Abstract
Regulatory CD25+CD4+ T cells are considered as important players in T cell homeostasis and self-tolerance. Here we report that the integrin αEβ7, which recognizes epithelial cadherin, identifies the most potent subpopulation of regulatory CD25+ T cells. Strikingly, CD25-negative αE+CD4+ T cells displayed regulatory activity. Both αE+ subsets, CD25+ and CD25−, express CTLA-4, suppress T cell proliferation in vitro, and protect mice from colitis in the severe combined immunodeficient model (SCID) in vivo. Whereas αE+CD25+ T cells produce almost no cytokines, αE+CD25− T cells represent a unique subset in which high IL-2, IFN-γ and T helper 2-cytokine production is linked with suppressive function. Thus, the integrin αEβ7 can be regarded as a novel marker for subsets of highly potent, functionally distinct regulatory T cells specialized for crosstalk with epithelial environments.
Besides central tolerance, several mechanisms of peripheral tolerance, including the activity of suppressive T cells, synergize in balancing the reactivity of the immune system and preventing autoimmunity. Results of the last few years indicated that subsets of CD4+ rather than CD8+ T cells might have a central role as regulatory cells (1, 2). In vivo reconstitution models as well as in vitro analyses provided evidence that CD4+ T cells with regulatory function were to be found among CD45RBlow and CD25+ cells (3–5). Recent studies focused on CD25 as the best marker for regulatory CD4+ T cells in mice and humans (6), though its function as an activation-induced cytokine receptor component is apparently unrelated to the regulatory function and does not allow a discrimination of the regulatory subset from activated T cells. In fact, it has not been clarified so far whether the whole population of CD25+ cells is regulatory or whether subsets exist which are distinct in potency or mechanisms used for suppression. Moreover, a regulatory function has been demonstrated in subsets negative for CD25 (7–9).
The mechanisms of action of CD25+CD4+ regulatory T cells remain controversial: in some studies, cytotoxic T lymphocyte antigen-4 (CTLA-4) has been found to be involved (10–12), whereas others have excluded a significant role (13–15). IL-10-deficient mice spontaneously develop colitis (16), and IL-10 was found to be required in control of autoreactivity in some models (9, 17, 18). Transforming growth factor (TGF)-β is another crucial cytokine mediating generalized control of autoimmunity (19–23), oral tolerance (24) and regulatory effects in vitro (25). However, most in vitro experimental data using regulatory CD25+CD4+ T cells point to cell contact-dependent mechanisms rather than soluble mediators (13, 26). Recently, evidence has been provided that surface-bound TGFβ might be a key mediator of suppression acting by means of direct T cell interaction (27), thereby resolving some of the discrepancies. Regulatory CD25+CD4+ T cells show a partially anergic phenotype, in that they proliferate poorly on T cell receptor (TCR) stimulation in vitro and their growth depends on exogenous IL-2 (28). Furthermore, a peculiar pattern of cytokine expression showing reduced IL-2, IL-4, IFN-γ and tumor necrosis factor (TNF)-α but high IL-10 production was found (13, 29, 30).
The origin of regulatory CD25+CD4+ T cells is not yet well defined. As CD25+CD4+ cells with suppressive capacities have already been found within the thymus, it has been proposed that they represent a distinct lineage (31–33). Others provided evidence that induction of oral tolerance induces CD25+CD4+ T cells in the mucosal compartment acting by means of TGFβ (24, 34, 35).
The integrin αEβ7 was initially described as a marker for intraepithelial T cells residing in the gut wall and other epithelial compartments such as skin or lung (36, 37). Whereas the related integrin α4β7 serves as a homing receptor for mucosa-seeking populations by recognizing mucosal addressin cell adhesion molecule-1 (MAdCAM-1) (38), the only ligand for αEβ7 clearly identified so far is E-cadherin, expressed on epithelial cells but not on endothelium (39). A role in homing is therefore unlikely (40), though the existence of a further ligand on endothelium has been postulated (41). Conceivable is a role of αEβ7 in retention of T cells within epithelial compartments (42). Early data demonstrated a costimulatory role of αEβ7 on T cells (43); yet the functional impact of interactions between T cells and epithelial cells has not been further investigated. Recently, global gene expression analysis has revealed αE expression on regulatory T cell populations (44–46). Data from αE-deficient animals suggest that the molecule might indeed be involved in the control of autoimmunity in the skin (47).
In the present study we report that CD4+ T cells from lymphoid tissues expressing the integrin αEβ7 represent previously uncharacterized subpopulations of regulatory T cells. Expression of CTLA-4, cytokine profiles, and suppressive properties in vitro and in vivo are distinct for αE-expressing CD4+ T cells, αE+CD25+ well as αE+CD25−, and identify them as unique subsets with highly potent regulatory function.
Materials and Methods
Mice.
Female BALB/c and C57Bl6 mice were bred in our animal facility and used at 6–12 weeks of age. For analysis of cytokine expression, approximately 10-month-old BALB/c mice were used. Perforin-deficient mice (48) were kindly provided by U. Steinhoff. Female C.B-17 severe combined immunodeficient (SCID) mice, obtained from Charles River Breeding Laboratories (Sulzfeld, Germany), were used at 5–9 weeks of age. All animal experiments were performed under specific pathogen-free conditions and in accordance with institutional, state, and federal guidelines.
Antibodies, Staining, and Sorting Reagents.
The following antibodies were purified and labeled in our laboratory: anti-FcR II/III (2.4G2), anti-CD3 (145.2C11), FITC- and Cy5-labeled anti-CD4 (GK1.5), biotinylated anti-αE (M290), anti-CTLA-4 (4F10), anti-IL-10 (JES5.2A5.7G4), phycoerythrin (PE)-labeled anti IL-13 (382.13.11) and rat IgG1 isotype control (1A5). The following antibodies and secondary reagents [FITC-, PE-, Cy5-, antigen presenting cell (APC)-labeled or biotinylated] were purchased from BD PharMingen (Heidelberg, Germany): anti-CD4 (RM4–5), anti-CD62L (Mel-14), anti-CD25 (PC61), anti-CD25 (7D4), anti-CD38 (90), anti-CD45RB (16A), anti-CD44 (IM7), anti-γδTCR (GL3), anti-FasL (MLF3), anti-IL-2 (JES6–5H4), anti-IL-4 (11B11), anti-IL-5 (TRFK5), anti-IL-10 (JES5–16E3), anti-TNFα (MP6-XT22), anti-IFN-γ (XMG1.2), streptavidin (SA), anti-hamster IgG1, and isotype controls. Polyclonal anti-TGFβ was purchased from R&D Systems (Wiesbaden, Germany). All microbeads were obtained from Miltenyi Biotec (Bergisch-Gladbach, Germany).
Cell Purification and Culture Conditions.
CD4+ T cell subsets were isolated from pooled spleens and peripheral and mesenteric lymph nodes from BALB/c mice. Briefly, CD4+ T cells were enriched by using the MACS MultiSort kit and the AutoMACS magnetic separation system (Miltenyi Biotec). The different regulatory subpopulations were separated as follows: after staining CD4+ T cells with biotinylated anti αE, PE-labeled SA and anti-PE MicroBeads, the αE+ cells were isolated by using AutoMACS technology (Miltenyi Biotec). Afterward, these αE-enriched cells were stained with APC-labeled anti-CD25, and the αE+CD25+ and αE+CD25− subsets were separated with the Vantage fluorescence activated cell sorter (FACS) (BD Biosciences, Heidelberg, Germany). αE−CD4+ T cells were stained with biotinylated anti-CD25 and anti-biotin MicroBeads, and the αE−CD25+ as well as the αE−CD25− subsets were isolated by AutoMACS. Naive CD4+CD62L+ cells were positively selected from the αE−CD25− fraction on a AutoMACS by using anti-CD62L microbeads. APCs were prepared by depletion of CD90+ cells from BALB/c spleen cells using anti-CD90 microbeads and irradiated (30 Gy) before culture. CD45RBhigh and CD45RBlow CD4+ T cells were FACS sorted. All sorted subsets were >95–99% pure on reanalysis (for data for the investigated subsets, see Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org). Cell cultures were done with RPMI medium 1640 (GIBCO/BRL) plus 10% FCS (Linaris, Wertheim-Bettingen, Germany).
Flow Cytometry.
Cytometric analysis was performed as described (49), using a FACS Calibur (BD Biosciences) and cellquest software. For CTLA-4 detection, cells were stained for surface expression of CD4, αE, and CD25, fixed, and permeabilized. Intracellular expression of CTLA-4 was detected with hamster anti-mouse CTLA-4 and Cy5-labeled anti-hamster IgG. To analyze the expression of cytokines of the particular subpopulations, MACS-sorted CD25+ and CD25− cells were stimulated for 4 h with phorbol 12-myristate 13-acetate (PMA) (10 ng/ml) plus ionomycin (250 ng/ml) or alternatively for 5 h with plate-bound anti-CD3 plus anti-CD28 (each 5 μg/ml) by the addition of BrefeldinA (10 μg/ml) after 2 h. Before fixation and intracellular staining, CD25+ and CD25− cells were stained for surface CD4 and αE, whereas CD25− cells were additionally stained with anti-CD45RB. PMA/ionomycin stimulation did not change the expression of αE and CD45RB (data not shown). Unspecific binding was blocked by addition of whole rat IgG (Jackson ImmunoResearch).
In Vitro Proliferation Assay.
Unlabeled CD4+ T cell subpopulations were mixed with CFSE-labeled naive T cells (5-carboxy-fluorescein diacetate succinimidyl ester; Molecular Probes; staining as described in ref. 50) at different regulator/target ratios and cultured with APC at a ratio of 1:2 in round-bottom microtiter plates with or without addition of anti-CD3 (1 μg/ml) for 80 h in duplicates. Blocking antibodies were used at final concentrations of 20 μg/ml. Transwell chambers (0.4 μm pore size) were obtained from Costar. After incubation cells were collected, stained for CD4 and analyzed by FACS; propidium iodide was added to exclude dead cells. Proliferation analysis is based on CFSE+CD4+ T cells. The mean number of cell divisions (d) was calculated with the formula: d = ∑(ni/nt × gi), where gi = generation number starting with 0 for nondivided cells, ni = number of cells within each generation, and nt = total cell number.
T Cell Reconstitution.
C.B-17 SCID mice were injected i.p. with sorted CD4+ T cell subpopulations in PBS. Mice received CD45RBhighCD4+ T cells alone or in combination with CD4+ T cell subpopulations (CD45RBlow, αE+CD25+, αE−CD25+, or αE+CD25−) at ratios and cell counts as indicated.
Clinical and Histological Examination.
Weight changes of reconstituted C.B-17 SCID mice were scored weekly and monitored with clinical symptoms (0: no signs of clinical progression; 1: hunched posture, impaired movement or bristeled fur; 2: liquid feces; 4: rectal prolapse). For histology, colon tissue samples were taken about 10 weeks after T cell reconstitution and fixed in 10% phosphate-buffered formalin. Paraffin-embedded sections were cut and stained with hematoxilin and eosin. The degree of inflammation of the colon was graded in a blinded fashion semiquantitatively from 0 to 4 (0: no signs of inflammation; 1: very low levels of and only focal infiltrating mononuclear cells; 2: low levels and only focal infiltration of mononuclear cells; 3: high levels of mononuclear cell infiltration, high vascular density, thickening of the colon wall; 4: transmural inflammatory infiltration, loss of goblet cells, high vascular density, vasculitis, thickening of the colon wall). The severity of colitis was assessed by both scores together with the body weight changes. An overall score ≥3 was considered as moderate or severe colitis. No animals showed inflammatory signs in the small intestine (data not shown).
Statistics.
Data were presented as mean ± SD. Significance was determined by Student's t test (CTLA-4), Wilcoxon test (proliferation assay), repeated measures analyses (body weight), and Mann–Whitney U test (severity of colitis). Differences were considered statistically significant with P ≤ 0.05 and highly significant with P ≤ 0.01.
Results
αEβ7 Expression on CD4+ T Cells in Lymphoid Tissues Is Correlated with CD25 and CD45RBlow Expression.
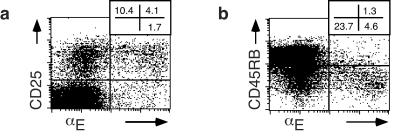
The presence of a small population of αE-expressing CD4+ T cells has been known for long time; however, their functional role remained elusive. We therefore characterized αE+CD4+ T cells from pooled lymphoid organs, which comprise between 2 and 6% of total CD4+ cells in spleen, peripheral lymph nodes, and mesenteric lymph nodes, and are almost exclusively αβTCR+ cells (>98%; data not shown). Flow cytometry analysis revealed that about 75% of αE-expressing CD4+ T cells are CD25+ (Fig. 1a). Furthermore, αE+CD4+ T cells were mainly found within the memory (CD45RBlow) compartment (Fig. 1b), regardless of their CD25 expression, and expressed CD38 and high levels of CD44 (data not shown).
Figure 1.
Expression of αEβ7 integrin on CD4+ T cells is correlated with CD25 and CD45RBlow. (a) FACS analysis of pooled lymphocytes (spleen and lymph nodes) shows the expression of αE and CD25 within the CD4+ T cell pool (3.6 ± 0.7% αE+CD25+, 9.9 ± 2.5% αE−CD25+, and 1.2 ± 0.3% αE+CD25−; mean ± SD from six independent experiments). (b) Expression of αE and CD45RB on gated CD4+ T cells (one representative out of four independent experiments). The memory (CD45RBlow) state of αE+ cells is not correlated to CD25 expression (data not shown).
Consistent with its inhibitory function, CTLA-4 has been found on regulatory CD4+ T cells (12) and proposed to be involved in their function (10, 11). We therefore examined the coexpression of CTLA-4 with αE and CD25 on CD4+ T cells (Fig. 8, which is published as supporting information on the PNAS web site). About 18% of cells within the αE+CD25+ and the αE+CD25− fraction of CD4+ T cells expressed high levels of intracellular CTLA-4 (18.8 ± 5.7% and 17.6 ± 5.7%, respectively), whereas only 6.8 ± 3.2% and 0.5 ± 0.3% did so within the αE−CD25+ and αE−CD25− subpopulation, respectively (P < 0.01, n = 5). Thus, the expression of CTLA-4 strongly correlates with that of αEβ7 integrin on CD4+ T cells.
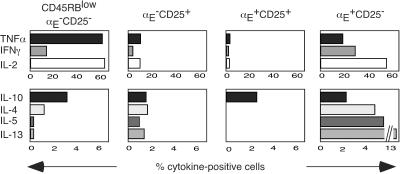
Distinct Cytokine Profiles of αE+ Subsets.
It has been reported that regulatory CD25+CD4+ T cells produce low amounts of proinflammatory cytokines as compared with other antigen-experienced CD4+ T cells (13, 26, 29, 30). Within the αE−CD25+ fraction, the frequencies of TNFα, IFN-γ, or IL-2 producing cells after restimulation were lower compared with the frequencies among CD45RBlow memory cells. T helper 2 (Th2) cytokines such as IL-4, IL-5, or IL-13 were less diminished or even higher (Fig. 2). In contrast, the αE+CD25+ population showed almost no IL-4, IL-5, and IL-13, and extremely small numbers of cells producing TNFα, IFN-γ, or IL-2. The only cytokine preserved in a reasonable frequency among αE+CD25+ cells was IL-10.
Figure 2.
αE+CD25+ cells do not produce proinflammatory cytokines on stimulation, and αE+CD25− cells display a unique pattern of Th2 cytokines. Sorted CD25+ and CD25− cells were restimulated with phorbol 12-myristate 13-acetate/ionomycin and stained for expression of CD4, αE, CD45RB, and intracellular cytokines. The frequencies of cytokine-producing cells were analyzed by gating on the following subpopulations: memory (CD45RBlow) αE−CD25−, αE−CD25+, αE+CD25+, and αE+CD25− CD4+ T cells (one representative out of three independent experiments). Similar results were obtained by triggering the cells with anti CD3 plus anti-CD28 (Fig. 10, which published as supporting information on the PNAS web site). The expression of αE and CD45RB was stable during restimulation (data not shown).
The αE single positive cells had a unique cytokine expression pattern. Whereas the frequencies of IL-2- and IFN-γ-positive cells were almost the same as in memory CD4+ T cells, the frequencies were lower for TNFα and IL-10, slightly higher for IL-4, and much higher for IL-5 and IL-13 (Fig. 2). The various cytokine producers and nonproducers among the αE+ single positive subset displayed similar levels of αE expresssion (Fig. 9, which is published as supporting information on the PNAS web site).
αE Expression Characterizes the Most Potent Regulatory CD25+CD4+ T Cells.
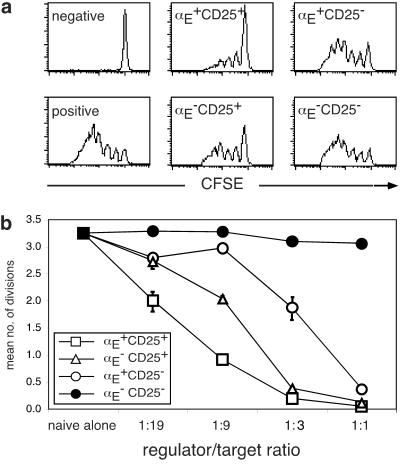
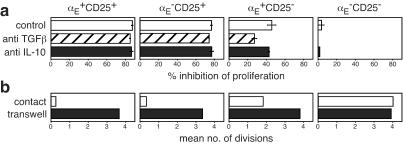
To analyze the regulatory capacity of CD4+ T cell subsets characterized by αE and CD25 expression, we separated four populations (αE+CD25+, αE−CD25+, αE+CD25−, and αE−CD25−, see Fig. 7) and tested their suppressive activity in in vitro proliferation assays. Naive CD4+ T cells were labeled with CFSE and stimulated for 80 h with anti-CD3 and APC. During this time the majority of naive T cells had undergone multiple proliferations indicated by the loss of CFSE content (positive control; Fig. 3a). Addition of αE+CD25+ cells at a regulator/target ratio of 1:9 showed an almost complete inhibition of naive T cell proliferation, whereas αE−CD25+ cells inhibited the proliferation only to a lesser degree. Remarkably, even αE+CD25− cells had a significant regulatory capacity resulting in a marked inhibition of naive T cell proliferation. αE−CD25− cells were used as a control population without any suppressive activity. The relative potencies of the subsets were studied in more detail by titrating the regulator/target ratios (Fig. 3b). At a 1:19 ratio only αE+CD25+ cells showed a significant inhibitory effect on naive T cell proliferation. Both αE−CD25+ and αE+CD25− cells displayed inhibitory effects at higher ratios (1:9 and 1:3), but remained inferior to the αE+CD25+ population (P < 0.005). At a ratio of 1:1, the proliferation of naive T cells was completely inhibited by all three subsets and CD45RBlow cells (Fig. 3b and data not shown). The αE expressing subsets remained positive during the culture time, whereas in the subset of αE−CD25+ a fraction of cells expressing low levels of αE emerged within 80 h. In contrast, CD25 was present on all T cells, regulatory subsets as well as naive target cells, after 80 h of stimulation (Fig. 11, which is published as supporting information on the PNAS web site). Thus, αE+CD4+ T cells from lymphoid tissues possess regulatory capacity, and the αE single positive subset represents a previously uncharacterized regulatory CD4+ T cell population outside the CD25+ compartment. To investigate whether the inhibitory effect of the regulatory subsets is mediated by soluble immunosuppressive cytokines, we blocked IL-10 and TGFβ in the in vitro proliferation assays. Addition of neutralizing antibodies against IL-10 or TGFβ had no major effect on the inhibition of the naive T cell proliferation by all regulatory subsets. Only the αE single positive cells showed a slightly reduced inhibitory capacity if TGFβ was blocked. (Fig. 4a). Moreover, we found that the inhibitory function of all regulatory subsets depends on cell contact as no suppression of naive T cell proliferation was observed if the regulatory cells and the targets were separated in culture by transwell chambers (Fig. 4b).
Figure 3.
αE+CD25+ cells show the strongest regulatory capacity in vitro. The regulatory capacities of the indicated CD4+ subpopulations were determined after 80 h of coculture with CFSE-labeled naive CD4+ T cells. As positive and negative controls naive T cells alone were cultured with or without anti CD3. The fluorescence intensities of CFSE+ T cells at a regulator/target ratio of 1:9 (a) and the comparison of the regulatory capacities of all subsets at different regulator/target ratios (αE+CD25+/□, αE−CD25+/▵, αE+CD25−/○ or αE−CD25−/●) (b) are shown. Shown is the mean number of cell divisions of the CFSE+ T cells (one representative out of six independent experiments; regulator/target ratio 1:1 from one experiment).
Figure 4.
All regulatory T cell subsets require cell contact to exert their regulatory function. (a) CFSE-labeled naive CD4+ T cells and the indicated CD4+ subpopulations were cocultured at a regulator/target ratio of 1:3 for 80 h together with control antibodies or neutralizing antibodies against IL-10 or TGFβ. The % inhibition of naive T cell proliferation (one representative out of two independent experiments) is shown. (b) CFSE-labeled naive CD4+ T cells were cocultured together with the indicated CD4+ subpopulations at a regulator/target ratio of 1:1 for 80 h or the CFSE+ cells and the regulatory T cells were separated by a Transwell insert. The mean number of cell divisions of the CFSE+ T cells (one representative out of two independent experiments) is shown.
αE+CD4+ T Cells Inhibit Development of Induced SCID Colitis.
To further analyze function and potency of the inquired CD4+ T cell subpopulations in vivo, their ability to inhibit the development of colitis after transfer of naive (CD45RBhigh) CD4+ T cells into SCID mice was tested. In a first experimental series, mice reconstituted with naive CD4+ T cells alone developed colitis in 88% of the cases (n = 17), whereas none of the mice reconstituted with naive cells plus either αE+CD25+, αE−CD25+, αE+CD25−, or CD45RBlow CD4+ T cells (ratio of 3:1) developed colitis. The body weights of all mice that in addition to naive cells received protective CD4+ T cells (αE+CD25+, αE−CD25+, αE+CD25−, or CD45RBlow) increased to a similar degree, whereas that of mice reconstituted with naive cells alone decreased (P ≤ 0.005; Fig. 5a). Histological examination of colonic tissue samples from mice reconstituted with naive cells alone revealed transmural and, in some cases, only focal mononuclear infiltrates and loss of goblet cells (Fig. 6a). These inflammatory signs were partly accompanied by ulceration or vasculitis (data not shown). In contrast, colons restored with a mixture of naive cells plus either αE+CD25+, αE−CD25+, or αE+CD25− cells exhibited no detectable pathological changes (Fig. 6 b–e). These results demonstrate that αE+ subsets both inside and outside the CD25+CD4+ compartment are effective regulators of intestinal inflammation in vivo.
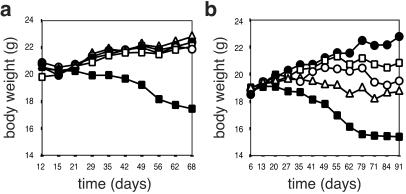
Figure 5.
αE+CD4+ T cells, CD25+ and CD25−, suppress induced colitis in SCID mice. (a) SCID mice received 3 × 105 CD45RBhighCD4+ T cells alone (■, n = 17) or in combination with 1 × 105 CD4+ T cells from either subpopulation (regulator/target ratio 1:3): CD45RBlow (●, n = 4), αE+CD25+ (□, n = 4), αE−CD25+ (▵, n = 4), or αE+CD25− (○, n = 4). The progression of colitis is monitored by the adjusted body weight loss. (b) SCID mice received 3 × 105 CD45RBhighCD4+ T cells alone (■, n = 7) or in combination with 5 × 104 CD4+ T cells from either subpopulation (regulator/target ratio 1:6): αE+CD25+ (□, n = 8), αE−CD25+ (▵, n = 8), or αE+CD25− (○, n = 8). Control mice receiving PBS only (●, n = 5) do not differ from mice receiving CD45RBhighCD4+ T cells plus CD45RBlow at a ratio of 3:1 (data not shown).
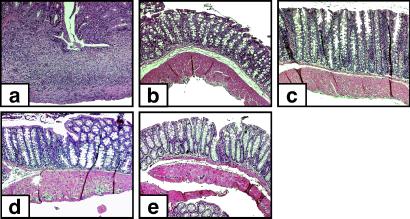
Figure 6.
Intestinal pathology in SCID mice reconstituted with CD45RBhighCD4+ T cells and subsets of regulatory CD4+ T cells. SCID mice received 3 × 105 CD45RBhighCD4+ T cells alone or in combination with 1 × 105 CD4+ T cells from the regulatory subpopulations (regulator/target ratio 1:3). After the investigation period, tissues were taken for histological examination. Representative photomicrographs show severe colitis (transmural infiltrations with ulceration) in mice that received CD45RBhighCD4+ T cells alone (a) and complete protection from colitis in mice given additionally CD45RBlow (b), αE+CD25+ (c), αE−CD25+ (d), or αE+CD25− (e) CD4+ T cells at the high regulator/target ratio used here.
To elucidate whether the inquired regulatory subpopulations differ in their suppressive capacity also in vivo, as we found in vitro, SCID mice were reconstituted in a second experimental series using lower regulator/target ratios. Analyses of the body weights of mice reconstituted with naive plus protective CD4+ T cells at the ratio of 6:1 revealed that αE+CD25+ cells had the highest regulatory potency also in vivo (Fig. 5b). Only one mouse of this group (n = 8) developed colitis. In contrast, reconstitutions with αE+CD25− or αE−CD25+ cells in addition to naive CD4+ T cells protected only 50% of the mice (n = 8) from ongoing colitis (Table 1). The comparison of the body weights of mice receiving either αE+CD25+ or αE−CD25+ cells in combination with naive CD4+ T cells revealed a nearly significant difference (P = 0.056), which was confirmed by the overall statistical analysis of body weight plus clinical and histological parameters (Table 1). These data clearly show that, also in vivo, αE+CD25+ have the highest regulatory capacity among subsets described so far. Interestingly, also the αE single positive subsets exerted a remarkable protective effect in vivo, despite their rather low efficiency in the in vitro assays.
Table 1.
αE+CD25+ T cells show the highest regulatory potency in vivo
| Phenotype | Ratio | Clinical score | Histological score | Incidence of colitis |
|---|---|---|---|---|
| CD45RBhigh alone (3 × 105) | – | 3.63 | 1.49 | 22 /24 |
| CD45RBhigh plus CD45RBlow | 3:1 | 0 | 0.42 | 0 /11 |
| CD45RBhigh plus αE+CD25+ | 3:1 | 0 | 0 | 0 /4 |
| plus αE−CD25+ | 3:1 | 0 | 0.38 | 0 /4 |
| plus αE+CD25− | 3:1 | 0 | 0.25 | 0 /4 |
| PBS | – | 0 | 0 | 0 /5 |
| CD45RBhigh plus αE+CD25+ | 6:1 | 0.37 | 0.69 | 1 /8 |
| plus αE−CD25+ | 6:1 | 1.75 | 0.94 | 4 /8 |
| plus αE+CD25− | 6:1 | 1.13 | 1.44 | 4 /8 |
SCID mice received 3 × 105 CD45RBhighCD4+ T cells alone or in combination with CD4+ T cell subpopulations as indicated; control mice received PBS only. The clinical and histological scores are shown as mean values. The incidence of colitis was determined by the overall analysis of body weights plus clinical and histological parameters. Data for mice receiving either CD45RBhigh or CD45RBhigh plus CD45RBlow CD4+ T cells are pooled from the two independent experiments.
Discussion
The integrin αEβ7 was first identified in the mucosa through its unique expression on more than 90% of CD8+ and approximately 40–50% of CD4+ intestinal T cells (36, 51). Whereas αEβ7 integrin expressing CD8+ T cells exert strong cytolitic activity (52), the precise role of αE+CD4+ T cells is currently not known. Here we report that αEβ7 integrin is a marker for regulatory CD4+ T cell subpopulations different from the regulatory subset described so far. The expression of αE subdivides the “classical” CD25+CD4+ regulatory T cells into two subsets with distinct properties, namely αE+CD25+ and αE−CD25+. Beyond that, it identifies another regulatory subpopulation localized outside the CD25+ compartment.
αE+CD25+ cells, comprising about 4% of all CD4+ and 25% of the CD25+CD4+ T cell pool isolated from lymphoid organs, turned out to be the most potent regulators in in vitro assays. At very low regulator/target ratios (1:19), approaching the relations in vivo, only αE+CD25+ cells showed suppressive effects. In contrast, CD25 single positive cells, comprising the majority of the regulatory T cells described so far, exhibited their suppressive potential only at higher ratios, as also described elsewhere recently (45). These data indicate that the “classical” CD25+ regulatory T cell pool is heterogeneous, as has been proposed by others (6), and that expression of αE allows a further subdivision. In addition, αE allows the identification of regulatory T cells regardless of their activation state, because αE, in contrast to CD25, is not induced on activation.
The regulatory potential of the investigated subsets was confirmed in vivo. Both αE+CD25+ and CD25 single positive cells prevented immunodeficient SCID mice from developing colitis after reconstitution with CD45RBhighCD4+ T cells at a high regulator/target ratio. When the ratio was lowered, the stronger suppressive potential of αE+CD25+ compared with CD25 single positive cells became obvious.
Interestingly, also the CD25-negative αE+CD4+ T cell subpopulation showed regulatory functions in vitro and in vivo. In vitro, this small subpopulation has only moderate suppressor capacity, which is comparable with that of total CD45RBlowCD4+ T cells, showing complete suppression at a regulator/target ratio of 1:1. However, αE single positive cells are potent suppressors in vivo. In the SCID model, αE+CD25− cells were as effective as αE−CD25+ cells in inhibiting colitis development. Some regulatory activity has been demonstrated previously in the compartment of CD25−CD45RBlow cells, but suppression was rather incomplete and not further related to a particular subpopulation (8–10). Because αE single positive cells are predominantly CD45RBlow and reveal complete protection of SCID mice from developing colitis at a 1:3 regulator/target ratio, this previously undescribed subset might indeed account for the minor suppressive activity within the CD25−CD45RBlow T cell pool. It is a challenging question whether αE contributes to the regulatory function in the colitis model, e.g., by interacting with inflamed gut epithelium, and whether the distinct cytokine profile of the αE single positive cells with high levels of Th2 cytokines plays a role in their protective effect.
αE+CD25+, αE−CD25+ and αE+CD25− CD4+ T cell subpopulations, which all have suppressive activity, cannot only be distinguished by the expression of αE and CD25, but also show differences with regard to CTLA-4 and cytokine expression, which suggests distinct modes of action. In recent publications, CTLA-4 expression was only detected within the CD25+ compartment of CD4+ T cells and frequencies in the range of 10% to 50% were described (10–12, 53). Intensities of CTLA-4 stainings are in general rather low, and this together with the largely varying staining protocols might explain the range of reported frequencies of positive cells. Subdivision of this compartment according to αE expression revealed a significantly higher expression of CTLA-4 in the αE+CD25+ subset. Additionally, αE single positive cells showed high frequencies of CTLA-4 expression. This finding leads to the conclusion that CTLA-4 is predominantly correlated with αE and not with CD25 in the regulatory subsets. Although it is discussed controversially whether CTLA-4 is functionally involved in the action of regulatory CD4+ T cells (10, 12, 13), the high expression of CTLA-4 in αE+ subsets might suggest a role in their suppressive function.
Although some reports assign a cytokine phenotype with reduced IL-2 and preferential IL-10 production to “classical” CD25+CD4+ T cells (13, 26, 29, 30, 54), the cytokine profile of regulatory CD4+ T cells is not well documented on the single cell level. Our data reveal a considerable heterogeneity within the regulatory T cell pool. Whereas TNFα, IFN-γ, and IL-2 frequencies were low but detectable within the CD25 single positive cells, αE+CD25+ cells produced almost no proinflammatory cytokines on restimulation. Beyond that, αE+CD25+ and αE−CD25+ cells differ in the production of Th2 cytokines. CD25 single positive cells, representing the majority of “classical” CD25+CD4+ regulatory T cells, produce IL-4, IL-5, and IL-13 in frequencies comparable to other memory CD4+ T cells. In contrast, the production of these Th2 cytokines is almost absent in the αE+CD25+ subset. Low cytokine production has been associated with regulatory T cells (13, 26, 29). Remarkably, αE+CD25+ cells display the lowest level of cytokine production, except for IL-10, which is only slightly less frequent in αE+CD25+ cells compared with memory CD4+ cells. On the mRNA level, αE+CD25+ cells produce seven times more IL-10 transcripts than αE−CD25+ cells (data not shown). So far, we could not demonstrate a functional role of IL-10 for the suppressive effect of αE+CD25+ cells, as neutralization of IL-10 in the in vitro proliferation assay had no influence on the inhibitory action of this as well as the other regulatory CD4+ T cell subpopulations. This finding is in agreement with in vitro experiments from other groups (13, 15, 27), although data from in vivo models regarding IL-10 are yet controversial (17, 18, 55). Additionally, blockade of TGFβ, another candidate for regulatory mechanisms (13, 26, 29), did not abrogate the suppressive effects of the investigated subpopulations. Only the inhibitory capacity of the αE single positive regulatory subset seems to be partially dependent on TGFβ. However, we cannot exclude that surface-bound TGFβ might be resistant to antibody blockade, as has been described recently (27). This might also apply for the blockade of IL-10, because this cytokine can be detected on the surface of CD4+ T cells as well (56). Indeed, suppressive effects of αE+ T cells were found to be cell contact-dependent, as has been described for other regulatory populations (13). We can exclude that the regulatory subsets inhibit naive T cell proliferation simply by killing the target cells, as none of subpopulations expressed FasL either on resting or on activated cells nor did regulatory subpopulations isolated from perforin-deficient mice show an defect in their suppressive capacity (Fig. 12, which is published as supporting information on the PNAS web site).
Interestingly, αE single positive CD4+ T cells, which also exhibit regulatory functions both in vitro and in vivo, display a peculiar cytokine phenotype. These cells show a high expression of all cytokines, including IL-2 and IFN-γ, and even elevated frequencies of IL-4 and IL-5, in contrast to other reports about regulatory T cells (13, 26, 29, 30). Most intriguing is the high frequency of IL-13 producing cells. Thus, αE single positive cells represent a unique subset, in which production of IL-2 and other cytokines is not conflicting with suppressive functions.
In conclusion, the data presented here indicate that expression of the integrin αEβ7 inside and outside the CD25 compartment correlates with regulatory function. The contrasting cytokine production profiles, differential expression of CTLA-4, and suppressive potencies suggest that these subsets represent distinct entities, possibly acting via different mechanisms. In addition, also their origin appears to differ from “classical” CD25+ regulatory T cells, as αE+CD4+ cells were found in significant numbers in the thymus only during adulthood and increased with age, whereas αE−CD25+ cells (CD4+CD8−) emerge within the first days after birth and do not change during aging (unpublished data). This finding suggests a role of environmental factors for the generation of the αE+ subsets.
The functional role of αEβ7 on the regulatory T cells is still elusive. Expression of αEβ7 is a hallmark of T cells residing in or near epithelial sites such as the gut mucosa. TGFβ is abundant at such sites and is able to induce the integrin (37). Concurrently, TGFβ has been proposed as a key factor in the development of regulatory cells (24). The presence of αE on highly effective regulators therefore might indicate their previous maturation in a TGFβ-rich milieu. The costimulatory function of αEβ7 could furthermore point toward a role in the development of this subset (43). Alternatively, presence of αEβ7 on regulatory cells predisposes them for retention within epithelial sites and might modify local responses. Thus, αE+ regulatory T cells are envisaged to fulfill a unique function as “heralds of tolerance” shuttling between epithelial and lymphoid sites and transfering tolerogenic potential.
Supplementary Material
Acknowledgments
We thank P. Kilshaw for providing the anti αE-Ab-producing hybridoma, K. Vogt and B. Sawitzky for mRNA analysis, T. Kaiser and K. Raba for FACS sorting, U. Haubold and S. Sommer for skillful technical assistance, U. Steinhoff for providing perforin-deficient mice, D. Huscher for help with statistical analysis, and M. Loehning, A. Radbruch, and H.-D. Volk for helpful discussions and comments. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 421, Ha1505/7) and the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (Germany) (BerlInFlame D5).
Abbreviations
- CFSE
5-carboxy-fluorescein diacetate succinimidyl ester
- SCID
severe combined immunodeficient
- CTLA-4
cytotoxic T lymphocyte antigen 4
- TNF
tumor necrosis factor
- TGF
transforming growth factor
- PE
phycoerythrin
- APC
antigen-presenting cell
- FACS
fluorescence activated cell sorter
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Sakaguchi S. Cell. 2000;101:455–458. doi: 10.1016/s0092-8674(00)80856-9. [DOI] [PubMed] [Google Scholar]
- 2.Shevach E M. J Exp Med. 2001;193:F41–F46. doi: 10.1084/jem.193.11.f41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powrie F, Leach M W, Mauze S, Caddle L B, Coffman R L. Int Immunol. 1993;5:1461–1471. doi: 10.1093/intimm/5.11.1461. [DOI] [PubMed] [Google Scholar]
- 4.Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. J Immunol. 1995;155:1151–1164. [PubMed] [Google Scholar]
- 5.Mason D, Powrie F. Curr Opin Immunol. 1998;10:649–655. doi: 10.1016/s0952-7915(98)80084-8. [DOI] [PubMed] [Google Scholar]
- 6.Maloy K J, Powrie F. Nat Immunol. 2001;2:816–822. doi: 10.1038/ni0901-816. [DOI] [PubMed] [Google Scholar]
- 7.Olivares-Villagomez D, Wensky A K, Wang Y, Lafaille J J. J Immunol. 2000;164:5499–5507. doi: 10.4049/jimmunol.164.10.5499. [DOI] [PubMed] [Google Scholar]
- 8.Stephens L A, Mason D. J Immunol. 2000;165:3105–3110. doi: 10.4049/jimmunol.165.6.3105. [DOI] [PubMed] [Google Scholar]
- 9.Annacker O, Pimenta-Araujo R, Burlen-Defranoux O, Barbosa T C, Cumano A, Bandeira A. J Immunol. 2001;166:3008–3018. doi: 10.4049/jimmunol.166.5.3008. [DOI] [PubMed] [Google Scholar]
- 10.Read S, Malmstrom V, Powrie F. J Exp Med. 2000;192:295–302. doi: 10.1084/jem.192.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salomon B, Lenschow D J, Rhee L, Ashourian N, Singh B, Sharpe A, Bluestone J A. Immunity. 2000;12:431–440. doi: 10.1016/s1074-7613(00)80195-8. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi T, Tagami T, Yamazaki S, Uede T, Shimizu J, Sakaguchi N, Mak T W, Sakaguchi S. J Exp Med. 2000;192:303–310. doi: 10.1084/jem.192.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thornton A M, Shevach E M. J Exp Med. 1998;188:287–296. doi: 10.1084/jem.188.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jonuleit H, Schmitt E, Stassen M, Tuettenberg A, Knop J, Enk A H. J Exp Med. 2001;193:1285–1294. doi: 10.1084/jem.193.11.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levings M K, Sangregorio R, Roncarolo M G. J Exp Med. 2001;193:1295–1302. doi: 10.1084/jem.193.11.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 17.Powrie F, Leach M W, Mauze S, Menon S, Caddle L B, Coffman R L. Immunity. 1994;1:553–562. doi: 10.1016/1074-7613(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 18.Asseman C, Mauze S, Leach M W, Coffman R L, Powrie F. J Exp Med. 1999;190:995–1004. doi: 10.1084/jem.190.7.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shull M M, Ormsby I, Kier A B, Pawlowski S, Diebold R J, Yin M, Allen R, Sidman C, Proetzel G, Calvin D, et al. Nature (London) 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powrie F, Carlino J, Leach M W, Mauze S, Coffman R L. J Exp Med. 1996;183:2669–2674. doi: 10.1084/jem.183.6.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bridoux F, Badou A, Saoudi A, Bernard I, Druet E, Pasquier R, Druet P, Pelletier L. J Exp Med. 1997;185:1769–1775. doi: 10.1084/jem.185.10.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seddon B, Mason D. J Exp Med. 1999;189:279–288. doi: 10.1084/jem.189.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorelik L, Flavell R A. Immunity. 2000;12:171–181. doi: 10.1016/s1074-7613(00)80170-3. [DOI] [PubMed] [Google Scholar]
- 24.Weiner H L. Nat Immunol. 2001;2:671–672. doi: 10.1038/90604. [DOI] [PubMed] [Google Scholar]
- 25.Yamagiwa S, Gray J D, Hashimoto S, Horwitz D A. J Immunol. 2001;166:7282–7289. doi: 10.4049/jimmunol.166.12.7282. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi T, Kuniyasu Y, Toda M, Sakaguchi N, Itoh M, Iwata M, Shimizu J, Sakaguchi S. Int Immunol. 1998;10:1969–1980. doi: 10.1093/intimm/10.12.1969. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura K, Kitani A, Strober W. J Exp Med. 2001;194:629–644. doi: 10.1084/jem.194.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papiernik M, de Moraes M L, Pontoux C, Vasseur F, Penit C. Int Immunol. 1998;10:371–378. doi: 10.1093/intimm/10.4.371. [DOI] [PubMed] [Google Scholar]
- 29.Asano M, Toda M, Sakaguchi N, Sakaguchi S. J Exp Med. 1996;184:387–396. doi: 10.1084/jem.184.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papiernik M, Banz A. Microbes Infect. 2001;3:937–945. doi: 10.1016/s1286-4579(01)01455-1. [DOI] [PubMed] [Google Scholar]
- 31.Seddon B, Mason D. Immunol Today. 2000;21:95–99. doi: 10.1016/s0167-5699(99)01559-5. [DOI] [PubMed] [Google Scholar]
- 32.Jordan M S, Boesteanu A, Reed A J, Petrone A L, Holenbeck A E, Lerman M A, Naji A, Caton A J. Nat Immunol. 2001;2:301–306. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- 33.Stephens L A, Mottet C, Mason D, Powrie F. Eur J Immunol. 2001;31:1247–1254. doi: 10.1002/1521-4141(200104)31:4<1247::aid-immu1247>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 34.Strober W, Kelsall B, Fuss I, Marth T, Ludviksson B, Ehrhardt R, Neurath M. Immunol Today. 1997;18:61–64. doi: 10.1016/s0167-5699(97)01000-1. [DOI] [PubMed] [Google Scholar]
- 35.Thorstenson K M, Khoruts A. J Immunol. 2001;167:188–195. doi: 10.4049/jimmunol.167.1.188. [DOI] [PubMed] [Google Scholar]
- 36.Cerf-Bensussan N, Jarry A, Brousse N, Lisowska-Grospierre B, Guy-Grand D, Griscelli C. Eur J Immunol. 1987;17:1279–1285. doi: 10.1002/eji.1830170910. [DOI] [PubMed] [Google Scholar]
- 37.Kilshaw P J, Murant S J. Eur J Immunol. 1991;21:2591–2597. doi: 10.1002/eji.1830211041. [DOI] [PubMed] [Google Scholar]
- 38.Hamann A, Andrew D P, Jablonski-Westrich D, Holzmann B, Butcher E C. J Immunol. 1994;152:3282–3293. [PubMed] [Google Scholar]
- 39.Cepek K L, Shaw S K, Parker C M, Russell G J, Morrow J S, Rimm D L, Brenner M B. Nature (London) 1994;372:190–193. doi: 10.1038/372190a0. [DOI] [PubMed] [Google Scholar]
- 40.Austrup F, Rebstock S, Kilshaw P J, Hamann A. Eur J Immunol. 1995;25:1487–1491. doi: 10.1002/eji.1830250602. [DOI] [PubMed] [Google Scholar]
- 41.Shieh C C, Sadasivan B K, Russell G J, Schon M P, Parker C M, Brenner M B. J Immunol. 1999;163:1592–1601. [PubMed] [Google Scholar]
- 42.Schon M P, Arya A, Murphy E A, Adams C M, Strauch U G, Agace W W, Marsal J, Donohue J P, Her H, Beier D R, et al. J Immunol. 1999;162:6641–6649. [PubMed] [Google Scholar]
- 43.Sarnacki S, Begue B, Buc H, Le Deist F, Cerf-Bensussan N. Eur J Immunol. 1992;22:2887–2892. doi: 10.1002/eji.1830221120. [DOI] [PubMed] [Google Scholar]
- 44.Gavin M A, Clarke S R, Negrou E, Gallegos A, Rudensky A. Nat Immunol. 2002;3:33–41. doi: 10.1038/ni743. [DOI] [PubMed] [Google Scholar]
- 45.McHugh R, Whitters M J, Piccorillo C A, Young D A, Shevach E M, Collins M, Byrne M C. Immunity. 2002;16:311–323. doi: 10.1016/s1074-7613(02)00280-7. [DOI] [PubMed] [Google Scholar]
- 46.Zelenika D, Adams E, Humm S, Graca L, Thompson S, Cobbold S P, Waldmann H. J Immunol. 2002;168:1069–1079. doi: 10.4049/jimmunol.168.3.1069. [DOI] [PubMed] [Google Scholar]
- 47.Schon M P, Schon M, Warren H B, Donohue J P, Parker C M. J Immunol. 2000;165:6583–6589. doi: 10.4049/jimmunol.165.11.6583. [DOI] [PubMed] [Google Scholar]
- 48.Kagi D, Ledermann B, Burki K, Seiler P, Odermatt B, Olsen K J, Podack E R, Zinkernagel R M, Hengartner H. Nature (London) 1994;369:31–37. doi: 10.1038/369031a0. [DOI] [PubMed] [Google Scholar]
- 49.Assenmacher M, Schmitz J A, Radbruch A. Eur J Immunol. 1994;24:1097–1101. doi: 10.1002/eji.1830240513. [DOI] [PubMed] [Google Scholar]
- 50.Lyons A B, Parish C R. J Immunol Methods. 1994;171:131–137. doi: 10.1016/0022-1759(94)90236-4. [DOI] [PubMed] [Google Scholar]
- 51.Kilshaw P J, Murant S J. Eur J Immunol. 1990;20:2201–2207. doi: 10.1002/eji.1830201008. [DOI] [PubMed] [Google Scholar]
- 52.Hadley G A, Bartlett S T, Via C S, Rostapshova E A, Moainie S. J Immunol. 1997;159:3748–3756. [PubMed] [Google Scholar]
- 53.Zhang X, Izikson L, Liu L, Weiner H L. J Immunol. 2001;167:4245–4253. doi: 10.4049/jimmunol.167.8.4245. [DOI] [PubMed] [Google Scholar]
- 54.Dieckmann D, Plottner H, Berchtold S, Berger T, Schuler G. J Exp Med. 2001;193:1303–1310. doi: 10.1084/jem.193.11.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suri-Payer E, Cantor H. J Autoimmun. 2001;16:115–123. doi: 10.1006/jaut.2000.0473. [DOI] [PubMed] [Google Scholar]
- 56.Scheffold A, Assenmacher M, Reiners-Schramm L, Lauster R, Radbruch A. Nat Med. 2000;6:107–110. doi: 10.1038/71441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.