Abstract
Duchenne muscular dystrophy is a lethal X-linked recessive disorder caused by mutations in the dystrophin gene. Delivery of functionally effective levels of dystrophin to immunocompetent, adult mdx (dystrophin-deficient) mice has been challenging because of the size of the gene, immune responses against viral vectors, and inefficient infection of mature muscle. Here we show that high titer stocks of three different gutted adenoviral vectors carrying full-length, muscle-specific, dystrophin expression cassettes are able to efficiently transduce muscles of 1-yr-old mdx mice. Single i.m. injections of viral vector restored dystrophin production to 25–30% of mouse limb muscle 1 mo after injection. Furthermore, functional tests of virally transduced muscles revealed almost 40% correction of their high susceptibility to contraction-induced injury. Our results show that functional abnormalities of dystrophic muscle can be corrected by delivery of full-length dystrophin to adult, immunocompetent mdx mice, raising the prospects for gene therapy of muscular dystrophies.
As the most common form of muscular dystrophy, Duchenne muscular dystrophy affects one of every 3,500 newborn males with one-third of cases arising from new mutations (1). Boys harbor the genetic mutation at birth but do not show signs of muscle impairment and, in the absence of a family history, are usually not diagnosed until early childhood (2). The mdx mouse carries a premature stop codon in exon 23 of the dystrophin gene and has no detectable levels of dystrophin protein in muscle tissue, except in rare revertant fibers (3, 4). The most notable functional deficit of mdx mouse limb muscles is a severe susceptibility to contraction-induced injury that increases significantly with age (5–7). Our laboratory has shown that transgenic restoration of dystrophin via the full-length 14-kb cDNA, in contrast to truncated versions, completely prevents the development of dystrophic abnormalities in mdx mice (8, 9). Similarly, delivery of dystrophin vectors to muscles of neonatal mdx mice can partially prevent the development of dystrophy (10–12). Although the prevention of pathologies in the mdx mouse has demonstrated the potential for dystrophin replacement in utero or neonatally, there exists little data on the ability to halt or reverse already established dystrophic characteristics.
Adenoviral (Ad) vectors are of interest for muscle gene therapy because they can be grown to high titer, efficiently infect muscle tissue, and have 8 kb of cloning capacity sufficient for truncated dystrophin cDNAs (13). However, transgene expression from first-generation Ad vectors in muscles of immunocompetent adult mice is generally lost 10–20 d after infection because of expression of viral late genes that elicit a host immune response (14, 15). Therefore, the overwhelming majority of studies to date report delivery of truncated forms of dystrophin transgenes by using first-generation Ad injection into either neonatal (11, 12), immunosuppressed adult (16, 17), or myotoxin-injected adult mdx mice (16, 18). Although these studies have shown potential for gene delivery to dystrophic muscles, they have not tested the functional consequences of viral delivery of full-length dystrophin to an unaltered, immunocompetent, adult host. Furthermore, truncated isoforms of dystrophin have been less successful than the full-length protein in rescuing the dystrophic phentoype of transgenic mdx mice (9, 19, 20).
In response to the limitations of cloning capacity and immunogenicity of first-generation Ad vectors, we and others (10, 21–23) have modified the Ad vector by deleting all viral sequences except those needed in cis for Ad genome replication and packaging. These “gutted” Ad vectors have a cloning capacity >30 kb, sufficient for the 14-kb dystrophin cDNA and highly active, muscle-specific regulatory cassettes. Expression of full-length dystrophin from a gutted Ad vector has been achieved in vitro (22–24) and in vivo in neonatal mice (10, 21, 24). In addition, prolonged persistence of viral DNA, a lack of associated toxicity, and avoidance of the host immune response against Ad proteins has been shown in studies that compared several gutted and first-generation Ad vectors (25–28). Nonetheless, the effectiveness of a dystrophin gutted Ad vector has not been tested in muscles of immunocompetent adult mice that have already developed a significant pathology.
Dystrophic muscles have been shown to contain elevated levels of immune effector cells, complicating successful viral gene transfer to adult animals (29). We have previously shown that muscle-specific expression from Ad vectors can avoid elicitation of a cellular immune response against immunogenic transgenes such as β-galactosidase (β-gal) (29). We also have found that Ad vectors infect regenerating mdx muscles at higher efficiency than C57BL/10 WT muscles in young adults, and at ages >12 mo and up to 2 yr (ref. 29; C.D. and J.S.C., unpublished data). Consequently, we asked here whether a gutted Ad vector carrying full-length human dystrophin (HDys) or a gutted Ad vector carrying full-length mouse dystrophin (MDys) dystrophin cDNAs regulated by a powerful muscle-specific promoter, could be used to deliver dystrophin to muscles of mature mdx mice and correct their high susceptibility to contraction-induced injury.
Materials and Methods
Virus Construction and Preparation.
Gutted Ad vectors were prepared in a modified pBluescript plasmid backbone (8) containing fused viral inverted terminal repeats and a viral packaging signal (ψ) isolated as a PvuI–BsgI fragment from pFG140 (30, 31), and blunt-end cloned into a KpnI site in the polylinker, which is flanked by BssHII sites. Mouse muscle creatine kinase (MCK) gene fragments were used to drive muscle-specific expression of dystrophin. GEβDys contained the 3.3-kb promoter plus enhancer, plus the simian virus 40 viral protein 1 intron (9). HDys and MDys carried the 6.5-kb MCK promoter plus enhancer plus intron 1 regulatory region (8) in a SmaI site. The full-length human dystrophin cDNA was assembled from a series of short overlapping cDNAs [kindly provided by L. Kunkel (32)] by conventional subcloning, recombinant PCR, and site-directed mutagenesis. The final cDNA was 12.1 kb, ending at the XbaI site in the 3′ untranslated region. The 13.9-kb full-length mouse dystrophin cDNA has been described (8). A 3′ genomic flanking region from the human dystrophin gene (3′ flank, a 5.5-kb PacI to XhoI fragment whose 5′ end is located 0.75 kb 3′ from exon 79) was used to increase MDys and HDys to a stably packagable size. GEβDys contains a reporter gene (lacZ) expression cassette driven by the ecdysone-inducible promoter (Ecdy).
Gutted Ad vectors were prepared as described by cotransfection of plasmid DNA containing a gutted virus genome together with viral DNA from a first-generation helper adenovirus into C7-Cre cells (30, 33). The helper virus genome contained loxP sites flanking the packaging signal and an ecdysone-inducible human placental alkaline phosphatase expression cassette cloned into the E3 region. High titer stocks of gutted vector were obtained after 4–5 serial passages on C7-Cre cells followed by purification on CsCl gradients (23, 30, 33, 34). Gutted Ad stocks were obtained at titers between 3 and 9 × 1012 particles per ml with 0.5–2% contamination by helper virus, as quantified by real-time PCR on a Prism 7700 sequence detector (Applied Biosystems). CNβ is a first-generation adenovirus that contains a lacZ gene driven by the cytomegalovirus promoter (35).
Experimental Design.
Mdx mice (C57BL/10ScSn-Dmdmdx/J, The Jackson Laboratory) 11–13 mo of age were anesthetized with Ketamine/Rompun (160 mg/kg), and the hair was shaved from the skin covering the tibialis anterior (TA) muscles. Then 30–60 μl of virus or storage buffer (sham; 20 mM Hepes/5% sucrose or PBS/3% sucrose) was injected directly into the TA muscles by using an insulin syringe. Four to eight mice were injected per viral or sham (often the contralateral leg) agent. The animals recovered and were maintained in a biohazard specific pathogen-free barrier facility for 5–30 d. At the desired time point, mice were anesthetized and the TA muscles were subjected to an in situ lengthening contraction protocol as described (6). In brief, after determination of optimal muscle length (Lo) and measurement of maximum isometric tetanic force, muscles were maximally stimulated and stretched from Lo through 40% of muscle fiber length (Lf) during two lengthening contractions (LC1 and LC2). Maximum isometric force was measured before and after each lengthening contraction, with the forces after LC1 and LC2 indicative of the ability of muscles to resist injury. At the conclusion of the protocol, muscles were dissected free of tendon and connective tissue, weighed, and either frozen in isopentane cooled with liquid nitrogen or processed for flow cytometry analysis. After 15 × 5- to 7-μm cryosections were cut, the remaining tissue was removed from OCT compound and was snap-frozen in liquid nitrogen for protein preparation and analysis by SDS/PAGE. Total muscle fiber cross-sectional area (CSA) was calculated by dividing muscle mass (milligram) by the product of fiber length (millimeter) and 1.06 mg/mm3, the density of mammalian skeletal muscle. Specific force was determined by normalizing maximum isometric tetanic force to CSA.
Flow Cytometry.
Flow cytometry was performed as described (29). In brief, muscles were digested with collagenase, stained with Abs against cell surface markers, and sorted. CD4+ and CD8+ T cells were identified and counted in the muscle homogenates.
Histology.
Frozen muscle cryosections of 5–7 μm were analyzed by immunofluoresence as described (36). Primary Abs used were an affinity purified rabbit anti-dystrophin Ab (1:600) specific for the N terminus of dystrophin (37) and FITC-conjugated mouse monoclonal anti-CD4+ and CD8+ cell Abs (1:100, Pharmingen). Secondary Abs used for dystrophin detection were either a goat anti-rabbit alexa 488 (green) or 594 (red) (1:1,200, Molecular Probes). Serial cryosections were stained with hematoxylin/eosin-phloxine as described (36).
Western Analysis.
SDS/PAGE was performed by using protein extract prepared from injected muscle samples and separated on a 5% gel (9). Each lane was loaded with 40 μg of protein. An identical gel was run simultaneously and stained with Coomassie brilliant blue. Proteins were transferred onto a 0.45-μm nitrocellulose membrane and incubated overnight with anti-dystrophin mouse monoclonal primary Ab (1:500, Dys-2, Novocastra, Newcastle, U.K.). An horseradish peroxidase-conjugated donkey anti-mouse secondary Ab (1:50,000, The Jackson Laboratory) was used to detect dystrophin by using an enhanced chemiluminescence (ECL-plus) kit (Amersham Biosciences).
Image Analysis and Quantitative Measures.
Photographs (×100 and ×200) were taken by using a Nikon E1000 microscope and a Spot-2 camera system (Diagnostic Instruments, Sterling Heights, MI). imagepro software (Media Cybernetics, Silver Spring, MD) was used to quantitate the dystrophin-positive area in photos of muscle cross sections. Ten 100× microscopic fields per muscle (10 muscles total) were analyzed by applying the same brightness and contrast thresholds in imagepro to exclude background and calculate the area surrounded by dystrophin immunofluoresence. Total dystrophin-positive area in 10 100× fields was normalized to the total area analyzed, which corresponded closely to the total muscle CSA. WT and sham-injected muscles were 100% and 0.2% dystrophin-positive, respectively. Montages were photographed by using montage explorer software (Syncroscopy, Frederick, MD).
Statistical Analysis.
Data are presented as the mean ± SEM. Statistical analyses were performed by using statview software (SAS Institute, Cary, NC). Each TA muscle was treated as an individual case. Differences between muscles injected with various agents (sham, HDys, MDys, GEβDys, and CNβ) and uninjected (WT and mdx) muscles for contractile and morphological measures were determined by one-way ANOVA. When a positive effect was observed, a Tukey/Kramer post hoc test was used to determine differences between group means. Significance was set at P ≤ 0.05.
Results
Injection with Full-Length Human Dystrophin-Gutted Ad Vector.
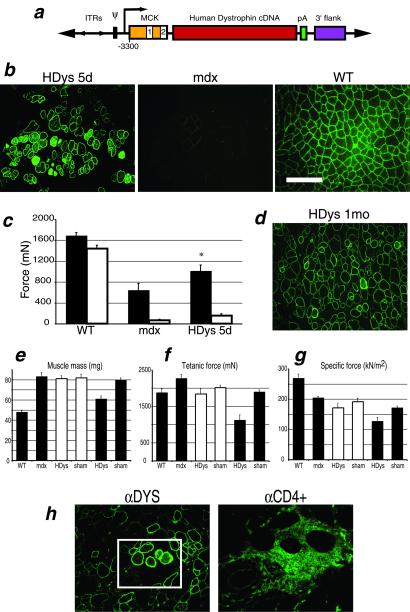
The plasmid used in generating HDys, a gutted Ad vector expressing full-length human dystrophin, is shown in Fig. 1a. We tested this virus by injection into the TA muscles of 1-yr-old mdx mice, which, in contrast to young adults, display more severe functional deficits (6). Immunolocalization of muscle cross sections by using an affinity-purified rabbit anti-dystrophin Ab revealed high levels of dystrophin expression properly localized to the muscle fiber sarcolemma (Fig. 1b). Overexpression of dystrophin was present in some of the fibers as evidenced by cytoplasmic staining.
Figure 1.
Analysis of mdx mouse muscles injected with HDys. (a) Plasmid vector used for generation of HDys gutted adenovirus: ITRs, inverted terminal repeats; Ψ, viral packaging signal; MCK gene regulatory element (the 6.5-kb MCK promoter/enhancer contains exon 1, intron 1, and a truncated exon 2) (8); 12.1-kb full-length human dystrophin cDNA; pA, simian virus 40 polyadenylation signal; and 3′-flank, 5.5-kb 3′-genomic flanking region of the human dystrophin gene. (b) Immunofluoresence (IF) analysis of cryosections from HDys-injected mdx, uninjected mdx, and C57BL/10J (WT) control mice. Anti-dystrophin Abs detected dystrophin expression localized to the sarcolemma (green). The scale bar applies to all IF photos and is equal to 100 μm. (c) TA muscles were analyzed in situ by measuring force production after two injury-inducing lengthening contractions, LC1 (filled bars) and LC2 (open bars). Muscles were tested 5 d after virus (HDys), or buffer (sham), injection into mdx mice. WT muscles were uninjected and age-matched. HDys-injected muscles showed significant recovery of force-producing capabilities after LC1 (P < 0.05 vs. mdx) and some protection after LC2. (d) After HDys injection (1 mo), IF analysis of TA muscles from mdx mice showed continuing high levels of dystrophin expression. However, e–g demonstrate adverse physiological effects in the injected muscles not observed at 5 d (injected samples; open bars = 5 d, filled bars = 1 mo). HDys-injected muscles revealed decreases in mass and force significantly different from sham-injected and WT control muscles (P < 0.01). Each bar represents the mean of 6–8 muscles ± SEM. (h) IF inspection of HDys-injected muscles showed the presence of CD4+ (Right, green) and CD8+ (not shown) T cells surrounding some of the muscle fibers highly expressing dystrophin (green, enclosed in box); [×200 (Left) and boxed area magnified ×4 (Right)].
An in situ assay was used to quantitate TA muscle susceptibility to contraction-induced injury by measuring muscle force production before and after two injury-inducing lengthening contractions (6). In muscles from mice injected with HDys, significant protection from injury was observed after one lengthening contraction (Fig. 1c). Sham-injected (buffer only) muscles did not show dystrophin-positive fibers or functional differences from muscles of uninjected mdx mice (data not shown).
We next tested the activity of HDys after a longer period. After 1 mo, injected TA muscles from 1-yr-old mdx mice continued to show high levels of dystrophin expression (Fig. 1d). However, two groups of muscles injected with independent preparations of HDys each demonstrated a decrease in mass and force-producing capabilities that was not apparent at 5 d (Fig. 1 e–g). Immunolocalization on serial muscle sections showed that some of the dystrophin-positive fibers were surrounded by immune effector cells (Fig. 1f). These results suggested that the human dystrophin protein had elicited a cellular immune response, as has been previously reported in mice after injection of naked DNA plasmids that express human dystrophin (38, 39).
Injection with Full-Length Mouse Dystrophin Gutted Ad Vectors.
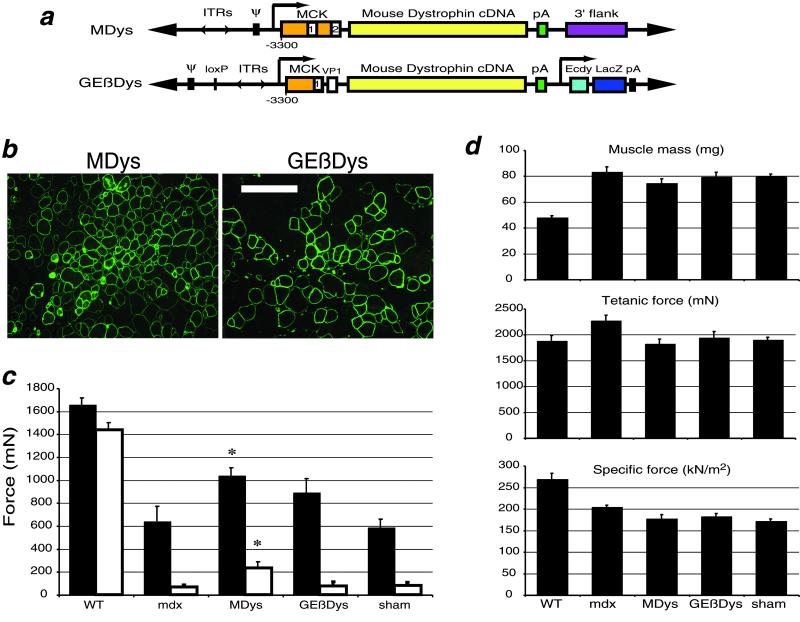
To avoid the immunogenicity of a human protein delivered to mouse tissue, we constructed two different gutted adenoviruses expressing full-length mouse dystrophin. GEβDys contains the 13.9-kb mouse dystrophin cDNA and an ecdysone-inducible β-gal reporter gene expression cassette (Fig. 2a). MDys is comprised of the full-length mouse dystrophin cDNA and a noncoding “stuffer” fragment. Injection of either of these viruses resulted in high levels of dystrophin expression in the TA muscles of 1-yr-old mdx mice after 1 mo (Fig. 2b).
Figure 2.
Mouse dystrophin protects against contraction-induced injury without adverse physiological consequences. (a) Plasmid constructs used to generate MDys and GEβDys gutted adenovirus. See Fig. 1 legend for abbreviations. MDys is identical to HDys with the exception of the species of the dystrophin cDNA. The promoter used in GEβDys is a 4.2-kb version of MCK that lacks the intron and exon 2 but contains an simian virus 40 viral protein 1 intron (9). (b) IF analysis of injected muscles show high levels of dystrophin expression 1 mo after injection. (Scale bar = 100 μm.) (c) Lengthening contractions in situ demonstrate significantly protected muscles 1 mo after injection of MDys gutted Ad. Protection is afforded after both LC1 (filled bars) and LC2 (open bars) (P < 0.05). GEβDys-injected muscles were protected after both contractions, but the effect was not significant. Control data from WT and mdx mice is the same as shown in Fig. 1c. (d) The protection against injury after virus injection was not accompanied by any loss in mass or force production. Control WT and mdx data are the same as shown in Fig. 1 e–g. Bars represent the mean of 6–8 muscles ± SEM.
Dystrophin expression from these viruses was associated with a decrease in the susceptibility to contraction-induced injury (Fig. 2c). MDys-injected muscles showed the most dramatic correction. After one lengthening contraction, muscles injected with MDys generated 62% more force than uninjected muscles in mdx mice. There was also a significant effect after the second lengthening contraction, after which MDys-injected muscles generated 250% more force than uninjected or sham-injected muscles in mdx mice. A trend toward protection was observed in GEβDys-injected muscles, but the effect was not significant, possibly because of lower dystrophin expression levels (see Fig. 3a).
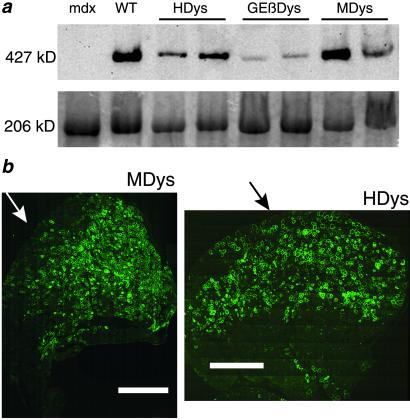
Figure 3.
Widespread, high level expression of dystrophin in adult mdx mice 1 mo after injection with gutted Ad. (a) Immunoblot analysis of protein extracts from control and injected TA muscles shows varying levels of the full-length 427-kDa dystrophin protein. Loading controls are shown below in an identical Coomassie-stained gel. Myosin is seen here at 206 kDa. (b) Montage photos of entire cross sections of TA muscles from 1-yr-old mdx mice injected with MDys (Left) or HDys (Right) demonstrate widespread expression of dystrophin from the approximate site of injection (arrows) 1 mo later. (Scale bar = 1.0 mm.)
Correction of the susceptibility to injury by the mouse dystrophin vectors was not correlated with any adverse effects on muscle mass or the ability to produce force (Fig. 2d), as had been observed in the HDys-injected muscles. Sham injections caused some reduction of mass and force-producing capacity, most likely because of the trauma of needle injection. The decreases in maximum isometric and specific force observed in mdx mice after injection with MDys and GEβDys were not different from those in sham-injected muscles.
Extent of Full-Length Dystrophin Expression.
We performed Western analysis to demonstrate the presence of the full-length dystrophin isoform in the injected muscles and to examine the relative levels of transgene expression from the three gutted Ad vectors. Fig. 3a shows expression of the 427-kDa dystrophin protein in muscles of mdx mice 1 mo after injection of HDys, GEβDys, or MDys compared to uninjected WT and mdx muscles. Although the amount of dystrophin varied between injections, MDys-transduced muscles generally displayed the highest protein expression. Lower levels of dystrophin in GEβDys-injected muscles may be related to leaky β-gal expression from GEβDys, which can elicit an immune response (29, 35). However, we also have noted that GEβDys expresses similar levels of dystrophin after injections of mdx or immune-deficient SCID/mdx mice for at least 4 mo (G.S. and J.S.C., unpublished data).
The extent of dystrophin expression throughout injected TA muscles is shown in immunofluorescently labeled montage images of whole cross sections in Fig. 3b. Widespread dystrophin expression from the site of injection demonstrates both the ability of our gutted Ad vectors to infect mature dystrophic muscle, and the potential of less-than-complete transduction to significantly correct the major functional deficiency of muscles in mdx mice. Quantitative analysis of immunofluorescently labeled frozen muscle sections revealed that our direct, i.m. injections achieved ≈25–30% dystrophin-positive total muscle CSA in both HDys- and MDys-injected TA muscles. When normalized to values obtained from muscles of WT mice, the susceptibility to contraction-induced injury of muscles in mdx mice was corrected by 40% with only ≈25% of the muscle CSA expressing dystrophin from MDys.
Presence of Immune Effector Cells in Virus-Injected Muscles.
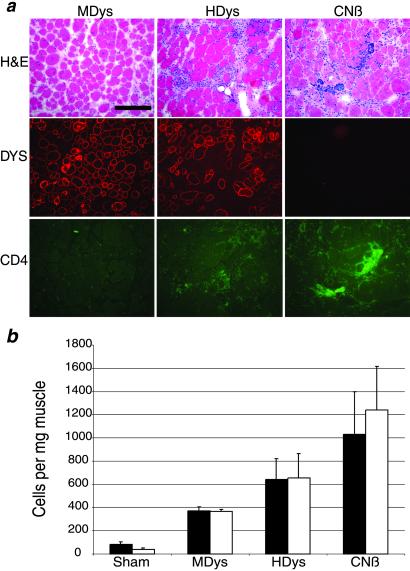
To explore further the adverse functional effects observed in muscles injected with HDys, we repeated the TA injections into 1-yr-old mdx mice with CNβ, a first-generation Ad vector that contains a lacZ gene driven by the cytomegalovirus promoter and expresses highly immunogenic β-gal (35). Shown in Fig. 4a are representative fields from muscles injected 1 mo earlier with MDys, HDys, and CNβ. Hematoxylin/eosin staining revealed the presence of mononuclear cell infiltrate in the muscles injected with HDys and CNβ. In serial sections, immunofluoresence against CD4+ and CD8+ T cells (Fig. 4a and data not shown) demonstrated the presence of a cellular immune response that is most notable in the CNβ-injected muscles. Injected muscles were subjected to flow cytometry analysis to quantify the extent of CD4+ and CD8+ immune cell infiltrates. Although there was a slight immune cell presence in the sham- and MDys-injected muscles, HDys- and CNβ-injected muscles revealed higher levels of CD4+ and CD8+ T cells.
Figure 4.
HDys induces less immune cell infiltration than a first-generation adenovirus expressing β-gal but more than MDys. (a) MDys-, HDys- and CNβ-injected muscle serial cryosections are shown stained with hematoxylin/eosin, anti-dystrophin antisera (Dys, red), or anti-CD4+ cell Ab (CD4+, green). Note the areas of mononuclear cell infiltration (hematoxylin/eosin) and CD4+ cells in the HDys- and CNβ- injected muscles. Results of flow cytometry analysis of injected muscles are shown in b. Filled bars = CD4+ cells; open bars = CD8+ cells. Each bar represents the mean of four muscles ± SEM. Mdx muscles injected with helper adenovirus at the levels present in our gutted Ad preparations contained the same number of infiltrating T cells as did sham-injected muscles (data not shown).
Discussion
Dystrophin “minigenes” have been extensively delivered to neonatal and immunosuppressed adult mdx mice via first-generation Ad vectors (11, 12, 14, 40), despite evidence that supports the use of gutted adenovirus technology (23, 25–28). In addition, although full-length dystrophin expression from a gutted Ad vector has been confirmed in neonates (10, 21, 24–26), dystrophin-carrying gutted viruses have not been tested for effectiveness in adult mdx animals with functional immune systems and more severe physiological deficits. In this study, we showed substantial progress in the development and feasibility of gene therapy for Duchenne muscular dystrophy by using the gutted adenoviral system, muscle-targeted transgene expression, and full-length, species-specific dystrophin cDNAs.
The ability of gutted adenoviruses to express high levels of dystrophin in old mdx mice 1 mo after injection was confirmed by both immunolocalization and Western analysis. We also showed that this high level of full-length dystrophin expression in injected muscles is correlated with a significant decrease in the high susceptibility to contraction-induced injury. Despite the relative inefficiency of the direct i.m. delivery system used here, >25% of the muscle CSA in 1-yr-old mdx mice was transduced. This level of dystrophin expression led to ≈40% correction of the functional difference between mdx and WT muscles, as measured by susceptibility to contraction-induced injury of MDys injected muscles in situ. These data reinforce previous suggestions that dystrophin-positive myofibers can provide functional support to neighboring, dystrophin-negative fibers (41), thereby affording a lower transduction efficiency necessary for functional improvements (19).
Together, these studies show that efficient dystrophin delivery to muscles of immunocompetent, noncompromised, adult mdx mice can be achieved with gutted Ad vectors and can lead to a significant correction of the major functional deficit of dystrophic muscle. Partial alleviation of susceptibility to injury in muscles of Duchenne muscular dystrophy patients may be successful in preventing further dystrophy, delaying the onset of lethal muscle degeneration in diseased muscles. We speculate that methods able to achieve more uniform vector delivery (16, 42) could result in a near complete correction of the dystrophic phenotype.
Acknowledgments
We thank Shana Kellogg for technical assistance. This work was supported by grants from the National Institutes of Health (to J.S.C. and S.V.B.) and the Muscular Dystrophy Association (to J.S.C.). C.D., G.S., and C.B. were partially supported by a predoctoral fellowship from the University of Michigan Rackham Graduate School, postdoctoral fellowships from the Center for Organogenesis at the University of Michigan and the Muscular Dystrophy Association, and a postdoctoral fellowship from the Association Française contre les Myopathies (France), respectively.
Abbreviations
- Ad
adenoviral
- β-gal
β-galactosidase
- IF
immunofluoresence
- TA
tibialis anterior
- MCK
muscle creatine kinase
- CSA
cross-sectional area
- LC
lengthening contraction
- HDys
gutted Ad vector carrying full-length human dystrophin
- MDys
gutted Ad vector carrying full-length mouse dystrophin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Emery A E H. Duchenne Muscular Dystrophy. Oxford: Oxford Medical Publications; 1993. [Google Scholar]
- 2.Miller R G, Hoffman E P. Neurol Clin. 1994;12:699–725. [PubMed] [Google Scholar]
- 3.Hoffman E P, Morgan J E, Watkins S C, Partridge T A. J Neurol Sci. 1990;99:9–25. doi: 10.1016/0022-510x(90)90195-s. [DOI] [PubMed] [Google Scholar]
- 4.Sicinski P, Geng Y, Ryder-Cook A S, Barnard E A, Darlison M G, Barnard P J. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 5.Brooks S V. J Muscle Res Cell Motil. 1998;19:179–187. doi: 10.1023/a:1005364713451. [DOI] [PubMed] [Google Scholar]
- 6.DelloRusso C, Crawford R W, Chamberlain J, Brooks S V. J Muscle Res Cell Motil. 2001;22:467–475. doi: 10.1023/a:1014587918367. [DOI] [PubMed] [Google Scholar]
- 7.Petrof B J, Shrager J B, Stedman H H, Kelly A M, Sweeney H L. Proc Natl Acad Sci USA. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox G A, Cole N M, Matsumura K, Phelps S F, Hauschka S D, Campbell K P, Faulkner J A, Chamberlain J S. Nature (London) 1993;364:725–729. doi: 10.1038/364725a0. [DOI] [PubMed] [Google Scholar]
- 9.Phelps S F, Hauser M A, Cole N M, Rafael J A, Hinkle R T, Faulkner J A, Chamberlain J S. Hum Mol Genet. 1995;4:1251–1258. doi: 10.1093/hmg/4.8.1251. [DOI] [PubMed] [Google Scholar]
- 10.Clemens P R, Kochanek S, Sunada Y, Chan S, Chen H H, Campbell K P, Caskey C T. Gene Ther. 1996;3:965–972. [PubMed] [Google Scholar]
- 11.Ragot T, Vincent N, Chafey P, Vigne E, Gilggenkrantz H, Couton D, Cartaud J, Briand P, Kaplan J, Perricaudet M, et al. Nature (London) 1993;361:647–650. doi: 10.1038/361647a0. [DOI] [PubMed] [Google Scholar]
- 12.Vincent N, Ragot T, Gilgenkrantz H, Couton D, Chafey P, Gregoire A, Briand P, Kaplan J C, Kahn A, Perricaudet M. Nat Genet. 1993;5:130–134. doi: 10.1038/ng1093-130. [DOI] [PubMed] [Google Scholar]
- 13.Hartigan-O'Connor D, Chamberlain J S. Microsc Res Tech. 2000;48:223–238. doi: 10.1002/(SICI)1097-0029(20000201/15)48:3/4<223::AID-JEMT10>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 14.Acsadi G, Lochmüller H, Jani A, Huard J, Massie B, Prescott S, Simoneau M, Petrof B J, Karpati G. Hum Gene Ther. 1996;7:129–140. doi: 10.1089/hum.1996.7.2-129. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y, Nunes F A, Berencsi K, Furth E E, Gonczol E, Wilson J M. Proc Natl Acad Sci USA. 1994;91:4407–4411. doi: 10.1073/pnas.91.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho W K, Ebihara S, Nalbantoglu J, Gilbert R, Massie B, Holland P, Karpati G, Petrof B J. Hum Gene Ther. 2000;11:701–714. doi: 10.1089/10430340050015608. [DOI] [PubMed] [Google Scholar]
- 17.Lochmuller H, Petrof B J, Pari G, Larochelle N, Dodelet V, Wang Q, Allen C, Prescott S, Massie B, Nalbantoglu J, Karpati G. Gene Ther. 1996;3:706–716. [PubMed] [Google Scholar]
- 18.Guibinga G H, Ebihara S, Nalbantoglu J, Holland P, Karpati G, Petrof B J. Mol Ther. 2001;4:499–507. doi: 10.1006/mthe.2001.0482. [DOI] [PubMed] [Google Scholar]
- 19.Chamberlain J S. Basic Appl Myol. 1997;7:251–255. [Google Scholar]
- 20.Harper S Q, Hauser M A, DelloRusso C, Duan D, Crawford R W, Phelps S F, Harper H A, Robinson A S, Engelhardt J F, Brooks S V, et al. Nat Med. 2002;8:253–261. doi: 10.1038/nm0302-253. [DOI] [PubMed] [Google Scholar]
- 21.Haecker S E, Stedman H H, Balice-Gordon R, Smith D, Greelish J P, Mitchell M M, Wells A, Sweeney H L, Wilson J M. Hum Gene Ther. 1996;7:1907–1914. doi: 10.1089/hum.1996.7.15-1907. [DOI] [PubMed] [Google Scholar]
- 22.Kochanek S, Clemens P R, Mitani K, Chen H H, Chan S, Caskey C T. Proc Natl Acad Sci USA. 1996;93:5731–5736. doi: 10.1073/pnas.93.12.5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar-Singh R, Chamberlain J S. Hum Mol Genet. 1996;5:913–921. doi: 10.1093/hmg/5.7.913. [DOI] [PubMed] [Google Scholar]
- 24.Gilbert R, Nalbantoglu J, Howell J M, Davies L, Fletcher S, Amalfitano A, Petrof B J, Kamen A, Massie B, Karpati G. Hum Gene Ther. 2001;12:1741–1755. doi: 10.1089/104303401750476249. [DOI] [PubMed] [Google Scholar]
- 25.Chen H H, Mack L M, Choi S Y, Ontell M, Kochanek S, Clemens P R. Hum Gene Ther. 1999;10:365–373. doi: 10.1089/10430349950018814. [DOI] [PubMed] [Google Scholar]
- 26.Chen H H, Mack L M, Kelly R, Ontell M, Kochanek S, Clemens P R. Proc Natl Acad Sci USA. 1997;94:1645–1650. doi: 10.1073/pnas.94.5.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morral N, Parks R J, Zhou H, Langston C, Schiedner G, Quinones J, Graham F L, Kochanek S, Beaudet A L. Hum Gene Ther. 1998;9:2709–2716. doi: 10.1089/hum.1998.9.18-2709. [DOI] [PubMed] [Google Scholar]
- 28.Schiedner G, Morral N, Parks R J, Wu Y, Koopmans S C, Langston C, Graham F L, Beaudet A L, Kochanek S. Nat Genet. 1998;18:180–183. doi: 10.1038/ng0298-180. [DOI] [PubMed] [Google Scholar]
- 29.Hartigan-O'Connor D, Kirk C J, Crawford R W, Mule J M, Chamberlain J S. Mol Ther. 2001;4:525–533. doi: 10.1006/mthe.2001.0496. [DOI] [PubMed] [Google Scholar]
- 30.Hartigan-O'Connor D, Barjot C, Salvatori G, Chamberlain J. Methods Enzymol. 2002;346:224–246. doi: 10.1016/s0076-6879(02)46058-2. [DOI] [PubMed] [Google Scholar]
- 31.Kunkel L M, Monaco A P, Middlesworth W, Ochs H D, Latt S A. Proc Natl Acad Sci USA. 1985;82:4778–4782. doi: 10.1073/pnas.82.14.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koenig M, Monaco A P, Kunkel L M. Cell. 1988;53:219–226. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- 33. Barjot, C., Hartigan-O'Connor, D., Salvatori, G., Scott, J. M. & Chamberlain, J. S. (2002) J. Gene Med., in press. [DOI] [PubMed]
- 34.Hartigan-O'Connor D, Amalfitano A, Chamberlain J S. J Virol. 1999;73:7835–7841. doi: 10.1128/jvi.73.9.7835-7841.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hauser M A, Robinson A, Hartigan-O'Connor D, Williams-Gregory D A, Buskin J N, Apone S, Kirk C J, Hardy S, Hauschka S D, Chamberlain J S. Mol Ther. 2000;2:16–25. doi: 10.1006/mthe.2000.0089. [DOI] [PubMed] [Google Scholar]
- 36.Crawford G E, Faulkner J A, Crosbie R H, Campbell K P, Froehner S C, Chamberlain J S. J Cell Biol. 2000;150:1399–1410. doi: 10.1083/jcb.150.6.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rafael J A, Cox G A, Corrado K, Jung D, Campbell K P, Chamberlain J S. J Cell Biol. 1996;134:93–102. doi: 10.1083/jcb.134.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Braun S, Thioudellet C, Rodriguez P, Ali-Hadji D, Perraud F, Accart N, Balloul J M, Halluard C, Acres B, Cavallini B, et al. Gene Ther. 2000;7:1447–1457. doi: 10.1038/sj.gt.3301261. [DOI] [PubMed] [Google Scholar]
- 39.Ferrer A, Wells K E, Wells D J. Gene Ther. 2000;7:1439–1446. doi: 10.1038/sj.gt.3301259. [DOI] [PubMed] [Google Scholar]
- 40.Deconinck N, Ragot T, Marechal G, Perricaudet M, Gillis J M. Proc Natl Acad Sci USA. 1996;93:3570–3574. doi: 10.1073/pnas.93.8.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rafael J A, Sunada Y, Cole N M, Campbell K P, Faulkner J A, Chamberlain J S. Hum Mol Genet. 1994;3:1725–1733. doi: 10.1093/hmg/3.10.1725. [DOI] [PubMed] [Google Scholar]
- 42.Greelish J P, Su L T, Lankford E B, Burkman J M, Chen H, Konig S K, Mercier I M, Desjardins P R, Mitchell M A, Zheng X G, et al. Nat Med. 1999;5:439–443. doi: 10.1038/7439. [DOI] [PubMed] [Google Scholar]