Abstract
The gene encoding CaCse4p, a homolog of the evolutionarily conserved histone H3-like kinetochore protein CENP-A, has been cloned from the human pathogenic diploid yeast Candida albicans. To study the phenotype of C. albicans diploid cells depleted of CaCse4p, we deleted one copy of CaCSE4 and brought the other copy under control of a regulated PCK1 promoter (repressed by glucose and induced by succinate). Inability of this strain to grow on glucose medium indicates that CaCse4p is essential for cell viability. Shutdown of CaCSE4 expression resulted in a sharp decline of CaCse4p levels with concomitant loss of cell viability. Examination of these CaCse4p-depleted cells revealed a mitosis-specific arrest phenotype with accumulation of large-budded cells containing single G2 nuclei at or near the bud neck along with short mitotic spindles. Subcellular localization of CaCse4p by anti-CaCse4p antibodies in both budding and filamentous C. albicans cells revealed an intense dot-like signal always colocalized with 4′,6-diamidino-2-phenylindole-stained nuclei. Unlike higher eukaryotes but similar to the budding yeast Saccharomyces cerevisiae, centromere separation in the budding yeast form of C. albicans occurs before anaphase, at a very early stage of the cell cycle. In the filamentous mode of cell division, however, centromere separation appears to occur in early anaphase. Coimmunostaining with anti-CaCse4p and antitubulin antibodies shows that CaCse4p localizes near spindle pole bodies, analogous to the localization pattern observed for kinetochore proteins in S. cerevisiae. CaCse4p promises to be a highly useful reagent for the study of centromere/kinetochore structure in C. albicans.
The most frequently isolated human fungal pathogen, Candida albicans, causes a wide variety of mucosal and systemic infections as an opportunistic organism in immunocompromised patients (1). It is a diploid, asexual polymorphic fungus that can switch between the unicellular yeast form into two different filamentous forms: pseudohyphae and true hyphae. Although the genome sequence of C. albicans is mostly complete, centromere DNA sequences have not been identified yet. A centromere is a cis-acting DNA locus, present uniquely on each eukaryotic chromosome, and is the site on which a kinetochore is formed. The kinetochore, a complex proteinaceous structure, is responsible for microtubule attachment and sister chromatid cohesion, thus enabling faithful segregation of chromosomes during mitosis and meiosis.
Functional centromere DNA sequences in budding yeasts such as Saccharomyces cerevisiae are contained within only 125–400 bp and are called “point” centromeres (2, 3). However, eukaryotes as diverse as humans, flies, and fission yeast have “regional” centromeres spanning 40–4,000 kb of reiterated DNA (2). Regional centromeres often are associated with short nucleosome-length satellite repeats (such as the α-satellite of humans) present within an untranscribed (AT)-rich region of DNA, whereas point centromeres have conserved DNA sequence elements and an (AT)-rich core (4).
Although centromere regions usually are (AT)-rich and present within untranscribed regions of DNA, no common sequence motif that specifies centromeres has been detected, which raises the possibility that transacting non-DNA determinants could be associated with centromere marking. A common element found to be associated with functional centromeres in all organisms examined to date is a histone H3-variant protein. These histone H3-variant proteins constitute the CENP-A family and are found in specialized nucleosomes at the centromeric regions in eukaryotes ranging from the baker's yeast to humans (5). Human CENP-A is present at all functional centromeres (native or neocentromere) but absent in nonfunctional (mutated or inactivated) centromeres. Inactivation of CENP-A proteins has been shown to severely affect chromosome segregation and cell-cycle progression in yeasts, worms, flies, and mammals (6).
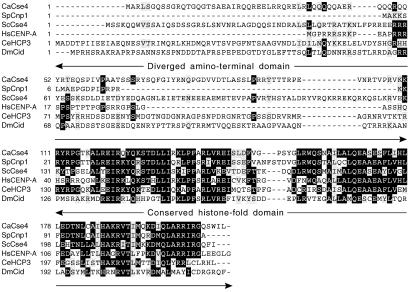
All proteins in the CENP-A family have a highly conserved C-terminal histone H3-like sequence (histone-fold domain) but a dissimilar N-terminal region (Fig. 1; ref. 7). The N terminus of S. cerevisiae CENP-A protein (ScCse4p) is localized outside the centromeric nucleosomal core and interacts with other kinetochore proteins (8). Mutational studies show that at least one essential domain exists in the N terminus of ScCse4p (8, 9). The C-terminal histone-fold domain is an evolutionarily conserved region shared by all four core histones and the CENP-A family members. In humans, the conserved histone-fold domain of CENP-A has been shown to be necessary for centromere targeting (10). ScCse4p directly interacts with histone H4, and mutations in the histone-fold domain of ScCse4p disrupt centromeric chromatin structure (11). Thus, ScCse4p has been proposed to replace at least one histone H3 molecule at the centromeric nucleosome. Moreover, CENP-A proteins are essential for the hierarchical assembly of other proteins at the centromere to initiate kinetochore formation (6).
Figure 1.
Amino acid sequence alignment of the histone H3-like proteins (Cse4p/CENP-A) of various organisms: C. albicans CaCse4, S. cerevisiae ScCse4, S. pombe SpCnp1, C. elegans CeHCP3, Drosophila melanogaster DmCid, and human HsCENP-A. Identical and similar amino acids between these proteins are shaded in black and gray, respectively.
Several recent studies indicate that a fundamental difference in the mechanism of chromosome segregation exists between budding yeasts and higher eukaryotes (12, 13). Centromere separation in S. cerevisiae occurs at a very early stage, probably at the S phase of the cell cycle, whereas in most eukaryotes including fission yeast, centromeres separate at anaphase (14–16). It has been postulated that because of the smaller size of an S. cerevisiae centromere compared with other eukaryotes, kinetochore microtubules, which normally are captured by centromeres to form bivalent attachment at metaphase followed by chromatid separation at anaphase, may remain attached to the centromere throughout the S. cerevisiae cell cycle (14). The mechanisms involved in chromosome segregation in dividing C. albicans cells are of special interest because of the ability of this organism to divide either by budding or in a true hyphal mode.
As a first step toward understanding centromere structure function and chromosome segregation in C. albicans, we have identified and characterized its CENP-A homolog, CaCse4p. In the present study, we report that, similar to other members of the CENP-A family, CaCse4p contains a divergent N-terminal region and a conserved C-terminal histone-fold domain. We show that CaCse4p is an essential protein and is required for proper chromosome segregation and completion of mitosis. Using CaCse4p as an effective centromere marker, we find that, as in S. cerevisiae, centromere separation in C. albicans budding cells occurs at a very early stage of the cell cycle but is delayed to early anaphase in dividing hyphal cells.
Materials and Methods
Yeast, Bacterial Strains, and Media.
The C. albicans strains used in this study were SC5314 and CAI4 (Δura3∷imm434/Δura3∷imm434) (17). C. albicans strains were grown in yeast extract/peptone/dextrose (YPD) or supplemented synthetic/dextrose (SD) minimal medium (18) at 30°C. The PCK1 promoter (19) was induced in yeast extract/peptone/2% sodium succinate (YPS) or B medium (0.67% yeast nitrogen base/2% succinate) and repressed in YPD or SD minimal medium. Yeast transformations were done as described (20).
Identification of CaCse4p and Sequence Analysis.
The C. albicans Cse4p/CENP-A homolog was identified by a BLAST search (www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html) with S. cerevisiae Cse4p (ScCse4p) as the query sequence against the C. albicans genome sequence database. This sequence analysis revealed two protein sequences with high homology to ScCse4p; one is the C. albicans histone H3, and the other is CaCse4p. Sequence data for C. albicans was obtained from the Stanford Genome Technology Center web site at www-sequence.stanford.edu/group/candida.
Gene Disruption.
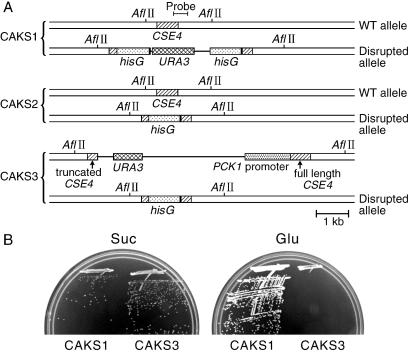
To disrupt the first CaCSE4 allele, we constructed a deletion cassette by using the URA-blaster (hisG-URA3-hisG) strategy (17). A PCR-amplified 630-bp DNA fragment comprising the 3′ region of the CaCSE4 ORF was first inserted at the NotI–SacI sites of pBluescriptII KS(+) to generate pDC1. A 4.0-kb BamHI–BglII fragment carrying the hisG-URA3-hisG cassette from pCUB6 (a gift from J. Pla Alonso, Universidad Complutense de Madrid, Madrid) then was cloned into the BamHI site of pDC1 to form pDC2. Finally, the first 230 bp of the CaCSE4 ORF was PCR-amplified and cloned into the SalI–PstI sites of pDC2 to form pDC3. In pDC3, an internal 30-bp sequence of the CaCSE4 ORF was replaced by hisG-URA3-hisG in a deletion cassette. A 4.9-kb SalI–SacI fragment carrying this cassette was transformed into C. albicans strain CAI4 selecting for uracil prototrophy. Genomic DNAs of transformants were analyzed on Southern blots by using as probe a 450-bp PCR-amplified fragment that contains the C-terminal 140 bp of the ORF plus 310 bp of the 3′-untranslated region of the gene (Fig. 2A). A Ura+ strain containing the desired disruption allele was termed CAKS1. Subsequently, Ura− strains resulting from intrachromosomal recombination between hisG repeats leading to loss of the URA3 marker were selected on medium containing 5-fluoroorotic acid (5-FOA). The correct revertant (CAKS2) was identified by genomic Southern blot analysis.
Figure 2.
CaCse4p is essential for growth of C. albicans. (A) Structural schematic of the diploid CSE4 loci in strains CAKS1, CAKS2, and CAKS3. These strains carry one disrupted and one full-length CSE4 allele, the latter under control of either a regulated (CAKS3) or wild-type promoter (CAKS1 and CAKS2). (B) Shutdown of CaCse4p expression on glucose medium (YPD) prevents CAKS3 cell growth, whereas induction of CaCse4p expression on succinate medium (YPS) allows CAKS3 cells to grow. Control CAKS1 cells grow on both media. Plates were incubated at 30°C for 3 days.
The single wild-type CSE4 allele in strain CAKS2 then was brought under control of the PCK1 promoter (Fig. 2A). To construct plasmid pPCK1-CSE4, a PCR-amplified 451-bp fragment containing the 5′ region of CaCSE4 was first cloned between the NotI and SacI sites of pBFG5 (a gift from R. Ballester, Univ. of California, Santa Barbara). This construct was digested with PstI, and the 0.6-kb PstI fragment carrying the first 451 bp of the CaCSE4 ORF was cloned between the NotI and SacI sites of pCaDis (21). The MET3 promoter region of this construct was removed by digestion with XbaI followed by a self-ligation to yield pKS1. A 1.4-kb BamHI–BglII fragment carrying the PCK1 promoter from pCA01 (19) was cloned into the BamHI site of pBluescriptII KS (+). The resulting construct was digested with BamHI and NotI, and the 1.5-kb fragment carrying the PCK1 promoter was cloned between the BamHI and NotI sites of pKS1 to yield pPCK1-CSE4. This vector was linearized by EcoRV and used to transform strain CAKS2, selecting transformants on B medium. The desired integrant (CAKS3) carrying the only full-length copy of CSE4 under control of the PCK1 promoter was identified by Southern hybridization.
Generation of Polyclonal Antibodies Against CaCse4p and Western Blot Analysis.
Polyclonal rabbit antibodies were generated against the peptide acetyl-MARLSGQSSGRQTGQGTSC-amide (Quality Control Biochemicals, Hopkington, MA), which includes amino acids 1–18 of CaCse4p. The antibody preparation was affinity-purified by using the same peptide as the affinity probe. This N-terminal region does not have similarity to histone H3. For Western immunoblots, strains CAKS1 or CAKS3 were grown overnight in YPS and subsequently transferred to both YPS and YPD media. Cells were collected at intervals after the media shift (0–8 h), and whole-cell lysates were prepared as described (9). CaCse4p was detected by anti-CaCse4p antibodies and goat anti-rabbit IgG conjugated to horseradish peroxidase (HRP) by chemiluminescence using ECL reagents (Amersham Pharmacia). Primary and secondary antibodies were diluted 1:400 and 1:1,000, respectively. Other solutions and reaction conditions were as recommended by the manufacturers.
Cell Viability, Flow Cytometry, and Cytological Analysis.
Strains CAKS1 and CAKS3 were grown overnight in YPS, and cells were spun down, washed, and inoculated in YPD at an A600 of 0.1. Samples were collected at 2-h intervals over a period of 10 h after inoculation and used for viability measurements, flow cytometry, and cytological analyses. Viability tests were conducted by counting cells on a hemacytometer and plating serial dilutions of a known number of cells on YPS plates. The plates were incubated at 30°C for 2–4 days, and the viability was calculated by dividing the number of colonies formed by the theoretical number of cells plated.
Flow-cytometry analysis was performed as described (22). Cell morphologies were analyzed by counting different types of cells (unbudded, small-, and large-budded) under a light microscope. The positions of nuclei of cells at different stages of growth were determined by staining with 4′,6-diamidino-2-phenylindole (DAPI; Roche Diagnostics) as described (18).
Immunofluorescence.
Intracellular CaCse4p was visualized by indirect immunofluorescence microscopy as described (18) with a few modifications. Cells were fixed with 3.7% formaldehyde at 25°C for 1 h at room temperature or overnight at 4°C. Antibodies were diluted as follows: 1:50 for anti-α-tubulin (YOL1/34; Harlan-Sera Lab, Leicestershire, England); 1:500 for affinity-purified rabbit anti-Cse4p; 1:30 for FITC-conjugated goat anti-rat IgG (Sigma); and 1:50 for goat anti-rabbit rhodamine-conjugated IgG (Molecular Probes). Cells were examined by using an Olympus BX 60 microscope and an ×100 objective. Digital images were captured by using an Optronics DEI 750 digital color camera and a Micron Mittenia computer. Images were processed by Adobe PHOTOSHOP software.
Results
Identification of the C. albicans CSE4 Gene.
With the objective of using the evolutionarily conserved kinetochore protein CENP-A as a tool to investigate centromere structure and the mechanism of chromosome transmission in the polymorphic yeast C. albicans, the gene encoding its CENP-A homolog was identified by sequence analysis (see Materials and Methods; Fig. 1). This gene, designated CaCSE4, encodes a predicted 24.4-kDa protein of 211 aa. The sequence analysis revealed that the conserved C-terminal region of CaCse4p is 64, 74, 57, and 61% identical to S. cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans, and human CENP-A proteins, respectively.
CaCSE4 Is Essential for Viability.
Because Cse4p/CENP-A proteins are essential for functional kinetochore formation in all organisms studied, we examined whether this gene plays a similar essential role in C. albicans. C. albicans is a diploid organism, and thus sequential deletion of both alleles is necessary to create a null mutant. We disrupted one allele of CaCSE4 with a hisG-URA3-hisG cassette (17), and the other allele was placed under control of the regulated promoter taken from the phosphoenolpyruvate carboxykinase (PCK1) gene of C. albicans (see Materials and Methods; Fig. 2A). The strain CAKS3 expresses CaCSE4 from the PCK1 promoter that is repressed by glucose and derepressed in succinate medium. The phenotype of a Cacse4 null mutant was studied by shifting CAKS3 cells grown in succinate medium to glucose medium. Strain CAKS1 grew well on plates containing either succinate or glucose. In contrast, however, strain CAKS3 cells grew normally on succinate but were unable to grow on glucose medium (Fig. 2B). This result confirmed that CaCSE4 is an essential gene for C. albicans.
Depletion of CaCse4p Affects Chromosome Segregation.
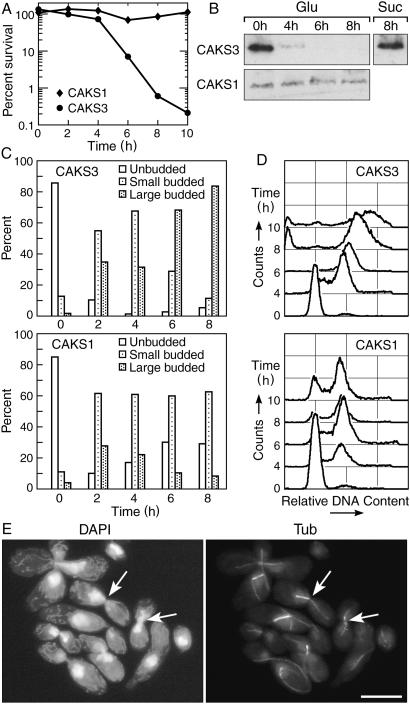
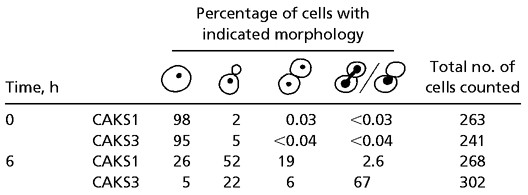
We again made use of the controllable PCK1 promoter to study the effect of depletion of CaCse4p on the cell cycle. CAKS1 and CAKS3 cells were grown overnight in succinate medium and transferred to fresh medium containing either glucose or succinate. After shifting the cells from succinate to glucose, CAKS3 cells experienced a dramatic loss of viability (from 72% at 4 h to 7% at 6 h after the media shift; Fig. 3A). Western blots revealed a concomitant drop in CaCse4p expression levels with progressively higher accumulation of large-budded cells over time, in contrast to the control CAKS1 cells grown under similar conditions (Fig. 3 B and C). Flow-cytometry analysis of glucose-repressed CAKS3 cells revealed a direct correlation between the accumulation of large-budded cells having 4C DNA content (because C. albicans is a diploid organism) and the time of incubation in glucose medium (i.e., depletion of the CaCse4p level; Fig. 3D Upper). As a control, CAKS1 cells grown under similar conditions did not show this phenotype (Fig. 3D Lower). A closer examination of the mutant phenotype was performed by staining the DNA and mitotic spindle of arrested CAKS3 cells incubated for 6 h in glucose medium. Most of the large-budded cells had an unsegregated nucleus either at or near the mother-bud neck or stretched through the neck region (Fig. 3E, DAPI; Table 1). The corresponding cells had a very short mitotic spindle (Fig. 3E, Tub). Taken together, these data suggest that CaCse4p plays an important role in the process of chromosome segregation and completion of mitosis in C. albicans.
Figure 3.
CaCse4p is required for proper chromosome segregation. (A) Viability of CAKS3 cells drops dramatically between 4 and 6 h after shutting down CaCse4p expression by media shift. CAKS3 cells were grown overnight in succinate (YPS) medium, washed, and inoculated into glucose (YPD) medium. Cells were harvested at 2-h intervals after the media shift, and cell viability was determined by spreading diluted aliquots of cultures at indicated time points on YPS plates and counting the number of colonies that appeared after incubating the plates for 3 days at 30°C. (B) Immunoblot analysis of whole-cell extracts prepared from CAKS3 cells shows a sharp decline of CaCse4p expression level 4 h after shifting from succinate to glucose medium. After 6 h, CaCse4p was undetectable (Upper). Succinate (YPS)-grown CAKS3 cells did not show any alteration of CaCse4p expression after 8 h of growth (Upper, right-most lane). As a control, whole-cell extract prepared from the same number of glucose-grown CAKS1 cells did not show any change in CaCse4p expression (Lower). (C) Distribution of different cell types of strains CAKS1 and CAKS3 after media shift from succinate to glucose. Depletion of CaCse4p in CAKS3 results in a progressively higher accumulation of large-budded cells with time (Upper), whereas the CAKS1 cells do not show any significant change of cell-type distribution over time (Lower). (D) Depletion of CaCse4p leads to a G2/M arrest. Flow-cytometry profiles of CAKS1 (Lower) and CAKS3 (Upper) cells incubated in glucose (YPD) for the indicated time intervals and stained with propidium iodide. The x axis is a measure of DNA content (intensity of propidium iodide staining), and the y axis represents the relative numbers of cells. (E) DAPI-stained CAKS3 cells incubated for 6 h in glucose (DAPI, Left) and their spindle morphology as determined by immunofluorescence microscopy using antitubulin antibodies (Tub, Right). Arrows indicate a typical G2/M arrest phenotype: large-budded cells having an unsegregated mass of DNA near the mother-bud neck and short mitotic spindles. (Bar = 10 μm.)
Table 1.
Phenotypes of CaCse4p-depleted cells
 |
Subcellular Localization of CaCse4p.
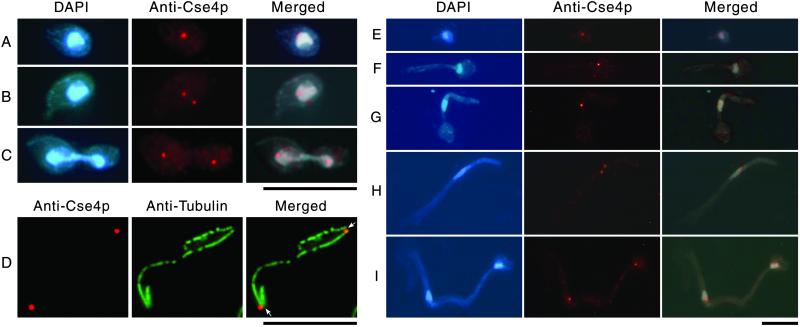
CaCse4p localization was examined separately in both C. albicans budding yeast and hyphal cells by using anti-CaCse4p antibodies. To examine C. albicans in the budding form of growth, CAI4 cells were harvested from YPD medium in the logarithmic phase of growth and fixed by formaldehyde. Indirect immunofluorescence microscopy using anti-CaCse4p antibodies revealed an intense dot-like signal in all cells examined, and the dots were always colocalized with the nucleus as visualized by DAPI staining (Fig. 4 A–C). Although a single dot always appeared in unbudded G1 cells (Fig. 4A), two clearly separated dots were seen even in cells at the early small-bud stage with a single mitotic nucleus (Fig. 4B). This observation indicates that in C. albicans, as in S. cerevisiae, centromere separation in the budding yeast form of growth occurs at a very early stage of the cell cycle. It also provides strong evidence that the protein we identified by sequence homology comparison with S. cerevisiae Cse4p is an authentic centromere marker in C. albicans. In late mitotic cells with a dividing nucleus, one dot corresponding to the position of CaCse4p near each daughter nucleus was observed (Fig. 4C). To confirm that the signals are CaCse4p-specific, we stained CAKS3 cells grown under conditions where they were either expressing CaCse4p (grown in succinate) or depleted of CaCse4p (incubated in glucose for 6 h) with this antibody preparation. Although clear dot-like signals were visible in succinate-grown cells, no detectable signal was observed when cells were incubated for 6 h in glucose (not shown). Coimmunostaining of fixed CAI4 cells with antitubulin antibodies and anti-Cse4p antibodies showed that in C. albicans, analogous to localization patterns of kinetochore proteins in S. cerevisiae, CaCse4p signals are located near the spindle pole bodies (Fig. 4D).
Figure 4.
Localization of CaCse4p at the kinetochore in C. albicans budding yeast and filamentous cells. Fixed CAI4 cells were stained by DAPI and anti-Cse4p antibodies. Dot-like CaCse4p signals were observed in unbudded (A), small-budded (B), and large-budded cells (C). Fixed CAI4 cells were coimmunostained with anti-Cse4p and anti-tubulin antibodies (D). Arrows show the positions of spindle pole bodies in a large-budded cell near the end of anaphase. SC5314 cells were grown in hyphae-inducing medium (see Materials and Methods) and fixed. Shown here are cells at different stages between G1/S and G2 with varied lengths of germ tubes (E–G) and cells at mitosis (H and I). (Bar = 10 μm.)
We also examined the mode of centromere separation in C. albicans cells growing in the filamentous form. Wild-type strain SC5314 was grown in YPD plus 10% horse serum at 37°C to induce hyphal filament formation. After incubation for 2 h, cells were fixed and immunostained with anti-CaCse4p and antitubulin antibodies. As described above, unbudded G1 cells had a dot-like Cse4p signal colocalized with the nucleus. Hyphal growth begins as a small protrusion from the mother cell (Fig. 4E), and cells at S/G2 had an elongated bud (germ tube) through which the nucleus migrated (Fig. 4 F and G). Cells at these stages of the filamentous cell cycle still showed only single dot-like signals for CaCse4p. Interestingly, mitotic cells having a more elongated but tapering bud with a stretched nucleus showed two distinct dots for CaCse4p (Fig. 4H). It is likely that these cells with a stretched nucleus have entered early anaphase. Subsequently, nuclear division occurred, and CaCse4p signals further separated in more elongated daughter cells. Finally, in late mitotic cells, one dot for each daughter nucleus was observed (Fig. 4I). Thus, in the filamentous form of growth, centromere separation probably first occurs at an early anaphase stage of the cell cycle.
Discussion
CENP-A Homolog CaCse4p Is a Centromere Protein and Is Essential for Chromosome Segregation in C. albicans.
We have identified a member of the important centromere-associated CENP-A protein family in the medically important human pathogen C. albicans. The lack of detectable sequence motifs between centromeres of different organisms has led to the speculation that non-DNA sequence determinants such as the histone H3-variant CENP-A, which has been found in centromeric heterochromatin in a wide variety of organisms, may epigenetically mark centromere DNA chromatin to distinguish it from noncentromeric regions (23, 24). Thus, identification of a CENP-A homolog in C. albicans could be a key first step in characterization of centromere structure function in this organism. As with all other known members of the CENP-A family, CaCse4p contains a highly homologous C-terminal histone-fold domain and a dissimilar N-terminal tail region. We used antibodies directed against CaCse4p to confirm centromere localization of this protein and placed the CSE4 gene under control of a regulated promoter to study the effect of depletion of CaCse4p on cell growth and chromosome transmission.
CENP-A proteins have been shown to be essential and required for kinetochore function in diverse systems. Injection of affinity-purified CENP-A-specific antibodies into the nucleus of human cultured cells at the early replicating stage results in a cell-cycle arrest in interphase before mitosis (25). CENP-A null mice show embryonic lethality, and affected embryos exhibit mitotic defects (26). Likewise, Drosophila and C. elegans cells depleted of CENP-A proteins by RNA interference or antibody injections show lethality as a result of defects in chromosome segregation and functional kinetochore formation (27–29). In yeasts, null mutations in the CENP-A family protein genes (cnp1Δ in S. pombe and cse4Δ in S. cerevisiae) are lethal, and depletion of these proteins in conditional mutants results in chromosome missegregation, generation of aneuploid cells, and accumulation of large-budded cells with an unsegregated nucleus in the neck (30, 31). In C. albicans, we find CaCse4p also to be an essential protein. Depletion of CaCse4p in cells in the budding mode of growth leads to loss of cell viability and formation of enlarged cells arrested in the large-budded stage with 4C DNA content. DAPI and tubulin staining reveal a significantly higher number of arrested cells with unsegregated nuclei and short mitotic spindles. The results of our experiments show that CaCse4p, similar to CENP-A proteins of other organisms, is an essential protein and is required for proper chromosome transmission and completion of mitosis in C. albicans.
Timing of Centromere Separation in Dividing C. albicans Cells.
Because C. albicans can grow in both budding yeast and filamentous cellular forms, studies of chromosome segregation in this organism are of special interest. Before this study, centromere-specific markers have been lacking for C. albicans; the centromere DNA and kinetochore proteins are yet to be characterized. We find CaCse4p to be an excellent centromere-specific marker to study centromere separation in C. albicans cells dividing in either growth mode. Affinity-purified antibodies generated against an N-terminal 18-aa peptide of CaCse4p specifically recognize a 25-kDa band on Western blots of whole-cell extracts. This band is depleted greatly in CAKS3 cells incubated in glucose to shut down expression of the CSE4 gene, confirming that these antibodies react highly specifically with CaCse4p. Subcellular localization of CaCse4p with this antibody preparation in budding or filamentous cells displays the clustered centromeres as a single bright dot in the nucleus of interphase or resting cells. Obvious early centromere separation is observed in cells at the small-bud stage with a short spindle and an undivided mass of DNA. The clustered centromere signals are localized very close to the ends of the spindle, near the spindle pole bodies as is seen in other yeasts (for example see ref. 14).
A similar early separation of centromeres has been reported in S. cerevisiae (14–16). It has been suggested that, unlike higher eukaryotes, kinetochore microtubules in S. cerevisiae may remain attached to the centromeres throughout the cell cycle. Centromere separation as early as S phase is made possible by a series of events: spindle pole body duplication occurs very early at the time of bud emergence (32), replication of centromere DNA occurs early in S phase (33), and nuclear microtubules are present throughout the cell cycle. Thus, the sequence of events in centromere separation in C. albicans cells dividing by budding appears to be very similar to that observed in S. cerevisiae.
The stage at which centromeres separate in dividing filamentous fungal cells is unknown. In the oomycete filamentous fungus, Saprolegnia ferax, short premetaphase spindles are observed, and kinetochores are always attached to nuclear microtubules throughout the cell cycle (34, 35). In our localization studies of centromeres in dividing filamentous C. albicans cells, centromere separation is observed first at a stage when the nuclear mass is beginning to elongate but is not yet divided, most likely in early anaphase. However, we cannot determine from these studies whether the centromeres are separating in a preanaphase stage. Live-cell imaging using a GFP-tagged centromere protein could be useful in this type of study. However, in our hands, a GFP-tagged CaCse4p fusion protein was nonfunctional when expression in C. albicans cells was attempted.
CaCse4p and Structure-Function Analysis of the C. albicans Kinetochore.
Although the C. albicans genome project is essentially completed, centromere DNA sequences are yet to be identified. Because CENP-A proteins are excellent centromere markers in all organisms examined to date, specific antibodies against CaCse4p will be useful in experiments directed toward the isolation and characterization of centromere DNAs from C. albicans. Immunoprecipitation of fragmented chromatin with these antibodies should yield DNA preparations highly enriched in centromeric sequences. A conventional mutational approach to identify other kinetochore protein genes would be difficult in C. albicans because of its diploid genome. The availability of authentic centromere DNAs will make possible the development of one-hybrid screens to identify kinetochore proteins (for example, see ref. 36). In addition, CaCse4p could be a useful reagent to identify kinetochore proteins via coimmunoprecipitation studies with anti-CaCse4p antibodies.
Previous studies indicate that despite having a few similar proteins (such as Cse4p/CENP-A and Mif2p/CENP-C), significant differences in kinetochore composition exist between budding yeast and higher eukaryotes. Several inner and outer kinetochore proteins of budding yeast seem unique and do not have any known counterparts in higher eukaryotes. Thus, certain C. albicans kinetochore proteins could be useful targets for the development of more specific, and potentially safer, antifungal drugs.
Acknowledgments
We thank R. Ballester, J. Berman, J. Ernst, W. Fonzi, D. Inglis, A. Johnson, H. Liu, J. Pla, P. Sudbery, and S. Stoler for providing strains and plasmids; B. Matsumoto, D. Thrower, and M. Maduro for help with microscopy; and D. McLaren for artwork. We also thank M. Baum, T. Stoyan, and L. Clarke for help with experiments, useful discussions, and critical comments on the manuscript. This work was supported by National Cancer Institute (National Institutes of Health) Research Grant CA-11034.
Abbreviation
- DAPI
4′,6-diamidino-2-phenylindole
References
- 1.Navarro-Garcia F, Sanchez M, Nombela C, Pla J. FEMS Microbiol Rev. 2001;25:245–268. doi: 10.1111/j.1574-6976.2001.tb00577.x. [DOI] [PubMed] [Google Scholar]
- 2.Clarke L. Curr Opin Genet Dev. 1998;8:212–218. doi: 10.1016/s0959-437x(98)80143-3. [DOI] [PubMed] [Google Scholar]
- 3.Clarke L, Carbon J. Nature (London) 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- 4.Choo K H A, editor. The Centromere. Oxford: Oxford Univ. Press; 1997. pp. 12–49. [Google Scholar]
- 5.Henikoff S, Ahmad K, Platero J S, van Steensel B. Proc Natl Acad Sci USA. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sullivan B A. Trends Biotechnol. 2002;3:89–92. doi: 10.1016/s0167-7799(02)01902-9. [DOI] [PubMed] [Google Scholar]
- 7.Malik H S, Henikoff S. Genetics. 2001;157:1293–1298. doi: 10.1093/genetics/157.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y, Baker R E, Keith K C, Harris K, Stoler S, Fitzgerald-Hayes M. Mol Cell Biol. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keith K C, Baker R E, Chen Y, Harris K, Stoler S, Fitzgerald-Hayes M. Mol Cell Biol. 1999;19:6130–6139. doi: 10.1128/mcb.19.9.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sullivan K F, Hechenberger M, Masri K. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meluh P B, Yang P, Glowcszewski L, Koshland D, Smith M M. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 12.Kitagawa K, Hieter P. Nat Rev Mol Cell Biol. 2001;2:678–687. doi: 10.1038/35089568. [DOI] [PubMed] [Google Scholar]
- 13.Winey M, O'Toole E T. Nat Cell Biol. 2001;3:E23–E27. doi: 10.1038/35050663. [DOI] [PubMed] [Google Scholar]
- 14.Goshima G, Yanagida M. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- 15.Goshima G, Yanagida M. Genes Cells. 2001;6:765–773. doi: 10.1046/j.1365-2443.2001.00464.x. [DOI] [PubMed] [Google Scholar]
- 16.He X, Asthana S, Sorger P K. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- 17.Fonzi W A, Irwin M Y. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaiser C S, Michaelis S, Mitchell A. In: Methods in Yeast Genetics. Kaiser C S, Michaelis S, Mitchell A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 207–217. [Google Scholar]
- 19.Leuker C E, Sonneborn A, Delbruck S, Ernst J F. Gene. 1997;192:235–240. doi: 10.1016/s0378-1119(97)00069-3. [DOI] [PubMed] [Google Scholar]
- 20.Hull C M, Johnson A D. Science. 1999;285:1271–1275. doi: 10.1126/science.285.5431.1271. [DOI] [PubMed] [Google Scholar]
- 21.Care R S, Trevethick J, Binley K M, Sudbery P E. Mol Microbiol. 1999;34:792–798. doi: 10.1046/j.1365-2958.1999.01641.x. [DOI] [PubMed] [Google Scholar]
- 22.Baum M, Clarke L. Mol Cell Biol. 2000;20:2852–2864. doi: 10.1128/mcb.20.8.2852-2864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henikoff S, Ahmad K, Malik H S. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 24.Karpen G H, Allshire R C. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- 25.Figueroa J, Saffrich R, Ansorge W, Valdivia M. Chromosoma. 1998;107:397–405. doi: 10.1007/s004120050323. [DOI] [PubMed] [Google Scholar]
- 26.Howman E V, Fowler K J, Newson A J, Redward S, MacDonald A C, Kalitsis P, Choo K H A. Proc Natl Acad Sci USA. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blower M, Karpen G. Nat Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buchwitz B J, Ahmed K, Moore L L, Roth M B, Henikoff S. Nature (London) 1999;40:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- 29.Oegema K, Desai A, Rybina S, Kirkham M, Hyman A A. J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoler S, Keith K C, Curnick K E, Fitzgerald-Hayes M. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi K, Chen E S, Yanagida M. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- 32.Byers B, Goetsch L. J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarroll R M, Fangman W L. Cell. 1988;54:505–513. doi: 10.1016/0092-8674(88)90072-4. [DOI] [PubMed] [Google Scholar]
- 34.Heath I B. J Cell Biol. 1980;84:531–546. doi: 10.1083/jcb.84.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heath I B, Rethoret K. J Cell Sci. 1981;49:353–367. doi: 10.1242/jcs.49.1.353. [DOI] [PubMed] [Google Scholar]
- 36.Ortiz J, Stemmann O, Rank S, Lechner J. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]