Abstract
Zmpste24 is an integral membrane metalloproteinase of the endoplasmic reticulum. Biochemical studies of tissues from Zmpste24-deficient mice (Zmpste24−/−) have indicated a role for Zmpste24 in the processing of CAAX-type prenylated proteins. Here, we report the pathologic consequences of Zmpste24 deficiency in mice. Zmpste24−/− mice gain weight slowly, appear malnourished, and exhibit progressive hair loss. The most striking pathologic phenotype is multiple spontaneous bone fractures—akin to those occurring in mouse models of osteogenesis imperfecta. Cortical and trabecular bone volumes are significantly reduced in Zmpste24−/− mice. Zmpste24−/− mice also manifested muscle weakness in the lower and upper extremities, resembling mice lacking the farnesylated CAAX protein prelamin A. Prelamin A processing was defective both in fibroblasts lacking Zmpste24 and in fibroblasts lacking the CAAX carboxyl methyltransferase Icmt but was normal in fibroblasts lacking the CAAX endoprotease Rce1. Muscle weakness in Zmpste24−/− mice can be reasonably ascribed to defective processing of prelamin A, but the brittle bone phenotype suggests a broader role for Zmpste24 in mammalian biology.
Keywords: metalloproteinase‖knockout mice‖brittle bones‖CAAX motif
The mammalian zinc metalloproteinase Zmpste24 has attracted attention because it shares a high degree of sequence identity with Ste24p, a Saccharomyces cerevisiae enzyme required for the maturation of the farnesylated mating pheromone a-factor (1–3). Ste24p plays two distinct roles in a-factor biogenesis (2, 4). First, it acts as a CAAX endoprotease, clipping off the C-terminal three amino acids from the protein (i.e., the −AAX of the CAAX motif) (3). Release of the −AAX from a-factor can also be mediated by Rce1p, the CAAX endoprotease involved in Ras processing (3). The removal of the −AAX exposes a carboxyl-terminal farnesylcysteine, which is methylated by Ste14p (5). Second, Ste24p clips the amino-terminal extension of a-factor, rendering it susceptible to a final endoproteolytic cleavage by Axl1p or Ste23p (6). Aside from a-factor, no other substrates for Ste24p have been identified, but other substrates likely exist because genetic screens in yeast have demonstrated that STE24 mutations can reverse the topological orientation of membrane proteins (7) and can affect the viability of yeast with mutations in genes encoding actin cytoskeleton proteins (8).
Zmpste24 faithfully carries out both of Ste24p's processing steps in a-factor biogenesis and thus is a bona fide Ste24p ortholog (2, 9). Although it would be tempting to speculate that Zmpste24 processes an “a-factor-like” peptide in mammals, no a-factor ortholog has yet been identified. We have previously speculated that prelamin A (a precursor to lamin A, a component of the nuclear lamina) might be a Zmpste24 substrate (2, 6) because prelamin A (like yeast a-factor) is a farnesylated CAAX protein that undergoes more than one proteolytic processing step (10). After the removal of the C-terminal −AAX, an additional 15 residues are removed from the C terminus of the protein, generating mature lamin A (10).
Three years ago, to assess the importance of Zmpste24 in mammals, we produced Zmpste24 knockout mice (Zmpste24−/−) by replacing the exon encoding the zinc-binding domain with a neomycin resistance cassette (neo) (9). We reported biochemical studies showing that tissues and cells lacking Zmpste24 were deficient in their ability to carry out the N- and C-terminal proteolytic processing steps in a-factor biogenesis. Although obvious abnormalities (including muscle weakness and spontaneous bone fractures) were plainly evident in independently derived lines of knockout mice, we were reluctant to report those data with the biochemical studies (9) because we were concerned that one or more of the phenotypes could have been due to effects of the neo cassette (particularly its promoter and enhancer elements) on the transcription of neighboring genes (11). Accordingly, we proceeded with the characterization of Zmpste24−/− mice lacking the neo cassette. In May 2002, Pendás et al. (12) reported a second mutant Zmpste24 allele in which several exons were replaced by a neo cassette. They reported that Zmpste24 deficiency was associated with a prelamin A processing defect as well as partial lipodystrophy, muscular dystrophy, and cardiomyopathy.
Here, we report the pathologic findings in Zmpste24-deficient mice, document a prelamin A processing defect in Zmpste24-deficient tissues and fibroblasts, and show that the prelamin A processing defect also occurs in cells lacking Icmt, the enzyme that carries out the carboxyl methylation of prenylated proteins.
Materials and Methods
Zmpste24−/− Mice.
Mice from two embryonic stem cell lines, each with a single integration event, were studied (9). Zmpste24−/− mice lacking the neo were generated by excising the neo with a deleter-Cre mouse strain (13). The phenotypes of the neo-positive and “neo-less” Zmpste24−/− mice were compared over a 2-year time frame. The mice had a mixed genetic background (≈50% C57BL/6 and ≈50% 129/SvJae), were fed a mouse chow diet, and were housed in a virus-free barrier facility.
Pathological Examination.
Full necropsies were performed on 16-, 20-, and 24-week-old Zmpste24−/− mice and littermate controls (n = 3–7) with histologic examination of >30 tissues, including all regions of the skeleton. In addition, necropsies and routine histology of the musculoskeletal system were performed on mice at 3, 6, 9, 16, 20, 24, and 30 weeks of age (n = 4–8). Whole-body radiographs were performed on 6- to 30-week-old Zmpste24−/− mice and littermate controls (n = 31) with a Faxitron specimen radiography system (Wheeling, IL).
Prelamin A Processing.
Extracts from Zmpste24+/+ fibroblasts, Zmpste24−/− fibroblasts, Rce1−/− fibroblasts (14), Icmt−/− fibroblasts (15), and wild-type fibroblasts treated with a farnesyltransferase inhibitor (SCH66336) were size-fractionated on SDS/4–15% polyacrylamide gels, and Western blots were performed with a C-terminal antilamin A goat antibody (sc-6214, Santa Cruz Biotechnology) and an N-terminal anti-lamin A/C mouse monoclonal antibody (sc-7293, Santa Cruz Biotechnology).
Microcomputed Tomography (μCT) Scans.
Zmpste24−/− mice (n = 3–5) were examined with a MicroCT 40 scanner (Scanco Medical, Bassersdorf, Switzerland) at 18 days, 2 months, and 3 months of age. The μCT scanner allows for imaging and quantification of cancellous bone microstructure, in both two and three dimensions. The sample was scanned in the axial plane mounted in a cylindrical sample holder with a current of 0.16 mA, a voltage of 50 kV, at an isotropic resolution of 20 μm (image matrix 1024 × 1024 pixels). Images of Zmpste24+/+ and Zmpste24−/− bones were generated at the identical threshold.
The three-dimensional trabecular structure of the 12th thoracic vertebral body was constructed with the internal software of the μCT system. Cortical bone and trabecular bone were manually separated on each slice by a cursor line. Several structural parameters, including the bone volume to total volume ratio and trabecular thickness, were measured. The central 50% (the diaphysis) of the tibia and fibula were also analyzed. The cortical bone volume and density were determined by application of an automated image segmentation algorithm and image analysis software libraries (AnalyzeDirect, Lenexa, KS). Histogram analysis and morphological filtering were used to extract the cortex volume and total tibia/fibula volume. A cortical bone threshold (0.77 gHA/cm3) was determined by histogram analysis of the data. The threshold was applied to extract cortex voxels in the image data. A series of morphological operations within the software package was then applied to link cortex voxels and remove voxels of similar density within trabeculae. The ratio of cortical bone volume to total diaphyseal volume and the cortical bone density were calculated.
Results and Discussion
Zmpste24−/− Mice.
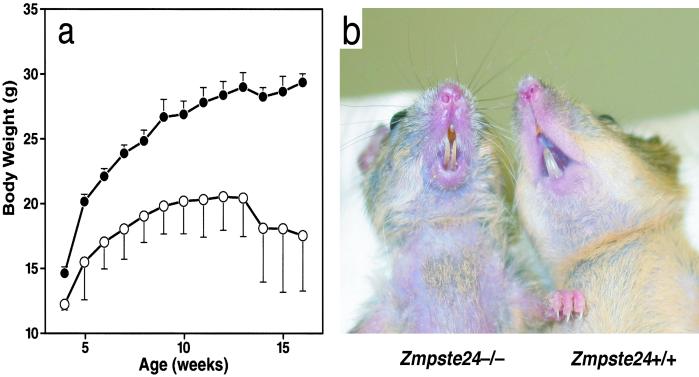
The phenotypes of neo-positive and neo-less knockout mice were identical over a 2-year period of study. Heterozygotes (Zmpste24+/−) appear normal for the first 12 months of life, but by 15 months of age, most heterozygotes were smaller than wild-type (Zmpste24+/+) littermates, appeared weak, and lost hair. Zmpste24−/− mice weigh slightly less than their littermates at weaning (3 weeks of age) and gain weight slowly (Fig. 1a). The mice also manifest muscle weakness (described below). By 6–8 weeks of age, they develop kyphosis of the spine, lose hair, and appear very malnourished. These phenotypes were progressive, and the mice either died or were killed by 6–7 months of age.
Figure 1.
(a) Retarded growth in male Zmpste24−/− mice (n = 4) vs. littermate male Zmpste24+/+ mice (n = 13). Similar results were obtained with female mice (not shown). (b) Photograph of a 3-month-old Zmpste24−/− mouse and littermate Zmpste24+/+ mouse.
Spontaneous Bone Fractures.
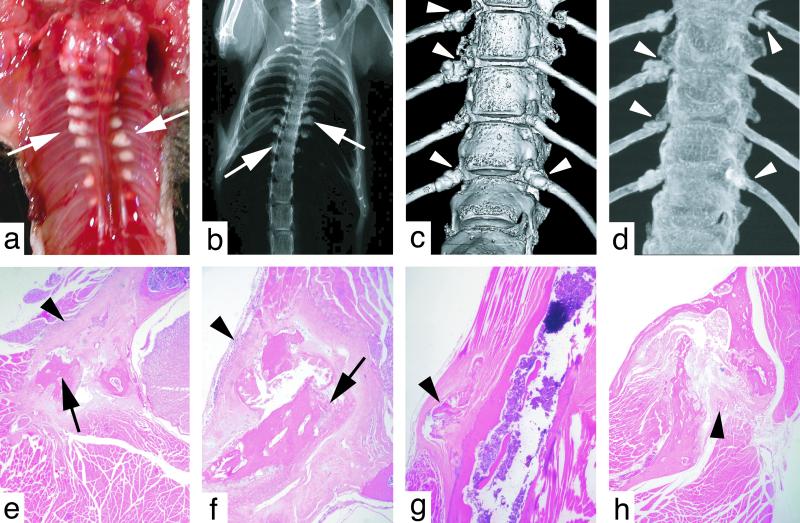
The lower incisors of Zmpste24−/− mice were splayed apart, thin, and long (Fig. 1b). This peculiar phenotype has been noted previously in a mouse model of osteogenesis imperfecta (16). Osteogenesis imperfecta in humans is characterized by brittle bones (17), and transgenic and gene-targeted mouse models of this disease develop spontaneous bone fractures (18–20). The incisor abnormalities in Zmpste24−/− mice led us to predict that the Zmpste24−/− mice might develop spontaneous bone fractures. Indeed, this proved to be the case. By 24–30 weeks of age, nearly every rib in Zmpste24−/− mice was broken in the vicinity of the costovertebral junction—obvious from hypertrophic calluses at the fracture sites (Fig. 2a). Callus surrounding broken ribs was also visible by routine x rays (Fig. 2b) and surface renderings of the vertebral spine from μCT scans (Fig. 2c). Penetrating views of the vertebral spine from the μCT scans (maximal intensity projections) revealed fractures of the tips of most ribs (Fig. 2d).
Figure 2.
(a) Thorax of a 24-week-old Zmpste24−/− mouse. Fibrotic lesions surrounding broken ribs are indicated by arrows. (b) Radiograph of a Zmpste24−/− mouse, revealing callus formation surrounding rib fractures (arrows). (c) Surface rendering of a μCT scan of the lower thoracic spine of a Zmpste24−/− mouse. Callus is visible at the tips of several ribs near the costovertebral joints (arrowheads). (d) Maximal intensity projection from a μCT scan of the lower thoracic spine of a Zmpste24−/− mouse, revealing multiple rib fractures (arrowheads). (e and f) Hematoxylin- and eosin-stained sections of rib fractures (arrows) and surrounding fibrosis (arrowheads). (g) Avulsion of a fragment of bone (arrowhead) from the cortex of the humerus. (h) Fracture of the mandible with fibrosis between bone fragments (arrowhead).
Microscopic sections also revealed broken ribs surrounded by exuberant fibrous tissue (Fig. 2 e and f). The fracture sites were surprisingly acellular, with little inflammatory infiltrate and minimal evidence of healing. Bone fragments within fracture sites were frequently necrotic, lacking viable osteocytes. Zmpste24−/− mice manifested bone fractures in multiple locations aside from the ribs, including the scapula, clavicle, sternum, zygomatic arch, mandible, and humerus. The avulsion of a fragment of bone from the humerus of a 24-week-old Zmpste24−/− mouse is shown in Fig. 2g, and a fracture of the mandible in a 24-week-old Zmpste24−/− mouse is shown in Fig. 2h. The epiphyseal plates of the large bones of the upper and lower extremities were invariably normal (not shown).
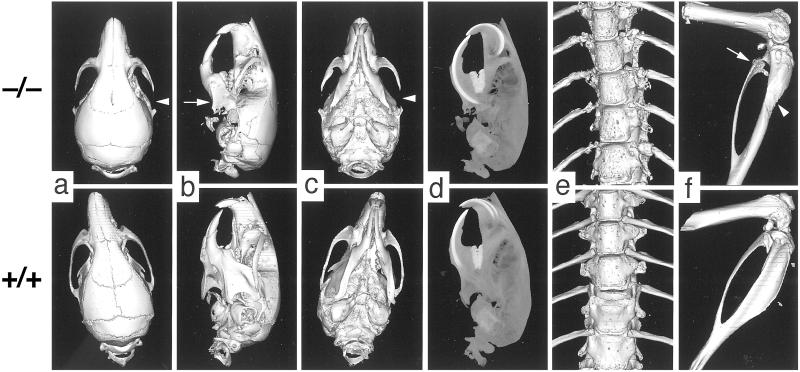
By 8 weeks of age, Zmpste24−/− mice invariably (n = 8 examined) had fractures of the posterior portion of the zygomatic arch where the masseter originates (Fig. 3 a–d). These fractures did not heal, and the posterior portion of the zygomatic arch was replaced by fibrous tissue containing spicules of bone and necrotic debris. Because the masseter is a crucial muscle for moving the jaw, it is likely that the nutritional status of the mice was adversely affected by the zygomatic arch fractures. Consistent with this idea, the plasma glucose and total protein levels tended to be lower in the Zmpste24−/− mice (Table 1). The μCT scans of 2- to 3-month-old mice also revealed striking micrognathia and a reduction in the zig-zag appearance of the cranial sutures. The micrognathia may have been secondary to the zygomatic arch fractures, as the zygomatic arch was usually normal at 18 days of age, and the size and shape of the mandible were fairly normal (not shown).
Figure 3.
μCT scans illustrating bony lesions in Zmpste24−/− mice. (a–c) Surface renderings of top, lateral, and bottom surfaces of the skulls of 8-week-old Zmpste24−/− and Zmpste24+/+ mice. Arrowheads indicate the destroyed zygomatic arch in a and c; an arrow indicates the small mandible in b. (d) Maximal intensity projection of the lateral view of the skulls of 8-week-old mice, revealing an increase in the length of the incisors and the size of other teeth. (e) Surface rendering of the thoracic spines of 8-week-old Zmpste24−/− and Zmpste24+/+ mice, revealing increased numbers of pock marks in the Zmpste24−/− mice. (f) Surface rendering of the femur, knee, tibia/fibula of 2-month-old Zmpste24−/− and Zmpste24+/+ mice, revealing increased pock marks in the proximal fibula of the Zmpste24−/− mouse as well as effacement of the tibial tuberosity.
Table 1.
Plasma and urine chemistries in Zmpste24−/− mice
| Zmpste24+/+ | Zmpste24−/− | |
|---|---|---|
| Glucose, mg/dl | 191 ± 61 | 176 ± 77 |
| Total protein, g/dl | 6.7 ± 1.0 | 5.8 ± 2.8 |
| Alanine aminotransferase, units/liter | 61 ± 37 | 44 ± 32 |
| Aspartate aminotransferase, units/liter | 141 ± 71 | 120 ± 83 |
| Alkaline phosphatase, units/liter | 83 ± 9 | 59 ± 9* |
| Calcium, mmol/liter | 9.22 ± 0.67 | 9.64 ± 0.70 |
| Phosphate, mmol/liter | 7.84 ± 4.47 | 9.78 ± 4.02 |
| Urinary deoxypyridinoline, nmol/mg creatinine (14-week-old mice) | 0.22 ± 0.04 | 0.25 ± 0.03 |
| Urinary deoxypyridinoline, nmol/mg creatinine (4-week-old mice) | 0.21 ± 0.03 | 0.21 ± 0.04 |
Data are presented as mean and SD; n = 6–10 adult mice per group, ages 15–24 weeks except as indicated.
, P < 0.01. Except for alkaline phosphatase, no differences were significant (P > 0.05).
The thoracic vertebrae of each Zmpste24−/− mouse had reduced bone density compared with Zmpste24+/+ controls. This was very obvious from the three-dimensional μCT reconstructions of the vertebral column (surface renderings generated at the same threshold), which showed the vertebral bodies from Zmpste24−/− mice (n = 4) to be more porous (“pock-marked”) than those of Zmpste24+/+ controls (n = 5) (Fig. 3e). The pock-marked areas identify regions of bone with a density below the threshold level. The three-dimensional analysis of trabecular bone within the 12th vertebral body revealed that the ratio of bone volume to the total volume was reduced by 34% in the Zmpste24−/− mice (P = 0.04). The thickness of the trabeculae in the vertebrae was reduced by 36% (P = 0.02). An increase in pock marking of bone was also evident in the lower extremities (note the proximal fibula in the Zmpste24−/− mouse) (Fig. 3f). To quantify bone abnormalities in this predominantly cortical area, we calculated the ratio of bone volume in the tibia and fibula to total volume of those bones. This ratio was lower in Zmpste24−/− mice (0.54 ± 0.09) than in littermate Zmpste24+/+ mice (0.67 ± 0.03, P = 0.02), which means that even when the values are corrected for bone size, there is thinner cortical bone in the Zmpste24−/− mice. We suspect that there might be a bone abnormality in heterozygous mice as well. We performed μCT scans on two 15-month-old heterozygous Zmpste24 knockout mice (exhibiting alopecia and weight loss) and age-matched Zmpste24+/+ controls and identified degraded transverse and spinous processes in the thoracic and lumbar vertebrae of the heterozygotes (not shown).
The bone abnormalities in Zmpste24−/− mice were not accompanied by perturbations in the plasma levels of calcium or phosphate (Table 1). Zmpste24−/− mice did not manifest evidence of increased bone turnover. Zmpste24−/− mice had lower alkaline phosphatase levels. Urinary levels of deoxypyridinoline, a bone collagen breakdown product (21), were not different in Zmpste24−/− and Zmpste24+/+ mice. Also, bones from Zmpste24−/− and Zmpste24+/+ mice contained similar numbers of osteoclasts, as judged by staining for tartrate-resistant acid phosphatase (not shown).
Gait and Grip Abnormalities.
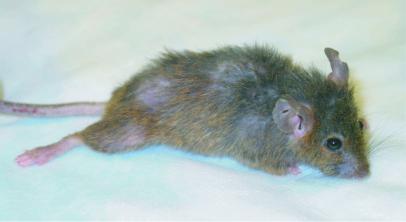
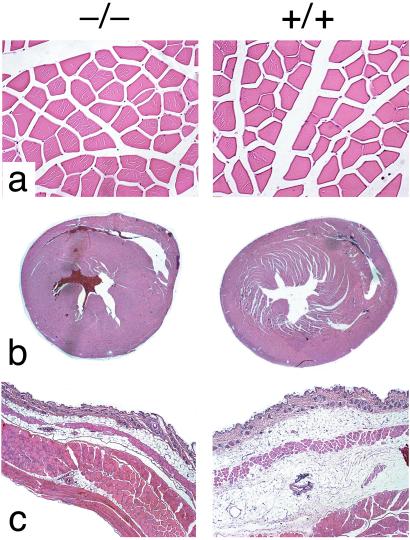
Zmpste24−/− mice also exhibited a slow hobbling gait, frequently dragging their hind limbs (Fig. 4). We suspect that the muscle weakness in the lower extremity contributed to the marked effacement of the anterior tibial tuberosity (the origin of the tibialis anterior muscle) (Fig. 3f). In addition, Zmpste24−/− mice (n = 11) were able to hang onto a grid after it was turned upside down for only a few seconds [males, 3.0 ± 1.5 s (n = 8); females, 4.5 ± 5.3 s (n = 6)], whereas littermate Zmpste24+/− and Zmpste24+/+ mice (n = 12 males; n = 13 females) were able to hold on almost indefinitely (>45 s). We examined muscles throughout the limbs and trunk of Zmpste24−/− mice. Despite exhaustive analysis of hematoxylin- and eosin-stained material, we identified no abnormalities in the skeletal muscle fibers of Zmpste24−/− mice (Fig. 5a). Further specialized neuromuscular studies are warranted to define the nature of the muscular defect in Zmpste24−/− mice.
Figure 4.
Photograph of a Zmpste24−/− mouse, illustrating a characteristic dragging of the hind limb.
Figure 5.
(a) Paravertebral muscles from 19-week-old male Zmpste24−/− and Zmpste24+/+ mice. (b) Sections through the hearts of 19-week-old male Zmpste24−/− and Zmpste24+/+ mice. (c) Sections through the skin of 15-week-old female Zmpste24−/− and Zmpste24+/+ mice, showing reduced adipose tissue in Zmpste24−/− mice.
Prelamin A Processing Defect.
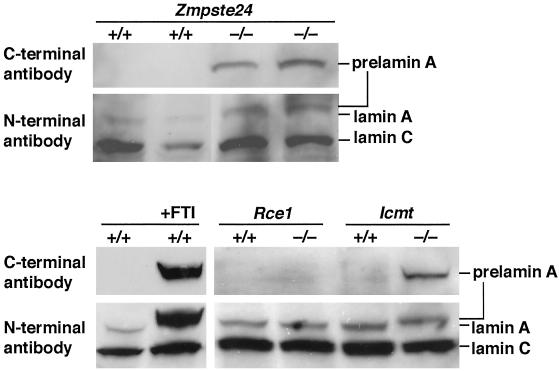
The inability to hold onto a grid while upside down is identical to a phenotype in mice lacking prelamin A (22). Accordingly, we examined prelamin A processing in Zmpste24−/− fibroblasts and found clear-cut abnormalities. Western blots of Zmpste24−/− fibroblast extracts with an antibody against the C terminus of prelamin A revealed a striking accumulation of prelamin A (Fig. 6). Western blots with an antibody against the N terminus of lamin A revealed fully processed lamin A (72 kDa) in Zmpste24+/+ fibroblasts and a larger unprocessed prelamin A (74 kDa) in Zmpste24−/− fibroblasts (Fig. 6). These findings are identical to those in a very recent paper by Pendás et al. (12). We asked whether prelamin A might accumulate in fibroblasts lacking the CAAX methyltransferase Icmt (15) or the CAAX endoprotease Rce1 (14). Interestingly, prelamin A accumulated in Icmt−/− but not in Rce1−/− fibroblasts (Fig. 6). As predicted from earlier studies (10), a farnesyltransferase inhibitor blocked prelamin A processing (Fig. 6).
Figure 6.
Western blots of Zmpste24−/−, Icmt−/−, and Rce1−/− fibroblast extracts with a C-terminal prelamin A antibody and an N-terminal lamin A/C antibody. Also shown are Western blots of extracts of wild-type fibroblasts and fibroblasts treated with 5 μM SCH66336, a farnesyltransferase inhibitor.
Given that CAAX endoproteolysis (i.e., removal of the −AAX) is required for carboxyl methylation by Icmt, we are forced to conclude that the release of the −AAX from prelamin A can proceed in the absence of Rce1. Because Ste24p can release the −AAX from a-factor, it seems reasonable to presume that Zmpste24 is capable of releasing the −AAX from prelamin A. The results with the Rce1−/− fibroblasts are intriguing because they are the first to suggest that a protease aside from Rce1 can remove the −AAX from a mammalian CAAX protein (6, 14, 23).
We do not yet know the precise role of Zmpste24 in prelamin A processing. An attractive possibility is that Zmpste24 carries out both prelamin A processing steps (the release of the −AAX and the release of the additional 15 residues), just as Ste24p has dual functions in a-factor biogenesis. On the other hand, it is possible that the only role of Zmpste24 is clipping the −AAX, allowing methylation to proceed. We do not yet know whether Rce1 retains the capacity to cleave the −AAX from prelamin A in Zmpste24+/+ cells. If Rce1 lacks that ability, one would be forced to consider the possibility that Zmpste24's only role in prelamin A processing is to release the −AAX, leaving the second proteolytic processing step to another enzyme. Consistent with the latter possibility are reports that the second prelamin A proteolytic processing step is carried out by a serine protease located in the cell nucleus (10, 24). [Zmpste24 is a zinc metalloproteinase, not a serine protease (1–3).] Additional biochemical and genetic studies over the next few years will undoubtedly define the precise role(s) of Zmpste24 in lamin A biogenesis.
The muscle weakness in Zmpste24−/− mice seems consistent with a defect in prelamin A processing (22, 25), but it is unclear whether the prelamin A processing defect lies at the root of the osteogenesis imperfecta-like phenotype. Osteogenesis imperfecta is an autosomal dominant syndrome generally caused by mutations in the type I procollagen genes. Most commonly, the procollagen mutations result in reduced amounts of type I procollagen secretion, but some mutations result in the secretion of a structurally abnormal procollagen (17). One possibility is that the prelamin A accumulation in the nuclear envelope of Zmpste24−/− mice indirectly affects the amount or quality of type I procollagen secretion, accounting for the bone phenotype in Zmpste24−/− mice. Somewhat against this possibility, although, is the fact that a heightened susceptibility to bone fractures has never been identified in humans with lamin A mutations. Another obvious possibility is that Zmpste24 has additional protein substrates and that defective processing of those additional substrates causes the bone abnormality. The fact that Zmpste24 might have additional substrates is not farfetched, as Zmpste24 orthologs clearly exist in organisms that lack type A nuclear lamins (e.g., yeast and nematodes). Also, STE24 mutations in yeast reverse the topological orientation of certain membrane proteins (7), and the stability of several misfolded proteins retained in the endoplasmic reticulum is enhanced in ste24Δ yeast (S.M. and Claude Jakob, unpublished observations). Both of the latter observations suggest that Ste24p could be required, directly or indirectly, in the recognition and removal of misfolded proteins within the endoplasmic reticulum. If Zmpste24 deficiency in mice were to interfere with the recognition and/or degradation of misfolded type I procollagen molecules, it is easy to imagine how an osteogenesis imperfecta-like syndrome might occur.
Comparisons with a Second Line of Zmpste24−/− Mice.
Very recently, Pendás et al. (12) characterized a second line of Zmpste24−/− mice. A large number of similarities (abnormal growth curves, early death, loss of fur, kyphosis, abnormal gait, muscle weakness, normal or low plasma glucose levels, and abnormal prelamin A processing) indicate that our Zmpste24−/− mice and theirs are fundamentally the same. However, there were some differences in our reports. Pendás et al. (12) claimed that their mice exhibited a “growth plate dysplasia” in the femur and tibia, although they did not show data. The growth plates of the femur and tibia in our mice were normal. They did not uncover a bone fracture phenotype, which can easily be overlooked unless one specifically looks for it. They reported that heterozygotes were healthy, whereas we found that older heterozygotes lose weight and develop alopecia. They also reported that their mice had a lamin A-related cardiomyopathy with thinning of the ventricles and secondary liver disease. Over the past 3 years, we have looked assiduously for heart or liver pathology over the life span of Zmpste24−/− mice, but have found none. Our Zmpste24−/− mice did not have ventricular thinning (Fig. 5b) nor did they have myocardial fibrosis as assessed by trichrome staining (not shown). Also, the plasma levels of alanine aminotransferase and aspartate aminotransferase were not elevated in our Zmpste24−/− mice (Table 1). Finally, they found reduced adipose tissue in Zmpste24−/− mice and concluded that they had a lamin A-related partial lipodystrophy. Subcutaneous adipose tissue was invariably reduced in our Zmpste24−/− but was always detectable, even in old and debilitated mice (Fig. 5c). It is possible that Zmpste24−/− mice do have a lamin A-related partial lipodystrophy, but in view of the low-normal glucose levels in our mice (Table 1) and the normal or low glucose and triglyceride levels in their mice, we suspect that the reduced fat stores may have had more to do with starvation secondary to the bone abnormalities (i.e., complete destruction of the zygomatic arch and other bone fractures limiting access to food).
Acknowledgments
We thank M. Sinensky for advice, R. Bishop (Schering–Plough, Kenilworth, NJ) for SCH66336, and S. Bergö for assistance with statistics. This work was supported in part by National Institutes of Health Grants HL41633 and AG15451 (to S.G.Y.) and GM41223 (to S.M.), by grants from the University of California Tobacco-Related Disease Research Program (to M.O.B. and S.G.Y.), and by the Swedish Cancer Foundation (to M.O.B.).
Abbreviation
- μCT
microcomputed tomography
References
- 1.Fujimura-Kamada K, Nouvet F J, Michaelis S. J Cell Biol. 1997;136:271–285. doi: 10.1083/jcb.136.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tam A, Nouvet F J, Fujimura-Kamada K, Slunt H, Sisodia S S, Michaelis S. J Cell Biol. 1998;142:635–649. doi: 10.1083/jcb.142.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyartchuk V L, Ashby M N, Rine J. Science. 1997;275:1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- 4.Boyartchuk V L, Rine J. Genetics. 1998;150:95–101. doi: 10.1093/genetics/150.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hrycyna C A, Sapperstein S K, Clarke S, Michaelis S. EMBO J. 1991;10:1699–1709. doi: 10.1002/j.1460-2075.1991.tb07694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young S G, Ambroziak P, Kim E, Clarke S. In: The Enzymes. Tamanoi F, Sigman D S, editors. Vol. 21. San Diego: Academic; 2000. pp. 155–213. [Google Scholar]
- 7.Tipper D J, Harley C A. Mol Biol Cell. 2002;13:1158–1174. doi: 10.1091/mbc.01-10-0488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tong A H Y, Evangelista M, Parsons A B, Xu H, Bader G D, Pagé N, Robinson M, Raghibizadeh S, Hogue C W V, Bussey H, et al. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 9.Leung G K, Schmidt W K, Bergo M O, Gavino B, Wong D H, Tam A, Ashby M N, Michaelis S, Young S G. J Biol Chem. 2001;276:29051–29058. doi: 10.1074/jbc.M102908200. [DOI] [PubMed] [Google Scholar]
- 10.Kilic F, Dalton M B, Burrell S K, Mayer J P, Patterson S D, Sinensky M. J Biol Chem. 1997;272:5298–5304. doi: 10.1074/jbc.272.8.5298. [DOI] [PubMed] [Google Scholar]
- 11.Olson E N, Arnold H-H, Rigby P W J, Wold B J. Cell. 1996;85:1–4. doi: 10.1016/s0092-8674(00)81073-9. [DOI] [PubMed] [Google Scholar]
- 12.Pendás A M, Zhou Z, Cadiñanos J, Freije J M P, Wang J, Hultenby K, Astudillo A, Wernerson A, Rodríguez F, Tryggvason K, Lopéz-Otín C. Nat Genet. 2002;31:94–99. doi: 10.1038/ng871. [DOI] [PubMed] [Google Scholar]
- 13.Lewandoski M, Martin G R. Nat Genet. 1997;17:223–225. doi: 10.1038/ng1097-223. [DOI] [PubMed] [Google Scholar]
- 14.Kim E, Ambroziak P, Otto J C, Taylor B, Ashby M, Shannon K, Casey P J, Young S G. J Biol Chem. 1999;274:8383–8390. doi: 10.1074/jbc.274.13.8383. [DOI] [PubMed] [Google Scholar]
- 15.Bergo M O, Leung G K, Ambroziak P, Otto J C, Casey P J, Gomes A Q, Seabra M C, Young S G. J Biol Chem. 2001;276:5841–5845. doi: 10.1074/jbc.C000831200. [DOI] [PubMed] [Google Scholar]
- 16.Pereira R, Khillan J S, Helminen H J, Hume E L, Prockop D J. J Clin Invest. 1993;91:709–716. doi: 10.1172/JCI116252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byers P H. In: The Metabolic and Molecular Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, Childs B, Kinzler K W, Vogelstein B, editors. Vol. 4. New York: McGraw–Hill; 2001. pp. 5241–5285. [Google Scholar]
- 18.Stacey A, Bateman J, Choi T, Mascara T, Cole W, Jaenisch R. Nature (London) 1988;332:131–136. doi: 10.1038/332131a0. [DOI] [PubMed] [Google Scholar]
- 19.Bonadio J, Saunders T L, Tsai E, Goldstein S A, Morris-Wiman J, Brinkley L, Dolan D F, Altschuler R A, Hawkins J E, Jr, Bateman J F, et al. Proc Natl Acad Sci USA. 1990;87:7145–7149. doi: 10.1073/pnas.87.18.7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forlino A, Porter F D, Lee E J, Westphal H, Marini J C. J Biol Chem. 1999;274:37923–37931. doi: 10.1074/jbc.274.53.37923. [DOI] [PubMed] [Google Scholar]
- 21.Cahoon S, Boden S D, Gould K G, Vailas A C. J Med Primatol. 1996;25:333–338. doi: 10.1111/j.1600-0684.1996.tb00025.x. [DOI] [PubMed] [Google Scholar]
- 22.Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart C L, Burke B. J Cell Biol. 1999;147:913–919. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otto J C, Kim E, Young S G, Casey P J. J Biol Chem. 1999;274:8379–8382. doi: 10.1074/jbc.274.13.8379. [DOI] [PubMed] [Google Scholar]
- 24.Kilic F, Johnson D A, Sinensky M. FEBS Lett. 1999;450:61–65. doi: 10.1016/s0014-5793(99)00482-2. [DOI] [PubMed] [Google Scholar]
- 25.Brown C A, Lanning R W, McKinney K Q, Salvino A R, Cherniske E, Crowe C A, Darras B T, Gominak S, Greenberg C R, Grosmann C, et al. Am J Med Genet. 2001;102:359–367. doi: 10.1002/ajmg.1463. [DOI] [PubMed] [Google Scholar]