Crowe, R. R. Dasari, and M. S. Feld. 2002. Identifying microcalcifications in benign and malignant breast lesions by probing differences in their chemical composition using Raman spectroscopy. Cancer Res. 62:5375–5380. [PubMed] [Google Scholar]
- Hawi, S. R., W. B. Campbell, A. Kajdacsy-Balla, R. Murphy, F. Adar, and K. Nithipatikom. 1996. Characterization of normal and malignant human hepatocytes by Raman microspectroscopy. Cancer Lett. 110:35–40. [DOI] [PubMed] [Google Scholar]
- Holman, H.-Y. N., M. C. Martin, E. A. Blakely, K. Bjornstad, and W. R. McKinney. 2000. IR spectroscopic characteristics of cell cycle and cell death probed by synchrotron radiation based Fourier transform IR spectromicroscopy. Biopolymers. 57:329–335. [DOI] [PubMed] [Google Scholar]
- Huang, Y.-S., T. Karashima, M. Yamamoto, and H. Hamaguchi. 2003. Molecular-level pursuit of yeast mitosis by time- and space-resolved Raman spectroscopy. J. Raman Spectrosc. 34:1–3. [Google Scholar]
- Kendall, C., N. Stone, N. Shepherd, K. Geboes, B. Warren, R. Bennett, and H. Barr. 2003. Raman spectroscopy, a potential tool for the objective identification and classification of neoplasia in Barrett's oesophagus. J. Pathol. 200:602–609. [DOI] [PubMed] [Google Scholar]
- Krishan, A. 1975. Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J. Cell Biol. 66:188–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz-Schughart, L. A., A. Simm, and W. Mueller-Klieser. 1995. Oncogene-associated transformation of rodent fibroblasts is accompanied by large morphologic and metabolic alterations. Oncol. Rep. 2:651–661. [DOI] [PubMed] [Google Scholar]
- Lau, D. P., Z. Haung, H. Lui, C. S. Man, K. Berean, M. D. Morrison, and H. Zeng. 2003. Raman spectroscopy for optical diagnosis in normal and cancerous tissue of the nasopharynx – preliminary findings. Lasers Surg. Med. 32:210–214. [DOI] [PubMed] [Google Scholar]
- Lawson, E. E., A. N. C. Anigbogu, A. C. Williams, B. W. Barry, and H. G. M. Edwards. 1998. Thermally induced molecular disorder in human stratum corneum lipids compared with a model phospholipid system: FT-Raman spectroscopy. Spectrochim. Acta A. 54:543–558. [DOI] [PubMed] [Google Scholar]
- Lieber, C. A., and A. Mahadevan-Jansen. 2003. Automated method for subtraction of fluorescence from biological Raman spectra. Appl. Spectrosc. 57:1363–1367. [DOI] [PubMed] [Google Scholar]
- Mahadevan-Jansen, A., M. F. Metchell, N. Ramanujam, S. Malpica, S. Thomsen, U. Utzinger, and R. Richards-Kortum. 1998. Near-infrared Raman spectroscopy for in vitro detection of cervical precancers. Photochem. Photobiol. 68:123–132. [DOI] [PubMed] [Google Scholar]
- Manoharan, R., J. J. Baraga, M. S. Feld, and R. P. Rava. 1992. Quantitative histochemical analysis of human artery using Raman scattering. J. Photochem. Photobiol B Biol. 16:211–233. [DOI] [PubMed] [Google Scholar]
- Mizuno, A., H. Kitajima, K. Kawauchi, S. Muraishi, and Y. Ozaki. 1994. Near-infrared Fourier transform Raman spectroscopic study of human brain tissues and tumors. J. Raman Spectrosc. 25:25–29. [Google Scholar]
- Molckovsky, A., L.-M. W. K. Song, M. G. Shim, N. E. Marcon, and B. C. Wilson. 2003. Diagnostic potential of near-infrared Raman spectroscopy in the colon: differentiating adenomatous from hyperplastic polyps. Gastrointest. Endosc. 57:396–402. [DOI] [PubMed] [Google Scholar]
- Motz, J. T., M. Hunter, L. H. Galindo, J. A. Gardecki, J. R. Kramer, R. R. Dasari, and M. S. Feld. 2004. Optical fiber probe for biomedical Raman spectroscopy. Appl. Opt. 43:542–554. [DOI] [PubMed] [Google Scholar]
- Mourant, J. R., M. Canpolat, C. Brocker, O. Esponda-Ramos, T. M. Johnson, A. Matanock, K. Stetter, and J. P. Freyer. 2000. Light scattering from cells: the contribution of the nucleus and the effects of proliferative status. J. Biomed. Opt. 5:131–137. [DOI] [PubMed] [Google Scholar]
- Mourant, J. R., S. Carpenter, K. W. Short, P. Kunapareddy, L. Coburn, and J. P. Freyer. 2005. Biochemical differences in tumorigenic and non-tumorigenic cells measured by Raman and infrared spectroscopy. J. Biomed. Opt. In press. [DOI] [PubMed]
- Mourant, J. R., R. R. Gibson, T. M. Johnson, S. Carpenter, K. W. Short, Y. R. Yamada, and J. P. Freyer. 2003b. Methods for measuring the infrared spectra of biological cells. Phys. Med. Biol. 48:243–257. [DOI] [PubMed] [Google Scholar]
- Mourant, J. R., Y. R. Yamanda, S. Carpenter, L. R. Dominique, and J. P. Freyer. 2003a. FTIR spectroscopy demonstrates biochemical differences in mammalian cell cultures at different growth stages. Biophys. J. 85:1938–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann, D. 2001. FT-infrared and FT-Raman spectroscopy in biomedical research. Appl. Spectrosc. Rev. 36:239–298. [Google Scholar]
- Nijssen, A., T. C. Bakker Schut, F. Heule, P. J. Caspers, D. P. Hayes, M. H. A. Neumann, and G. J. Puppels. 2002. Discriminating basal cell carcinoma from its surrounding tissue by Raman spectroscopy. J. Invest. Dermatol. 119:64–69. [DOI] [PubMed] [Google Scholar]
- Nunes, L. O., A. A. Martin, L. Silveira, and M. Zampieri. 2003. FT-Raman spectroscopy study for skin cancer diagnosis. Spectrosc. Int. J. 17:597–602. [Google Scholar]
- Omberg, K. M., J. C. Osborn, S. L. Zhang, J. P. Freyer, J. R. Mourant, and J. R. Schoonover. 2002. Raman spectroscopy and factor analysis of tumorigenic and non-tumorigenic cells. Appl. Spectrosc. 56:813–819. [Google Scholar]
- Petrich, W. 2001. Mid-infrared and Raman spectroscopy for medical diagnostics. Appl. Spectrosc. Rev. 36:181–237. [Google Scholar]
- Römer, T. J., J. F. Brennan III, M. Fitzmaurice, M. L. Feldstein, G. Deinum, J. L. Myles, J. R. Kramer, R. S. Lees, and M. S. Feld. 1998. Histopathology of human coronary atherosclerosis by quantifying its chemical composition with Raman spectroscopy. Circulation. 97:878–885. [DOI] [PubMed] [Google Scholar]
- Salenius, J. P., J. F. Brennan III, A. Miller, Y. Wang, T. Aretz, B. Sacks, R. R. Dasari, and M. S. Feld. 1998. Biochemical composition of human peripheral arteries examined with near-infrared Raman spectroscopy. J. Vasc. Surg. 27:710–719. [DOI] [PubMed] [Google Scholar]
- Schuster, K. C., I. Reese, E. Urlaub, J. R. Gapes, and B. Lendl. 2000b. Multidimensional information on the chemical composition of single bacterial cells by confocal Raman microspectroscopy. Anal. Chem. 72:5529–5534. [DOI] [PubMed] [Google Scholar]
- Schuster, K. C., E. Urlaub, and J. R. Gapes. 2000a. Single-cell analysis of bacteria by Raman microscopy: spectral information on the chemical composition of cells and on the heterogeneity in culture. J. Microbiol. Methods. 42:29–38. [DOI] [PubMed] [Google Scholar]
- Shafer-Peltier, K. E., A. S. Haka, M. Fitzmaurice, J. Crowe, J. Myles, R. R. Dasari, and M. S. Feld. 2002. Raman microspectroscopic model of human breast tissue: implications for breast cancer diagnosis in vivo. J. Raman Spectrosc. 33:552–563. [Google Scholar]
- Stone, N., C. Kendall, N. Shepherd, P. Crow, and H. Barr. 2002. Near-infrared Raman spectroscopy for the classification of epithelial pre-cancers and cancers. J. Raman Spectrosc. 33:564–573. [Google Scholar]
- Sutherland, R. M., W. A. Ausserer, B. J. Murphy, and K. R. Laderoute. 1996. Tumor hypoxia and heterogeneity: challenges and opportunities for the future. Semin. Radiat. Oncol. 6:59–70. [DOI] [PubMed] [Google Scholar]
- Tannock, I. F. 2001. Tumor physiology and drug resistance. Cancer Metastasis Rev. 20:123–132. [DOI] [PubMed] [Google Scholar]
- Tannock, I. F., and R. P. Hill. 2004. The basic science of oncology. McGraw-Hill, New York.
- Utzinger, U., D. L. Heintzelman, A. Mahadevan-Jansen, A. Malpica, M. Follen, and R. Richards-Kortum. 2001. Near-infrared Raman spectroscopy for in vivo detection of cervical precancers. Appl. Spectrosc. 55:955–959. [Google Scholar]
- Venkatakrishna, K., J. Kurien, K. M. Pai, M. Valiathan, N. Nagesh Kumar, C. Murali Krishna, G. Ullas, and V. B. Kartha. 2001. Optical pathology of oral tissue: a Raman spectroscopy diagnostic method. Curr. Sci. 80:665–669. [Google Scholar]
- Wolthuis, R., T. C. Bakker Schut, P. J. Caspers, H. P. J. Buschman, T. J. Römer, H. A. Bruining, and G. J. Puppels. 1999. Raman spectroscopic methods for in vitro and in vivo tissue characterization. In Fluorescent and Luminescent Probes, 2nd ed. W. T. Mason, editor. Academic Press, London. 433–455.
- Yazdi, Y., N. Ramanujam, R. Lotan, M. Follen Mitchell, W. Hittelman, and R. Richards-Kortum. 1999. Resonance Raman spectroscopy at 257nm excitation of normal and malignant cultured breast and cervical cells. Appl. Spectrosc. 53:82–85. [Google Scholar]
- Zhang, W., B. Asztalos, P. S. Roheim, and L. Wong. 1998. Characterization of phospholipids in pre-α HDL: selective phospholipid efflux with apolipoprotein A-1. J. Lipid Res. 39:1601–1607. [PubMed] [Google Scholar]