Abstract
The p53 tumor suppressor gene product is a transcription factor involved in cell-cycle regulation, apoptosis, and DNA repair. We and others have shown that p53 is required for efficient nucleotide excision repair (NER) of UV-induced DNA lesions. p53-deficient cells are defective in the repair of UV photoproducts in genomic DNA but proficient for transcription-coupled repair. Therefore, we examined whether p53 regulates the expression of genes required for global genomic repair. In this study, we demonstrate that the mRNA and protein products of the xeroderma pigmentosum group C (XPC) gene are UV-inducible in a time- and dose-dependent manner in human WI38 fibroblasts and HCT116 colorectal cancer cells wild type for p53. However, no significant induction of XPC was observed in p53-deficient counterparts to these cells. Furthermore, regulated expression of wild-type p53 in p53 null Li–Fraumeni syndrome human fibroblasts significantly augmented the expression of XPC protein. Analysis of the human XPC gene sequence revealed a putative p53 response element in the XPC promoter that was capable of mediating sequence-specific DNA binding to p53 in vitro. These results provide strong evidence that the NER gene XPC is a DNA damage-inducible and p53-regulated gene and likely plays a role in the p53-dependent NER pathway.
The p53 tumor suppressor gene is a critical mediator of the cellular response to DNA damage. Its function as a tumor suppressor has been attributed to its role as a transcription factor regulating expression of genes involved in DNA damage-response pathways affecting apoptosis, DNA repair, and cell-cycle regulation (1). Recently, we demonstrated that wild-type (wt) p53 is also required for efficient nucleotide excision repair (NER) of UV irradiation-induced cyclobutane pyrimidine dimers (CPDs) from genomic DNA (2–5) and regulates an inducible DNA repair response (6). The mechanism of action of NER in eukaryotes has been well characterized and involves a complex network of proteins that recognize DNA adducts, excise the lesion, and catalyze DNA resynthesis and ligation (7). NER is subdivided into two genetically distinct pathways: global genomic repair (GGR), which functions to repair lesions over the entire genome, and transcription-coupled repair (TCR), which preferentially removes transcription-blocking lesions in the transcribed strand of RNA polymerase II-expressed genes (8, 9). Absence of functional p53 results in deficient GGR of CPDs but has no effect on TCR after UV-C irradiation (2–5, 10, 11).
We postulate that the mechanistic role of p53 in NER is through transcriptional up-regulation of genes, the products of which are involved in the NER pathway. Of particular interest are those gene products that serve a DNA damage recognition function in GGR but are not required for TCR and include xeroderma pigmentosum group C (XPC) (12), hHR23B (13), DDB1, and DDB2 (6). In support of this hypothesis, we recently demonstrated that expression of DDB2, the gene that encodes the p48 protein and when mutant results in xeroderma pigmentosum group E, is regulated transcriptionally by p53 and is required for p53-dependent GGR activity (6, 14, †). Similarly, the p53-regulated gadd45 gene contributes to GGR activity but is not required for TCR (11), although its mechanistic role in GGR remains unclear.
XPC has been identified as an early factor in the NER-reaction mechanism (12, 15). XPC cells are defective in removal of CPDs and 6-4 pyrimidine pyrimidone photoproducts (6-4PPs) from the global genome but repair CPDs selectively from the transcribed strand of active genes (16–18). Cheo et al. demonstrated a similar defect in repair of 6-4PPs from the nontranscribed strand of the p53 gene in murine embryonic fibroblasts lacking functional XPC (19). Activity of XPC in NER requires its coactivating protein hHR23B (20), and the hHR23B interaction domain has been localized to a highly conserved region in the C-terminal half of XPC (21). Although the DNA lesion binding (12, 15, 22) and repair (16, 18) properties of XPC have been characterized in some detail, much remains unknown about its regulation and mode of activation after DNA damage.
In the current study, we examined the DNA damage and p53-dependent induction of XPC protein and mRNA expression in isogenic HCT116 colon cancer cells wt and null for p53, in WI38 normal and human papillomavirus 16 E6-transformed primary human fibroblasts, and in a p53-deficient human fibroblast cell line that allows for regulated p53 induction. This report provides conclusive evidence that the XPC DNA damage recognition factor is regulated transcriptionally by p53 and lends further support to the idea that p53 mediates the DNA damage recognition step in GGR through the coordinate transcriptional control of a set of damage recognition NER factors.
Experimental Procedures
Cell Lines.
HCT116 colon adenocarcinoma cells and a derivative with a homozygous disruption of the p53 gene were a generous gift from Bert Vogelstein (Johns Hopkins University, Baltimore) and were grown in McCoy's 5A modified medium supplemented with 10% FBS and antibiotics. WI38 normal human fibroblasts and a p53-deficient derivative created by the expression of the human papillomavirus 16 E6 gene were obtained from G. Wahl (The Salk Institute, La Jolla, CA) and were cultured in 5% CO2 in DMEM supplemented with 20% serum, antibiotics, l-glutamine, MEM vitamins, and nonessential amino acids. TR9-7 cells, containing a stably transfected tetracycline (tet)-regulated system for expression of human wt p53 cDNA, were obtained from George Stark (Cleveland Clinic Foundation, Cleveland) (23). These cells were grown at 37°C and 10% CO2 in DMEM containing 400 μg/ml G418 sulfate, 150 μg/ml hygromycin B, and 1 μg/ml tet. To obtain maximal p53 expression, tet was removed 12 h before UV irradiation. The above-mentioned cell lines were chosen for this study, because we previously demonstrated that HCT116, WI38, and TR9-7 cells after wt p53 induction all exhibit proficient TCR and GGR of CPDs and 6-4PPs but that matched cell lines deficient for wt p53 are deficient in the GGR of CPDs and, to a lesser extent, 6-4PPs (3, 4, 24).
Western Blotting.
Protein levels were assessed by immunoblotting with antibodies against human p53, XPC, and tubulin. Total protein isolation, quantitation, and Western analysis were performed as described (24). Expression of proteins was detected by using mouse monoclonal antibodies to p53 (DO-1, Santa Cruz Biotechnology) (1:1,000) and XPC (a generous gift from Eva Lee, University of Texas Health Science Center, San Antonio, TX) (1:5) followed by a 1-h incubation with a horseradish peroxidase-conjugated anti-mouse secondary antibody (Pierce) (1:1,000). Anti-tubulin antibodies (B512, Sigma) (1:200,000) were used to control for protein loading. Protein bands were detected by using the Supersignal chemiluminescent substrate (Pierce) and autoradiography (Eastman Kodak) and quantitated by using QUANTITY ONE (Bio-Rad).
mRNA Expression.
To determine the levels of XPC transcripts at various times after UV irradiation, total RNA was isolated by using Trizol (Life Technologies, Grand Island, NY). 10–15 μg of total RNA was separated on a 1% glyoxal gel (Ambion, Austin, TX), transferred overnight onto Hybond N+ Nylon membrane (Amersham Pharmacia) and fixed by UV crosslinking. The membranes were prehybridized at 68°C for ≥2 h in Ultrahyb solution (Ambion). For Northern hybridizations, α-32P-labeled RNA probes were generated by in vitro transcription using the Strip-EZ RNA kit (Ambion). The T7 promoter-containing template for XPC was constructed by PCR using primers designed to add on the T7 promoter sequence to the XPC DNA template. The primer sequences were as follows: XPCFOR, 5′-ATA AAA ACT GGA GTT TGA GAC-3′; and XPCT7, 5′-GGA ATT AAT ACG ACT CAC TAT AGG AGA GCT GCA GAG CCC GGA GAA T-3′. The PCR product then was purified and quantitated, and 1 μg was used as template for the in vitro transcription reaction. pTRI-GAPDH vector (Ambion) was used as a template to generate a GAPDH RNA probe to control for loading. The labeled RNA probes were purified and hybridized to the membranes at 68°C overnight. Low- and high-stringency washes were performed by using the NorthernMax-Gly kit (Ambion) wash buffers according to manufacturer instructions. Band signal intensity was analyzed by using phosphorimager densitometry (Bio-Rad) and quantitated by using QUANTITY ONE (Bio-Rad).
Sequence Analysis.
The human genome draft was used to obtain XPC intronic sequence information as well as promoter sequences up to 10 kb upstream of the XPC start site. String searches were performed to locate potential p53-binding sites [consensus sequence (25): two RRRCWWGYYY motifs separated by 0–13 bp] as well as binding sites for other UV-inducible transcription factors (26–28) in the promotor and intronic sequences.
Electrophoretic Mobility-Shift Assays.
The p53 response elements in the p21 and XPC promoters and XPC intron were used as probes to detect sequence-specific p53 binding in vitro. The probes used were as follows: p21, 5′-TGG CCA TCA GGA ACA TGT CCC AAC ATG TTG AGC TCT GGC A-3′; XPC, 5′-TTA GCC GGG CAT GGT GGC ACA TGC CTG TAG TC-3′; XPCMUT, 5′-TTA GCC GGG GAT CGT GGC AGA TCC CTG TAG TC-3′; XPCL, 5′-TGA AAG CAC TGG CTT AGC CGG GCA TGG TGG CAC ATG CCT GTA GTC CCA GCT ACT CCA AGG G-3′; and XPCintron3, 5′-GAC CAC AGA TAA GGT TGT ACT AGG GAC TTG CTT TGA TAT ATA TGG AAA AAC TCA TGG CCA-3′. p53 response elements are underlined, and mutants are indicated in bold. Briefly, activation of purified recombinant p53 (1 μg) (Santa Cruz Biotechnology) was achieved by a 20-min preincubation with 500 ng of pAb421 anti-p53 antibody at room temperature in a buffer containing 10 mM Tris, pH 7.5, 50 mM KCl, 1 mM DTT, 2.5% glycerol, 5 mM MgCl2, and 0.05% Nonidet P-40 followed by another 20-min incubation with the addition of 20 fmol of biotin end-labeled DNA probe. Biotin labeling of probes was carried out by using the Biotin 3′-end DNA-labeling kit (Pierce) per manufacturer instructions. Single-stranded complementary oligos were labeled individually and annealed for 1 h at room temperature to produce double-stranded probes. For specific or nonspecific competition or antibody supershifts, the respective unlabeled DNA probes (50-fold excess) or antibody (2 μg) (DO-1, Santa Cruz Biotechnology) was added to the reaction mixture before the addition of the labeled probes. The free probe and protein–DNA complexes were separated on a 4% neutral polyacrylamide gel run in 0.5× TBE at 120 V and 4°C. Transfer to nylon N+ membranes was carried out in 0.5× TBE (50 mM Tris/45 mM boric acid/0.5 mM EDTA, pH 8.3) at 380 mA, 4°C for 1 h, and the DNA crosslinked, and detected by the Lightshift electrophoretic mobility-shift assay kit (Pierce) by using streptavidin-horseradish peroxidase binding and chemiluminescent detection.
Results
XPC Is Induced After DNA Damage in a p53-Dependent Manner.
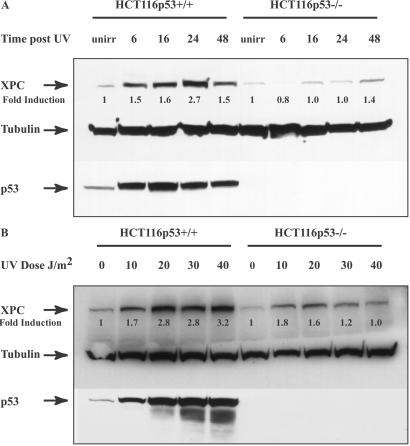
Expression of the XPC gene product after UV irradiation was studied in HCT116 human colon carcinoma cells wt or null for the p53 gene. As expected, in p53 wt HCT116 cells, UV irradiation resulted in increased p53 protein levels, reaching maximum levels by 16 h post-UV (Fig. 1A). XPC protein levels also were induced after UV irradiation, increasing by 1.5-fold by 6 h and reaching a maximum of 2.7-fold by 24 h (Fig. 1A). XPC levels were evaluated also in HCT116 cells with a homozygous disruption of the p53 gene. A very slight induction was observed (up to 1.4-fold) when p53 was absent, but it was not nearly as dramatic as in the presence of p53. The effects of various UV doses on XPC protein levels also were examined in both cell lines (Fig. 1B). A UV dose-dependent induction of XPC was observed both with and without p53. However, both relative and overall levels were significantly higher (up to 3.2 vs. 1.8-fold) when p53 was present.
Figure 1.
XPC and p53 expression in HCT116p53+/+ and HCT116p53−/− cells. (A) HCT116 wt p53 cells as well as cells with homozygous disruptions of the p53 gene were treated with 15 J/m2 UV and lysed at the indicated times thereafter. Total protein was extracted and quantitated as described in Experimental Procedures. Anti-XPC and anti-p53 mouse monoclonal antibodies were used to evaluate XPC and p53 levels after UV irradiation. Tubulin (Sigma) was used as a loading control. unirr, unirradiated. (B) The cells were treated with the varying doses of UV indicated and harvested 24 h later. Antibodies used were as described above. Fold inductions at the various times post-UV were quantified relative to the respective unirradiated levels by using QUANTITY ONE software (Bio-Rad).
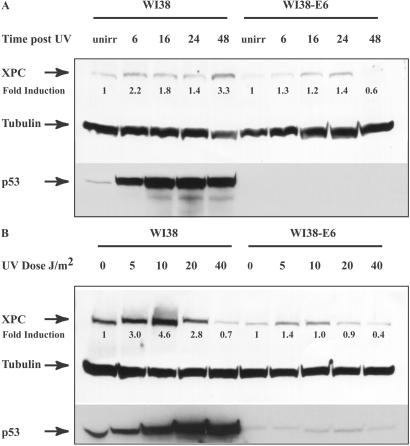
We next measured XPC protein levels in WI38 normal human fibroblasts as well as in a human papillomavirus 16 E6-transformed, p53-dysfunctional counterpart. The E6 protein, in association with the E6AP factor, promotes the ubiquitination and degradation of p53 (29). In the p53 wt fibroblasts, XPC levels increased by 2.2-fold 6 h after UV, reaching a maximum of 3.3-fold by 48 h (Fig. 2A). Minimal induction was observed in the absence of functional p53. The effect of varying UV doses on XPC expression in WI38 cells was also evaluated. Unlike the HCT116 cells, where XPC levels increased with increasing doses up to 40 J/m2, in WI38 cells XPC levels displayed a maximum of 4.6-fold induction with 10 J/m2 UV irradiation and then tapered off at higher doses. An identical pattern was noted in the WI38-E6 cell line, although the induction was not as pronounced with nonfunctional p53. In both sets of cell lines, basal levels of uninduced XPC protein were higher (1.5–2.0-fold) in p53 wt than p53 null cells (Figs. 1 and 2). hHR23B levels remained unchanged after UV treatment in both cell lines (data not shown).
Figure 2.
XPC and p53 expression in WI38 and WI38-E6 cells. (A) Expression of XPC and p53 was evaluated in WI38 normal human fibroblasts as well as in the E6-transformed p53 null counterpart. The times indicated are incubation times after treatment with 15 J/m2 UV irradiation. Antibodies used are as described above. unirr, unirradiated. (B) UV dose response of p53 and XPC expression was studied by using WI38 and WI38-E6 cells. Cells were harvested 48 h after UV treatment. Fold inductions were calculated as described in the Fig. 1 legend.
A DNA-Damage Signal Is Required for the p53-Dependent Activation of XPC Gene Expression.
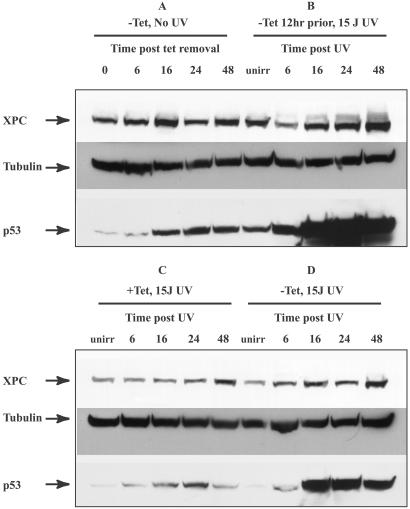
We next examined the effects of expression of wt p53 in a p53-deficient background on XPC levels both in the presence and absence of DNA damage. For this purpose we used the TR9-7 human Li–Fraumeni fibroblast cell line null for p53 and containing a stably integrated tet-regulatable human p53 cDNA (23). This cell line enabled us to evaluate systematically whether XPC induction requires both p53 and DNA damage. Four conditions were tested: (i) induction of p53 in the absence of UV treatment (Fig. 3A), (ii) induction of p53 12 h before UV treatment (Fig. 3B), (iii) UV irradiation in the absence of p53 (Fig. 3C), and (iv) simultaneous p53 induction and UV irradiation (Fig. 3D). p53 induction patterns under these conditions were as expected. The removal of tet resulted in an increase in p53 levels (Fig. 3A), and when the cells were UV-irradiated, further stabilization of p53 was noted (Fig. 3D). When tet was removed 12 h before UV irradiation, higher levels of p53 expression were noted (Fig. 3B). When tet was present in the medium to suppress p53 expression, slight p53 induction was observed (Fig. 3C), possibly due to leakiness of the tet-regulatable promoter. No significant induction of XPC was noted when p53 was induced in the absence of UV treatment (Fig. 3A) or when p53 was absent (Fig. 3C). When p53 was induced simultaneously with UV treatment and tet withdrawal, a slight induction of XPC by 48 h was noted (Fig. 3D). However, in the presence of high levels of p53 and UV-induced DNA damage, a dramatic induction of XPC protein was noted, starting by 16 h and reaching high levels by 48 h (Fig. 3B). Thus, it seems that induction of XPC protein is both p53- and DNA damage-dependent.
Figure 3.
XPC and p53 levels in Li–Fraumeni p53−/− cells containing a tet-regulatable human p53 cDNA. Levels of XPC and p53 proteins were examined after reintroduction of p53 into a p53 mutant background by using the p53-tet regulatable cell line. (A) Tet was withdrawn from the medium, and the cells were sampled at the times indicated after tet removal. Cells were not treated with UV irradiation (unirr). (B) Tet was removed from the medium 12 h before UV irradiation and treated with 15 J/m2. The times indicated are incubation times post-UV irradiation. (C) Medium was supplemented with 2 μg/ml tet, and cells were irradiated with 15 J/m2 UV. The times indicated are post-UV irradiation. (D) Tet was removed at the time of UV irradiation. The times indicated are times following UV and tet withdrawal. The antibodies used were as described in the Fig. 1 legend.
p53-Dependent Regulation of XPC Expression Is Mediated at the Level of Transcription.
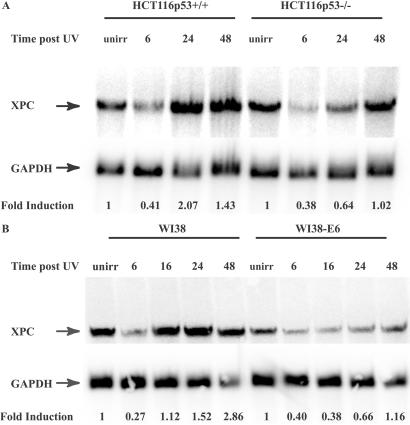
Northern blotting using an XPC RNA probe generated by transcription in vitro was used to examine XPC transcript levels after DNA damage in both HCT116 and WI38 cells ± p53. Induction of XPC mRNA was observed in HCT116 cells after UV irradiation primarily in the presence of p53, reaching maximum levels by 24 h (Fig. 4A). In WI38 cells with wt p53, maximum induction was observed 24 h post-UV, whereas no significant induction was observed in the WI38-E6 cells with deficient p53 (Fig. 4B). hHR23B message levels were also examined, and a decrease post-UV was observed in all cell lines (data not shown), in agreement with previously published results (30). Quantification of XPC levels yielded a 2.07-fold induction of XPC mRNA in wt HCT116 cells vs. a 1.02-fold induction when p53 was absent. In WI38 cells, XPC mRNA was induced 2.86-fold in the presence of p53, compared with a 1.16-fold induction in the E6-transformed WI38 cells.
Figure 4.
Northern blot analysis of XPC mRNA expression in HCT116p53+/+, p53−/−, and WI38 and WI38-E6 cells. (A) Analysis of XPC mRNA expression in HCT116 cells. Times indicated are incubation times after 15 J/m2 UV irradiation. XPC and GAPDH riboprobes were generated as described in Experimental Procedures. GAPDH was probed to control for loading. Fold inductions at the various times post-UV were quantified relative to the respective unirradiated (unirr) levels by using phosphorimager densitometry and QUANTITY ONE software (Bio-Rad). (B) Northern blot of XPC levels in WI38 and WI38-E6 cells. The procedure used was as described in A.
p53 Binds to the XPC Gene p53 Response Element in Vitro.
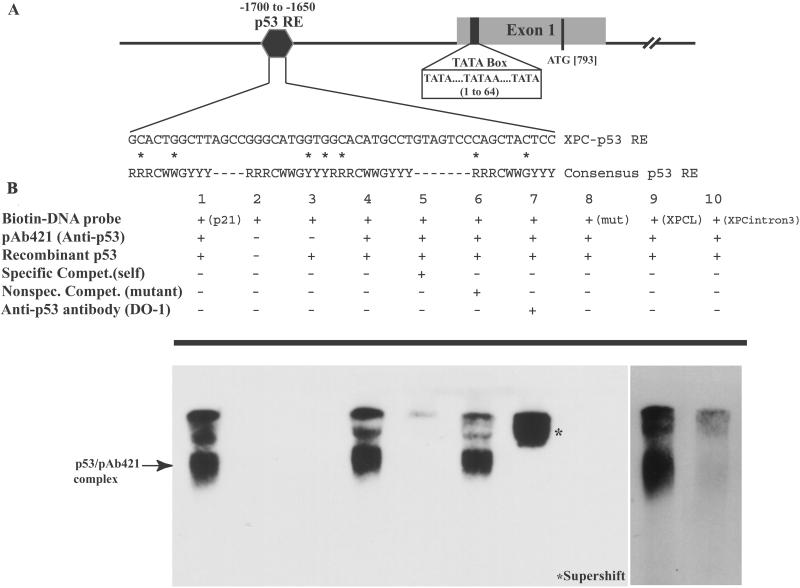
The promoter and intronic sequences of the XPC gene were analyzed to identify potential p53-binding sequences based on the consensus p53 response element (25). A stretch of 50 nucleotides in the XPC promoter at positions 1,650–1,700 bp upstream from the TATA box contained a putative p53 response element (Fig. 5A). One full p53 response element (three mismatches from consensus) and two adjacent half sites (each with two mismatches) were identified within this region. Gel-retardation assays were performed to determine whether p53 bound to this response element in vitro. Fig. 5B demonstrates specific binding of p53 to both the XPC probe encompassing the full element plus the two half sites (XPCL) as well as to the full element (XPC) alone. Because bacterially purified p53 was used, which contains no posttranslational modifications, no sequence-specific binding was noted with recombinant p53 alone (lane 3). With the addition of the monoclonal antibody anti-p53 pAB421, which activates p53 for sequence-specific DNA binding (32), a band shift was noted with the p53 response element in the XPC gene (lane 4) but not to a mutant oligo (lane 8). This binding is inhibited by excess unlabeled XPC probe (lane 5), whereas a nonspecific mutant probe (lane 6) is unable to compete for p53 binding. Furthermore, the addition of a p53-specific antibody (DO-1) induces a supershift (lane 7), strengthening the specificity of the band shift observed. Binding to the XPC full plus two half elements (XPCL probe) was noted also (lane 9), whereas no binding was detected to another potential p53 response element in intron 3 of the XPC gene (XPCintron3, lane 10), containing three overlapping response elements within 60 bp (each with two mismatches from consensus). Binding to the p53 response element of the p21 gene was included as a positive control (lane 1).
Figure 5.
Electrophoretic mobility-shift assay illustrates binding of p53 to the XPC gene p53 response element in vitro. (A) Shown is a schematic (not drawn to scale) of a portion of the promoter and exon 1 of the XPC gene. The numbers represented indicate the nucleotide position relative to the upstream TATA element in the promoter. Three TATA signal elements have been located previously and collectively are referred to here as the TATA box. A candidate p53 response element was identified between nucleotides 1,650 and 1,700 upstream from the TATA box. As indicated, one full p53 response element (three deviations from the consensus) along with two half sites (two mismatches from consensus), one on either side of the full element, have been identified. The mismatched nucleotides are indicated by the asterisk (*). (B) An electrophoretic mobility-shift assay demonstrates sequence-specific DNA binding of bacterially purified p53 to the identified XPC-p53 response element in vitro. Lane 1 indicates binding of p53 to a probe containing the p53 response element in the p21 gene. Lane 2 is free biotinylated probe containing the XPC p53 response element, and lane 3 is the XPC probe with recombinant p53. No band shift was observed and required the activating p53 antibody, pAB421, to observe a sequence-specific mobility shift (lane 4). Fiftyfold unlabeled oligo was used to compete out the band shift (lane 5), whereas a mutant oligo containing a 4-base interchange (see Experimental Procedures for sequence) was unable to achieve this competition (lane 6). Furthermore, addition of another p53 antibody (DO-1) was able to supershift the complex further (lane 7), adding strength to the specificity of the complex. Lane 8 shows absence of binding to the mutant oligo. Binding is noted also when the probe comprises both the full and two half p53-binding sites (XPCL, lane 9), whereas a similar length probe containing another potential p53-binding site in intron 3 (XPCintron3) is not bound by p53 (lane 10). In lanes where band shifts were observed, larger complexes were noted also, potentially because of further oligomerizations. The top-most band is the well, and the excess free probe at the bottom of the gel is not shown here.
To explain the minimal p53-independent induction observed, the XPC promoter was analyzed to identify potential binding sites for other UV-inducible transcription factors (26–28). Three potential OCT-1-binding sites (ATGCAAAT) were located (two in the promoter and one in intron 1) at positions −4,491, −4,435, and +3,614 relative to the XPC TATA element (Fig. 6). Similarly, two possible binding sites for AP-1 (TGACTCA) in intron 1 (+4,662) and intron 2 (+8,298) and one possible EGR-1 element [GCGG(G/T)GGCG] in the promoter (+722) were identified.
Figure 6.
Putative binding sites for UV-inducible transcription factors in the XPC gene. The sequence positions are indicated relative to the TATA box in the XPC promoter. The shaded boxes designate the potential binding sequences for the OCT-1, AP-1, and EGR-1 transcription factors. The EGR-1 putative binding sequence in the XPC gene contains one deviation from the consensus.
Discussion
Although the enzymatic functions of most human NER proteins have been identified, their mode of regulation and activation is still not clearly understood. We have focused on elucidating the molecular mechanism of p53-dependent NER. We and others have shown that functional p53 is required for proficient NER of UVC-induced CPDs in the overall genome (2–5, 10, 32, 33) but is dispensable for TCR of lesions in the transcribed strand of expressed genes (2–5). It has been suggested that p53 may regulate NER through protein–protein interactions (33), protein-nonspecific DNA binding (34), transcriptional control of downstream NER genes (6, 11), or even indirectly through cell-cycle checkpoint functions (35). We previously demonstrated that p53 transcriptionally regulates the basal and UV-inducible expression of the known NER gene DDB2 (6), and that ectopic expression of a DDB2 cDNA allows for complementation of the NER defect associated with p53 deficiency in rodent and human cells (14, †). These results strongly suggested that p53 regulates NER after DNA damage by functioning as a transcriptional activator of repair genes.
To identify other p53-regulated NER genes, we studied the DNA damage- and p53-dependent regulation of various NER factors. We and others have observed the induction of several additional genes involved in NER after UV and/or ionizing radiation including XPC, ERCC1, and PCNA (refs. 36–38 and unpublished results). However, when p53 regulation of these genes was evaluated, we determined a strong bias for the induction of XPC after UV in the presence of p53. Therefore, we evaluated XPC protein and mRNA levels after UV irradiation in a variety of human cell lines including both primary fibroblasts and tumor cells, matched for the presence or absence of wt p53, as well as cells allowing for regulated expression of p53. In all cases we observed a clear time- and dose-dependent induction of XPC mRNA and protein expression in cells expressing wt p53. Significantly less XPC induction was seen in cells absent for p53. The XPC gene spans a region of ≈30 kb, and the initial decrease in levels of XPC mRNA likely is due to UV-induced transcription-blocking lesions in the XPC gene. Because TCR is intact in these cells (3, 4, 24), recovery of mRNA expression is noted by 6–12 h after UV irradiation. The fact that, at least in HCT116 and WI38 cells, XPC protein levels have increased by 1.5–2.0-fold 6 h after UV treatment, before the transcriptional induction of XPC mRNA, suggests an additional, posttranscriptional level of regulation, especially at the early time points after DNA damage. In fact, recent work has demonstrated that XPC (as well as p48) is targeted for degradation by the proteasome pathway (40, ‡), implying a secondary posttranslational mechanism of regulation. Basal XPC mRNA and protein levels before UV irradiation also were significantly higher in p53 wt cells when compared with their p53 null counterparts. Therefore, p53 transcriptionally regulates both the basal and DNA damage-inducible levels of XPC. Furthermore, the p53-dependent induction of XPC seems to require DNA damage, suggesting that damage-specific activation of p53, possibly through phosphorylation or other changes, enhances its ability to induce XPC transcriptionally.
The human genome project has enabled us to examine promoter and intronic sequence information for the XPC gene. Using the p53 consensus binding sequence, we located a strong candidate p53 response element in the promoter of the XPC gene and in vitro gel-retardation assays indicate sequence-specific binding of p53 to this element. Another interesting candidate for p53 binding was identified within the third intron of XPC, containing three overlapping elements, each with two mismatches to the consensus sequence, but no binding of p53 protein to this element was observed experimentally. We also scanned the XPC promoter and intronic sequences for other UV-inducible transcription factor-binding sites such as OCT-1, EGR-1, AP-1, etc. Several candidate binding sites were identified (Fig. 6) and may play a role in the slight p53-independent induction of XPC that we observed after UV irradition.
The precise order of events leading to DNA lesion recognition is unclear. Volker et al. (15) have suggested that NER factors sequentially assemble at the site of DNA lesions in vivo with XPC together with its heterodimer partner hHR23B binding first and XPA involved at later steps. Recent work from our lab suggests that the p48 gene product may be the initial CPD recognition protein in vivo and facilitates XPC binding (M. Fitch and J.M.F, unpublished data). Our current studies on XPC, along with our previously published results with p48 (6), imply that the recognition step of GGR is a p53-mediated process. p53 regulates both the basal and inducible levels of XPC (Figs. 1 and 2) and p48 (6), with protein and mRNA levels significantly higher in p53 wt cells. The basal levels of p48 and XPC are likely critical for immediate damage recognition after UV irradiation. The kinetics of repair of 6-4PPs vs. CPDs is also potentially governed by the varying levels of repair factors. Because XPC has a significantly higher affinity for 6-4PPs over CPDs (22), the basal XPC levels most likely are involved in 6-4PP recognition, as further highlighted by their rapid-repair profiles.
The fate of XPC in the subsequent steps of GGR has not been elucidated. The dramatic increase in XPC levels after damage implies a significant role for XPC in NER in addition to the rapid initial lesion binding. Induction of XPC after DNA damage in p53 wt cells seems to occur between 6 and 48 h post-UV, which parallels the kinetics of repair of CPDs. It is plausible that the induced expression of XPC (and p48) assists in recognition of CPDs less accessible to the repair complex, possibly through chromatin remodeling (40).
The requirement by GGR for two damage-recognition factors regulated by p53 suggests distinct nonoverlapping but potentially synergistic roles for XPC and p48 in UV-induced lesion recognition. Comparative binding kinetics of p53 to the respective p53 response elements in the DDB2 and XPC gene is in progress in our laboratory and should shed some light on the potential differential regulation of these genes by p53. Specific posttranslational modifications of p53 also may control the selectivity in p53 binding to response elements and therefore different modifications may affect gene expression differentially.
The results reported here demonstrate that XPC is a DNA damage-inducible gene and that its expression requires the presence of functional p53. We have illustrated this effect in several cell lines and also identified a strong candidate for a p53 response element in the promoter of the XPC genomic sequence that is recognized specifically and bound by p53. Identification of XPC as a p53-regulated gene, along with our previous observations of p53 regulation of the DDB2 NER gene, provide strong evidence that by directing expression of GGR-specific genes, p53 coordinately controls the damage-recognition step in GGR. Kinetics of p53 binding to the respective response elements in vivo will shed light on the timeline of events regulated by p53 in the initial steps of GGR.
Acknowledgments
We thank Drs. Phil Hanawalt and Ann Ganesan for helpful discussions and critical reading of the manuscript and Irina V. Cross for technical assistance. This work was supported by American Cancer Society Postdoctoral Fellowship PF-0122901-MGO (to S.A.) and National Institutes of Health Award RO1 CA83889, a Sidney Kimmel Foundation for Cancer Research Scholar Award, and a Burroughs Wellcome Fund New Investigator Award in Toxicological Sciences (to J.M.F.).
Abbreviations
- wt
wild type
- NER
nucleotide excision repair
- CPD
cyclobutane pyrimidine dimer
- GGR
global genomic repair
- TCR
transcription-coupled repair
- XPC
xeroderma pigmentosum group C
- 6-4PP
6-4 pyrimidine pyrimidone photoproduct
- tet
tetracycline
Footnotes
Ford, J. M. (2000) Proc. Am. Assoc. Cancer Res. 41, 1131 (abstr.).
Chen, M. C., Whitfield, M., Brown, P. O., Botstein, D. & Ford, J. M. (2002) Proc. Am. Assoc. Cancer Res. 43, 149 (abstr.).
References
- 1.Balint E E, Vousden K H. Br J Cancer. 2001;85:1813–1823. doi: 10.1054/bjoc.2001.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ford J M, Hanawalt P C. Proc Natl Acad Sci USA. 1995;92:8876–8880. doi: 10.1073/pnas.92.19.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ford J M, Hanawalt P C. J Biol Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- 4.Ford J M, Baron E L, Hanawalt P C. Cancer Res. 1998;58:599–603. [PubMed] [Google Scholar]
- 5.Bowman K K, Sicard D M, Ford J M, Hanawalt P C. Mol Carcinog. 2000;29:17–24. [PubMed] [Google Scholar]
- 6.Hwang B J, Ford J M, Hanawalt P C, Chu G. Proc Natl Acad Sci USA. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wood R D. Biochimie. 1999;81:39–44. doi: 10.1016/s0300-9084(99)80036-4. [DOI] [PubMed] [Google Scholar]
- 8.Mellon I, Spivak G, Hanawalt P C. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 9.de Laat W L, Jaspers N G, Hoeijmakers J H. Genes Dev. 1999;13:768–785. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 10.Wani M A, Zhu Q Z, El Mahdy M, Wani A A. Carcinogenesis. 1999;20:765–772. doi: 10.1093/carcin/20.5.765. [DOI] [PubMed] [Google Scholar]
- 11.Smith M L, Ford J M, Hollander M C, Bortnick R A, Amundson S A, Seo Y R, Deng C X, Hanawalt P C, Fornace A J., Jr Mol Cell Biol. 2000;20:3705–3714. doi: 10.1128/mcb.20.10.3705-3714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugasawa K, Ng J M, Masutani C, Iwai S, van der Spek P J, Eker A P, Hanaoka F, Bootsma D, Hoeijmakers J H. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 13.Sugasawa K, Ng J M, Masutani C, Maekawa T, Uchida A, van der Spek P J, Eker A P, Rademakers S, Visser C, Aboussekhra A, et al. Mol Cell Biol. 1997;17:6924–6931. doi: 10.1128/mcb.17.12.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang J Y, Hwang B J, Ford J M, Hanawalt P C, Chu G. Mol Cell. 2000;5:737–744. doi: 10.1016/s1097-2765(00)80252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volker M, Mone M J, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers J H, van Driel R, van Zeeland A A, Mullenders L H. Mol Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 16.Venema J, van Hoffen A, Karcagi V, Natarajan A T, van Zeeland A A, Mullenders L H. Mol Cell Biol. 1991;11:4128–4134. doi: 10.1128/mcb.11.8.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venema J, van Hoffen A, Natarajan A T, van Zeeland A A, Mullenders L H. Nucleic Acids Res. 1990;18:443–448. doi: 10.1093/nar/18.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emmert S, Kobayashi N, Khan S G, Kraemer K H. Proc Natl Acad Sci USA. 2000;97:2151–2156. doi: 10.1073/pnas.040559697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheo D L, Ruven H J, Meira L B, Hammer R E, Burns D K, Tappe N J, van Zeeland A A, Mullenders L H, Friedberg E C. Mutat Res. 1997;374:1–9. doi: 10.1016/s0027-5107(97)00046-8. [DOI] [PubMed] [Google Scholar]
- 20.Sugasawa K, Masutani C, Uchida A, Maekawa T, van der Spek P J, Bootsma D, Hoeijmakers J H, Hanaoka F. Mol Cell Biol. 1996;16:4852–4861. doi: 10.1128/mcb.16.9.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L, Lu X, Peterson C, Legerski R. Mutat Res. 1997;383:197–203. doi: 10.1016/s0921-8777(97)00002-5. [DOI] [PubMed] [Google Scholar]
- 22.Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F. Genes Dev. 2001;15:507–521. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agarwal M L, Agarwal A, Taylor W R, Stark G R. Proc Natl Acad Sci USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adimoolam S, Lin C X, Ford J M. J Biol Chem. 2001;276:25813–25822. doi: 10.1074/jbc.M102240200. [DOI] [PubMed] [Google Scholar]
- 25.el Deiry W S, Kern S E, Pietenpol J A, Kinzler K W, Vogelstein B. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 26.Zhao H, Jin S, Fan F, Fan W, Tong T, Zhan Q. Cancer Res. 2000;60:6276–6280. [PubMed] [Google Scholar]
- 27.Devary Y, Gottlieb R A, Lau L F, Karin M. Mol Cell Biol. 1991;11:2804–2811. doi: 10.1128/mcb.11.5.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Chen S. Exp Cell Res. 2001;266:21–30. doi: 10.1006/excr.2001.5196. [DOI] [PubMed] [Google Scholar]
- 29.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 30.van der Spek P J, Visser C E, Hanaoka F, Smit B, Hagemeijer A, Bootsma D, Hoeijmakers J H. Genomics. 1996;31:20–27. doi: 10.1006/geno.1996.0004. [DOI] [PubMed] [Google Scholar]
- 31.Gu W, Roeder R G. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 32.Smith M L, Chen I T, Zhan Q, O'Connor P M, Fornace A J., Jr Oncogene. 1995;10:1053–1059. [PubMed] [Google Scholar]
- 33.Wang X W, Yeh H, Schaeffer L, Roy R, Moncollin V, Egly J M, Wang Z, Freidberg E C, Evans M K, Taffe B G, et al. Nat Genet. 1995;10:188–195. doi: 10.1038/ng0695-188. [DOI] [PubMed] [Google Scholar]
- 34.Jayaraman J, Prives C. Cell. 1995;81:1021–1029. doi: 10.1016/s0092-8674(05)80007-8. [DOI] [PubMed] [Google Scholar]
- 35.Lane D P. Nature (London) 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 36.Hall P A, McKee P H, Menage H D, Dover R, Lane D P. Oncogene. 1993;8:203–207. [PubMed] [Google Scholar]
- 37.Amundson S A, Do K T, Shahab S, Bittner M, Meltzer P, Trent J, Fornace A J., Jr Radiat Res. 2000;154:342–346. doi: 10.1667/0033-7587(2000)154[0342:iopmbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 38.van Laar T, van der Eb A J, Terleth C. Mutat Res. 2002;499:53–61. doi: 10.1016/s0027-5107(01)00291-3. [DOI] [PubMed] [Google Scholar]
- 39.Rapic-Otrin V, McLenigan M P, Bisi D C, Gonzalez M, Levine A S. Nucleic Acids Res. 2002;30:2588–2598. doi: 10.1093/nar/30.11.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baxter B K, Smerdon M J. J Biol Chem. 1998;273:17517–17524. doi: 10.1074/jbc.273.28.17517. [DOI] [PubMed] [Google Scholar]