Abstract
The hematopoietic microenvironment provides a complex molecular milieu that regulates the self-renewal and differentiation activities of stem cells. We have characterized a stem cell supportive stromal cell line, AFT024, that was derived from murine fetal liver. Highly purified in vivo transplantable mouse stem cells are maintained in AFT024 cultures at input levels, whereas other primitive progenitors are expanded. In addition, human stem cells are very effectively supported by AFT024. We suggest that the AFT024 cell line represents a component of an in vivo stem cell niche. To determine the molecular signals elaborated in this niche, we undertook a functional genomics approach that combines extensive sequence mining of a subtracted cDNA library, high-density array hybridization and in-depth bioinformatic analyses. The data have been assembled into a biological process oriented database, and represent a molecular profile of a candidate stem cell niche.
Stem cells hold great promise for regenerative medicine and cell-based therapies (1). Many somatic tissues, such as liver, intestine, skin, and muscle are replenished from their respective stem cell compartments; however, the best characterized stem cells are those responsible for hematopoiesis (2). The fundamental property shared by all stem cells is an ability to balance the cell fate decision of self-renewal vs. differentiation. The mechanisms that regulate cell fate choices in all stem cell systems have cell-autonomous (stem cell intrinsic) and non-cell-autonomous (microenvironmental) components. Traditionally, it was held that the differentiation potential of somatic stem cells is limited to their tissue of origin. This view has been challenged by recent observations that suggest a high degree of plasticity or transdifferentiation for stem cells (3–5). Little is known about the cellular and molecular features of cell fate mechanisms; however, if stem cells do transdifferentiate, then signals emanating from the microenvironment must play a crucial role.
To generate a sufficient number of stem cells for life-long blood cell production, the hematopoietic stem cell population undergoes a period of extensive expansion in the mid-gestation fetal liver (6). The molecular signals that promote self-renewal are likely to be produced by the cellular components of the fetal liver microenvironment. To characterize the specific cell types that mediate stem cell expansion, we dissected this microenvironment into a panel of over 200 cloned stromal cell lines (7, 8). These were tested for their ability to support primitive hematopoietic stem cells in ex vivo cultures. A broad distribution of supportive abilities was observed, and effective stem cell supporting cell lines were very rare. The most potent stromal cell line, AFT024, maintains highly purified fetal and adult stem cell populations in vitro, for at least 4–7 weeks. These cultured stem cells retain the ability to regenerate in vivo hematopoiesis after transplantation in a manner that is indistinguishable from their freshly purified counterparts (7). Other studies have shown that AFT024 is a potent supporter of human hematopoietic stem cells (9–11). Our data show that (i) AFT024 provides a cellular milieu where stem cell self-renewal and differentiation are balanced; (ii) cells with the properties of AFT024 are rare, many other fetal liver-derived cell lines do not support stem cells; (iii) signals from AFT024 cells alone are sufficient to support stem cells without acting through intermediate microenvironmental elements; and (iv) the mechanisms that maintain this balance are conserved, at least between mouse and man. Therefore, we maintain that AFT024 represents a crucial cellular component of an in vivo stem cell niche.
We reasoned that molecules responsible for the stem cell supportive phenotype would be preferentially expressed in AFT024 compared with nonsupporting lines derived from the same tissue source. An RNA expression survey for known stem cell regulatory cytokines, both positive and negative, did not reveal significant differences among supportive and nonsupportive cell lines (8). We hypothesize that there are yet to be discovered regulators, and that the stem cell supportive ability of AFT024 comes from many interacting molecules. In support of these hypotheses, we have shown that one molecule, delta-like (dlk) (12)/preadipocyte factor-1 (pref-1) (13) is preferentially expressed in AFT024, and functions as a positive stem cell regulator (14). We have also demonstrated that the expression of dlk/pref-1 accounts for only a portion of the supportive activity of AFT024 (14).
Herein we describe a functional genomics approach toward elucidating the microenvironmental signals that mediate the cell-fate choices of stem cells. The data have been assembled into a biological process oriented database, and represent the first molecular profile of a stem cell niche. We show that this molecular profile correlates with the documented stem cell supportive properties of other stromal cell lines, and suggest that it may predict stem cell supportive ability in general.
Materials and Methods
Cell Lines and Culture Conditions.
The fetal liver stromal cell lines used in this study were derived as described (8). Cells were cultured at 31–33°C, 5% CO2 in DMEM containing 10% FBS and 50 μM β-mercaptoethanol (BME). For long-term cultures (LTC) with hematopoietic stem cells, confluent monolayers were irradiated (20 Gy), placed in modified Dexter media (DMEM/10% FBS/10% horse serum/50 μM BME, 0.1 μM hydrocortisone) and maintained at 37°C, 5% CO2.
Hematopoietic Stem Cell Isolation and in Vitro Hematopoietic Assays.
Hematopoietic stem cell populations were derived from 2- to 4-month-old, Ly5.2-C57BL/6J (Jackson Laboratories). Bone marrow (BM) stem cells were isolated essentially as described (15) with the following modifications. Sca-1 expressing lineage depleted (Linneg) cells were first isolated by fluorescence activated cell sorting (FACS) using an enrichment mode. Viable, Sca-1, and cKit expressing cells were then further purified by three-color FACS. After rhodamine-123 staining and efflux, the lower 10% of the rhodamine-retaining Linneg, Sca-1pos, cKitpos (Rholo) cells was collected. Cell sorting and data analyses were performed with a Becton Dickinson FACS Vantage using cellquest software. Antibodies were obtained from BD-PharMingen and rhodamine-123 was obtained from Molecular Probes. Long term-cobblestone area forming cell (LT-CAFC) frequency was determined by limiting dilution analysis (LDA) for each stromal cell line after 4–5 weeks culture (16). Typically Rholo cells were plated on irradiated monolayers in microtiter trays, using 12–24 replicas of 8 different cell numbers. Cells maintained in LTC on each stromal cell line were collected and replated into LDA onto AFT024 monolayers to assess the frequency of secondary CAFC. CAFC frequency was determined by Poisson statistics at 37% negative wells (17). Colony forming cell (CFC) content was determined on the freshly purified and cultured hematopoietic cells. All CFC assays were performed in cytokine enriched semisolid media obtained from Stem Cell Technologies.
Competitive Repopulating Cell Transplantation Assay.
Freshly purified or cultured stem cells from Ly5.2-C57BL/6J mice were harvested, combined with 2 × 105 fresh, nonfractionated, competitor BM cells obtained from congenic Ly5.1-C57BL/6 mice (Charles River Breeding Laboratories) and transplanted into lethally irradiated (10 Gy) recipient mice of the latter genotype. Long term-competitive repopulating stem cell (LT-CRSC) frequency in the starting populations was determined by LDA of cohorts of mice (10 animals per cell dose group). Cell doses ranged from 10–100 purified cells per recipient. Mice were bled and analyzed for Ly5.2pos cells and multilineage reconstitution at intervals for 6 months after transplantation as described (7).
Library Construction.
Two cDNA libraries were constructed by using commercially available vectors (pSport 1 and pSport 2) from Life Technologies, and subtractive hybridization was performed as recommended by the manufacturer. The AFT024–pSport1 library contained 2.5 × 107 clones and the 2018–pSport2 library contained 1.7 × 107 clones. After subtraction the AFT024 library size was reduced to 4.2 × 105 clones. The average insert size for both libraries was 2,000 base pairs and ranged from 500–4,000 base pairs. Library clones were initially sequenced by chain termination using the SEQUENASE VERSION 2.0 kit (U.S. Biochemicals). The majority of single-pass sequences were generated by Incyte Genomics (Palo Alto, CA) or by Commonwealth Biotechnologies (Richmond, VA).
Database Architecture.
The Stromal Cell Database (StroCDB) is available at http://stromalcell.princeton.edu. StroCDB is a relational database with MYSQL software as the database manager (www.mysql.com). It contains all primary sequence data, results of bioinformatic analyses, and is described in detail below. The bioinformatic analyses follow the same general schema as previously outlined (18).
Stromal Cell cDNA Microarray (StroChip) Development.
The Stromal Cell cDNA microarray (StroChip) was developed and printed by Incyte Genomics Inc. from nonredundant cDNAs in the AFT024-subtracted library. To determine the nonredundant clone set, individual sequences were compared by blast (19) against the mouse and human UniGene databases (www.ncbi.nlm.nih.gov/UniGene). Highly significant matches (e ≤ 10−15) were assembled into the same cluster. For entries with multiple representatives, the one containing the most 5′ sequence was selected. PCR-amplified DNAs (n = 3,600) were arrayed on glass slides and correspond to an annotated entry in StroCDB. Controls on the cDNA arrays included housekeeping genes (α tubulin, 23-kDa heat shock protein, and ribosomal subunit S9), a complex target to measure probe complexity, as well as sensitivity controls and fluorescence intensity controls, spotted in triplicate on each chip.
Expression Array Analyses.
Total and poly(A)+ RNA were prepared from stromal cell monolayers by using commercial reagents (Life Technologies, Rockville, MD, and Ambion, Austin, TX). The quantity and quality of the mRNA was evaluated by Northern blotting (data not shown). Cy3 and Cy5 fluorescent labels were incorporated into cDNA transcribed from RNA templates, hybridized to microarrays, and quantitated. Probe labeling, microarray hybridizations, and image quantitation were performed by Incyte Genomics. The stromal cell lines, 2012, 2018, 2058, and BFC012 were each compared with AFT024. Each comparison was performed in both labeling orientations, allowing compensation for different dye incorporation efficiencies and an experimental replicate. The resulting data were processed as follows. Cy5 signal was normalized relative to the Cy3 signal by whole-chip balancing to account for differences in the intensity of each fluorochrome. To assess data set quality, each type of control was grouped and asked, by t test, if the means of each signal (Cy3 and Cy5, after balancing) were significantly different. In no case was there a significant difference. The data were filtered as log 2 ≥ 1 across all four comparisons; 381 cDNAs passed this filter. Log2-transformed, balanced signals were expressed as a ratio (AFT024 vs. the compared cell line) and averaged. These results were grouped into 18 clusters by using a k means clustering algorithm (adapted by J.A.H. from the cclust r package written by E. Dimitriadou, www.r-project.org), by using Euclidean distance as the similarity metric. Eight clusters containing 143 cDNAs were chosen with patterns of expression that correlated with a biological phenotype, i.e., levels of expression that correlated with AFT024 compared with known nonsupporting stromal cell lines.
Approximately 2,000 clones from the subtracted library were arrayed on nylon membranes. Each duplicate spot corresponds to an entry in StroCDB. The membranes were hybridized with [α-33P]dCTP-labeled subtracted probes produced by using the PCR Select technology (CLONTECH). Probes were generated from AFT024, 2012, 2018, and 2058, where material from each of these test lines was subtracted with BFC012. We have previously shown that BFC012 cells do not support stem cells in long-term culture (14). Image data were collected by a Molecular Probes PhosphoImager and quantitated by Incyte Genomics by using arrayvision software. Raw signal/background intensities were log2 transformed and filtered for log2 ≥ 1; values for each element were averaged. A total of 1,178 cDNAs passed this filter. These values were also subjected to the same k means clustering algorithm and 20 clusters generated. Of these, six representative clusters containing 201 cDNAs were selected. The log-transformed, averaged data were used to generate red to green pseudocolored images of the signal/background intensities.
Results
AFT024 Supports Quiescent BM Cells.
We extended our studies of the stem cell supporting ability of AFT024 to include the highly enriched Rhodaminelo cKitpos, Sca-1pos, Linneg (Rholo) fraction of adult murine BM (15, 20). Our data show that the transplantable activity of Rholo cells is quantitatively maintained in long-term cultures supported by AFT024 (Fig. 1). The cultured equivalent of 25 Rholo cells competitively repopulated mice equally as well as freshly purified cells. Therefore, AFT024 functions as a supportive microenvironment for both actively cycling fetal stem cells and largely quiescent adult stem cells. The in vitro clonogenic activities of the purified BM stem cells were also evaluated before and after LTC on AFT024. We observed a 100-fold increase in the number of CFC after AFT024 coculture and demonstrated a strong correlation between the frequencies of in vitro LT-CAFC and LT-CRSC (Fig. 1). A correlation between LT-CAFC and repopulating activity has been suggested (21, 22). We have confirmed and extended these observations by using a highly enriched stem cell population.
Figure 1.
Comparative analysis of cKitpos Sca-1pos Linneg Rhodaminelo BM (Rholo) stem cell activity before (day 0) and after LTC on AFT024. CFC content in 100 fresh, day 0 (n = 6), or the equivalent of 100 Rholo cells after 35 days of culture (n = 8). The frequency of LT-CRSC of freshly purified Rholo cells, 1 in 14, compared with the frequency of CAFC, 1 in 13 ± 1.7 (n = 11), after 28–35 days culture on AFT024. Limiting-dilution CRSC and CAFC frequency was determined by Poisson statistics at 37% negative mice or wells. Rholo cells from the same purification were used to determine both CRSC and CAFC frequencies. %Ly5.2 peripheral blood cells from 25 day 0 Rholo cells (n = 5) compared with the 35 day cultured equivalent of 25 Rholo cells (n = 5) from the same purification; both transplanted in CRSC assay.
A Functional Genomics Approach to the Hematopoietic Microenvironment.
To identify previously uncharacterized stem cell regulatory molecules expressed in the hematopoietic microenvironment, we adopted a global approach that integrates several individual strategies. We have combined high-throughput sequence acquisition and bioinformatics with high-density array and other expression analyses (Fig. 2A). We first made a subtracted cDNA library enriched for gene products preferentially expressed by AFT024, but not by 2018, the nonsupporting cell line. We then obtained and analyzed more than 6,000 forward-reading frame sequence tags. The range of computational analyses is described in detail below.
Figure 2.
Construction of an annotated database that profiles gene expression in a hematopoietic stem cell-supporting microenvironment. (A) Flow diagram depicting the subtracted cDNA library strategy and essential elements comprising StroCDB. (B) Categorization of informative sequences by homology (Left) or by putative or known protein type and function (Right).
A central feature of our efforts is the establishment of the on-line StroCDB (http://stromalcell.princeton.edu). The design of StroCDB is oriented toward the biological process of stem cell regulation by the microenvironment. The database contains all of the primary sequence data, as well as the bioinformatic analyses in a readily retrievable format. The front page presents the experimental overview as a clickable map, embedded with links to Northern and array expression analyses; it also provides links to additional pages that include (i) an introduction, (ii) expanded methods, (iii) supplementary biological and molecular data, (iv) additional tables and enhanced figures, (v) the bioinformatic classification of all entries, (vi) an explanation of the fields in each entry, (vii) a query page for multiple types of queries, (viii) a blast interface page to search the database by sequence, and (ix) five preset queries for interesting categories of gene products. The primary sequence data can be downloaded, and all sequence tags have been deposited into the GenBank EST database. Each individually annotated entry contains a summary that is a distillation of the relevant features of each gene-product. To compare the hematopoietic microenvironment with other tissues and biological systems, StroCDB contains “virtual expression” data that use gene and expressed sequence tag databases in the public domain. StroCDB is highly interactive, and is intended to serve as a central resource for the scientific community. StroCDB will be continually updated and annotated as data become available.
Internal redundancies within StroCDB itself, together with Poisson statistical analyses, suggest that the sequence set represents approximately 35% of the total gene expression complexity represented in the subtracted library. As expected, the sequence set is depleted of housekeeping genes and contains a high percentage of differentially expressed gene products (StroCDB: Flow Diagram, Expression Analysis). To identify sequence homologies and to classify the individual sequence entries, public databases were queried by using the blast algorithm (19). Bioinformatic profiles for each sequence were constructed, analyzed, and annotated as described (18). Homology analyses of a nonredundant and informative sequence set of 2600 cDNAs are depicted in Fig. 2B Left. Of note is the high percentage (25%) of gene products with no homologies in any of the public databases, and the high percentage (33%) with homologies only in the expressed sequence tag databases. Collectively, over 1,000 gene products were assigned to functional categories. These are presented in Fig. 2B Right. To categorize the known, previously defined molecules, we used published data. For the previously undescribed gene products, peptide motifs diagnostic for known protein families and homologies to genes from other species were identified.
Gene Products in StroCDB.
Of particular interest are cell surface, secreted, and extracellular matrix proteins, molecules likely to function in direct communication between stem cells and stroma. AFT024 expresses a battery of known cytokines and other molecules that are implicated in the regulation of stem cells, but many of these are not specific to AFT024 and are also expressed in nonsupporting lines. As such, few of these are found in the subtracted library (StroCDB: Cytokine Signaling Query). These observations suggest that additional signaling pathways play a role in the stem cell supporting activity of AFT024. In this regard, the expression of Wnt-5a, as well as other proteins in this signaling pathway, is noteworthy, and consistent with previous suggestions of their hematopoietic function (23, 24). Numerous other developmentally important signaling molecules, such as components of the bone morphogenic protein and Notch pathways are also represented in the subtracted library (StroCDB: Morphogen Signaling Query).
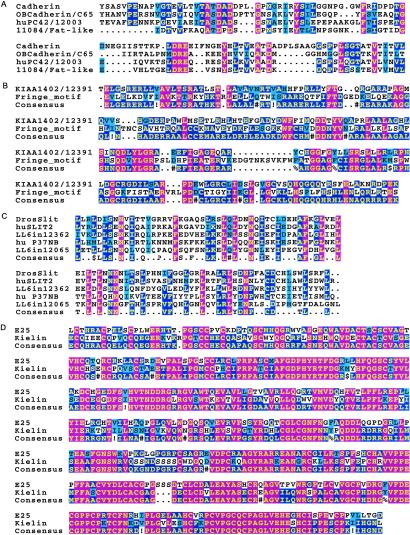
Selected examples of previously uncharacterized gene products with predicted peptide motifs and sequence characteristics suggestive of extracellular function are listed in Table 1. Fig. 3 presents sequence alignments of novel members of several relevant protein families. Among the more interesting molecules are those homologous or similar to Cadherin repeat-containing polypeptides, one related to Drosophila FAT, a morphogenic protein (25) (Fig. 3A); sugar transferases, including one that resembles Drosophila Fringe, a Notch signaling regulator (26) (Fig. 3B). Drosophila Slit, an axon repelling protein (27) (Fig. 3C) (recently, a different Slit-related molecule has been implicated in leukocyte chemotaxis; ref. 28); and Kielin, a secreted protein that is an inducer of midline structures in Xenopus (29) (Fig. 3D). In addition, other molecules with domains implicated in extracellular signaling, such as epidermal growth factor-like repeats, Ig domains, fibronectin repeats, and others involved in adhesion or protein–protein interactions have been identified. The sequence features of these and other gene products are contained in StroCDB. A particularly interesting observation is the identification of numerous molecules previously implicated in neuronal patterning, development, and migration (StroCDB: Neuronal Protein Query). A recent report suggests that stem cell intrinsic regulatory mechanisms may be shared by the neural and hematopoietic systems (30). We suggest that these systems also share many cell extrinsic regulatory mechanisms that facilitate interactions of stem cells with their microenvironments. Emerging similarities between hematopoietic and neural systems are of particular interest in light of several reports that suggest the cross-tissue plasticity of their respective stem cells (31–33).
Table 1.
Candidate stem cell regulatory molecules
| StroCDB ID | Characteristics |
|---|---|
| Cell surface | |
| LL6in11084 | Cadherin family member, related to Drosophila Fat |
| B46 | Cancer associated surface antigen RCAS1 |
| LL6in21904 | Epidermal growth factor-like repeat family; related to MEGF6 |
| LL6in10792 | FLJ13063, 2–3 TM domains and 3 Armadillo/beta-catenin-like repeats |
| LL6in20800 | FLJ22293, Immunoglobulin superfamily and Fibronectin domain containing |
| LL6in12229 | KIAA0657, Immunoglobulin superfamily |
| LL6in10039 | Laminin epidermal growth factor-repeat protein |
| LL6in10980 | Scavenger receptor superfamily |
| LL6in11670 | Xenopus homologue; previously uncharacterized MAM domain-containing adhesion molecule |
| Secreted | |
| LL6in10471 | Epidermal growth factor-like repeat family; homology to secreted hamster HT protein |
| E25 | Homologue of Xenopus Kielin |
| LL6in12391 | KIAA1402 protein; contains fringe-like motif |
| LL6in12065 | Leucine-rich repeat family, homology to Slits |
| LL6in12362 | Homology to human p37NB, homology to Slits |
| LL6in21519 | Putative secreted protein ZSIG37 |
| LL6in10388 | SRPUL; sushi-repeat upregulated in leukemia |
| Extracellular matrix or matrix remodeling | |
| LL6in10447 | Collagen family |
| LL6in10451 | Zinc-metalloproteinase family |
| LL6in11087 | Serine protease inhibitor/serine carboxypeptidase family |
| LL6in10953 | Cystatin domain family |
| LL6in11258 | Thrombospondin family |
| LL6in12280 | Cysteine protease family, with Somatomedin B domain repeats |
| LL6in11839 | Cysteine protease family, OTU-like (ovarian tumor Drosophila) motif |
Expanded tables containing additional categories of gene products (secreted, cell surface, matrix, cytoskeletal, and neuronal-related proteins) are available in StroCDB: Tables.
Figure 3.
Previously undescribed cell surface or secreted proteins contain peptide motifs and/or sequence homology to interesting developmental regulators. (A) One previously undescribed and 2 known cadherin domain-containing proteins are aligned with Cadherin. The LL6in11084 protein is related to the Drosophila Fat morphogenic protein. (B) LL6in12391 is homologous to KIAA1402 which contains a Fringe motif. (C) Two leucine-rich repeat-containing proteins are shown, LL6in120165 and LL6in12362. LL6in12362 is homologous to a human gene product, p37NB. These proteins are most related to proteins homologous to the chemokinetic protein Slit. (D) Keilin is a secreted, Xenopus laevis, signaling protein that mediates inductive activities in the embryonic midline. E25 represents the mouse homolog.
Correlating Gene Expression with Function.
While analyzing individual gene-products from the subtracted library, we noticed a strong correlation between their expression and stem cell support that extended beyond AFT024 and 2018 (StroCDB: Expression Analysis). Expression in AFT024 and 2012, cells that support highly enriched stem cells (7), correlated well. Conversely, expression in 2018 correlated with expression in BFC012, an additional nonsupporting line (14). These observations suggested that a gene expression profile might predict the stem cell supportive abilities of other stromal cell lines. Furthermore, it may be possible to identify a molecular profile that would be the “signature” of a stem cell supportive microenvironment, in general. To investigate these possibilities, we have developed a cDNA microarray, the stromal cell chip (StroChip), and a cDNA membrane array that contain the gene-products represented in StroCDB. This set contains the entire sequence complexity thus far identified in our efforts. We elected to investigate the comparative gene expression profile of 2058, a cell line present in our panel whose ability to support purified stem cells was unknown and whose Northern expression profile tended to correlate more highly with AFT024 and 2012 than with 2018 and BFC012 (StroCDB: Expression Analysis). The StroChip was hybridized with Cy3 and Cy5 labeled material from AFT024, 2012, 2018, BFC012, and 2058 cells. The gene expression profile for each line was evaluated relative to AFT024. Material from each cell line was hybridized in duplicate, using both combinations of Cy3 and Cy5 to control for labeling efficiency. A Pearson correlation coefficient analysis of all elements on the microarray where is line was compared The data were analyzed by using a k means clustering algorithm (34). Clusters with an expression pattern that correlate with known stem cell supporting ability (defined as correlated expression in AFT024 and 2102 compared with 2018 and BFC012) were selected for further analysis (StroCDB: Flow Diagram, Microarray Analysis).
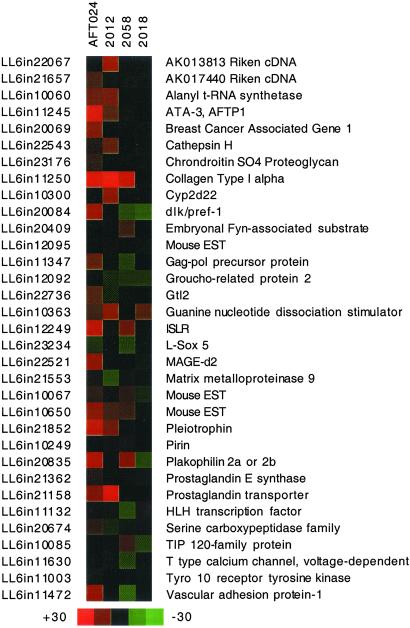
The membrane arrays were hybridized with subtracted probe populations from AFT024, 2012, 2018, and 2058 cells. These populations were depleted of transcripts expressed by BFC012 cells. Because the membranes are arrayed with the AFT024(−)2018 subtracted library, this strategy removed an additional set of transcripts in common with a different nonsupporting line, effectively increasing the stringency of the analysis. These data were also assembled into clusters and those with patterns that showed correlated expression in AFT024 compared with 2018 were selected for further analysis (StroCDB: Flow Diagram, Microarray Analysis). To reveal the most stringent subset of transcripts, the results of the StroChip and membrane array analyses were compared and those in common are presented in Fig. 4. Analysis of the data set suggests that 2058 more closely resembles supporting lines in the expression of this subset of genes. An F test analysis of the membrane array data, where all probe populations were subtracted with BFC012, shows that only 2018 is significantly different from AFT024 (P < 0.001).
Figure 4.
A gene expression profile may correlate with a stem cell-supporting phenotype in stromal cell lines. Expression arrays developed from the AFT024-subtracted library were interrogated with probes derived from stromal cell lines. A clustering algorithm was used to profile the data and informative clusters were selected. The contents of the clusters revealed in the cDNA microarray (StroChip) analysis were compared with those selected in the custom filter array analysis. The overlapping gene products are displayed as their relative signal to background ratios in red to green coloration for the filter arrays. The Pearson correlation coefficient (r) of this subset of gene products for each line compared with AFT024 is; 2012 (0.32), 2058 (0.42), and 2018 (0.04). An expanded version of the figure and the correlation coefficients of all array elements and comparisons is available in StroCDB: Figures and Microarray Analysis. StroCDB identifier (LL6inXXXXX) and the protein identities are listed.
We also determined the stem cell support behavior of the five different stromal cell lines by assaying for (i) primary LT-CAFC frequency on each line and (ii) for the ability of each line to maintain stem/progenitor cells in primary LTC that would generate secondary CAs on AFT024 monolayers. The results of these studies, presented in Fig. 5, suggest a stem cell supporting activity for 2058 that is similar to 2012.
Figure 5.
Correlating gene expression and stem cell support. The ability of stromal cell lines to support enriched stem cell populations was studied. (A) Limiting-dilution CAFC assay on each stroma at 4 weeks. (B) Stem cells were cultured on each stroma, and then after 4 weeks, were replated into limiting-dilution CAFC assay onto fresh AFT024 monolayers. This assay determines the ability of each stroma to maintain stem cells with a capacity to generate secondary CAFC. Limiting-dilution frequency was determined at 37% negative wells according to Poisson statistics and is shown in parentheses for both graphs. The ability of each line to maintain stem cells in the secondary CAFC assay relative to AFT024 is; 2012 (32%), 2058 (31%), and 2018 (1%). These data correlate well with the results from the gene expression analysis in Fig. 4.
Both the array expression data and the biological support data suggest that 2058 cells elaborate a molecular milieu that is similar to supporting lines but still lacking in some essential elements. Northern blot analysis supports this suggestion (StroCDB: Flow Diagram, Expression Analysis). Cell line 2058 does not express the dlk/pref-1 protein. We have shown initial, but not sustained stem cell maintenance by nonsupporting BFC012 cells engineered to express dlk (14). In addition, 2058 cells do not express the “slit-like” molecule described in Fig. 3C. We have begun to express these “missing” gene products in 2058. Very preliminary data suggest that expression of these proteins in 2058 cells enhances its stem cell support capacity (K.A.M., unpublished observations).
Discussion
The picture of hematopoietic stem cell regulation by the microenvironment that emerges from our studies is complex. We have identified a large number of interesting and previously undescribed gene products, suggesting that hematopoietic stem cell-fate choices will be controlled by multicomponent molecular networks. As such, the balance of self-renewal and differentiation is not likely to be governed by single or few “stem cell factors,” but rather by the integration of many interacting signal inputs. The overall behavior of hematopoietic stem cells may be a property of regulatory networks that owes its essence to the interactive architecture of the network rather than to its individual components. Indeed, many years of effort have failed to identify a defined in vitro system capable of more than a severalfold hematopoietic stem cell expansion. Even in situations where signaling systems such as Wnt or Notch have been suggested as hematopoietic stem cell regulators, their observed in vitro effects have been modest (24, 35). Our current data support a role for these signaling pathways (and other molecules, such as dlk/pref-1 and a previously uncharacterized “slit-like” protein) in the global regulation of hematopoietic stem cells.
Why might the regulation of hematopoietic stem cells appear to be more complex than that of other stem cells? For a tissue with the dramatic proliferative potential of hematopoiesis, relying on “simple” combinations of few factors for regulating key cell-fate decisions may present an unacceptable risk in terms of developing systemic pathologies such as leukemias. In contrast, neural stem cells, whose daily neuropoietic activity in the rodent is limited to relatively few mature cell progeny, can be extensively expanded in cultures supported by the single factors epidermal growth factor or basic-fibroblast growth factor (36).
The properties and beneficial aspects of network-based regulation have been described in other biological decision systems such as bacterial chemotaxis (37), patterning in Drosophila (38), and signaling by the T cell receptor (39). In particular, the ability to respond to systemic needs for enhanced blood cell production and at the same time resist deleterious perturbations in a robust manner are necessary features of the hematopoietic stem cell system. Network-based, distributed regulation can accommodate both of these properties.
Our studies provide the first global, molecular definition for a stem cell supportive niche. As such, they represent the necessary first step for the precise elucidation of microenvironmental regulatory functions. In a parallel effort, we have recently described the intrinsic genetic program of hematopoietic stem cells; that is, the global panel of gene products expressed preferentially in primitive but not in mature blood cells (18). These data and analyses reside in the Stem Cell Database (SCDb; http://stemcell.princeton.edu). The information in StroCDB and SCDb is complementary. It is reasonable to propose that functionally important extracellular molecules present in StroCDB will have an interacting partner in SCDb. In addition to the StroChip, we have developed custom stem cell microarrays from the gene products characterized in SCDb. These arrays will be used to explore the molecular cross-talk between stem cells and stromal cells as they interact in vitro and perhaps even in vivo. The StroChip will provide an ideal platform for comparative analyses of other rigorously tested stem cell supporting stromal cell lines, particularly those from adult and early fetal tissue. Initial data, presented here, indicate that these types of studies may be useful in defining key players in the gene expression space of the stem cell niche. It will also be interesting to compare molecular profiles of stem cell microenvironments from other tissues. Perhaps the basic mechanisms that mediate stem cell self renewal and differentiation will be conserved both in different stem cell compartments and in their corresponding microenvironments. We anticipate that the StroCDB will function as a central on line resource for the entire stem cell community. It is anticipated that this evolving molecular toolbox of a well-defined stem cell-supporting microenvironment will lead to discovery driven hypotheses that, in turn, will unravel general and tissue specific regulatory networks in stem cell niches.
Acknowledgments
This work is dedicated to the memory of G. Christian Overton, our colleague and friend. We thank the following for valuable contributions to these studies: H. Ema, R. Phillips, N. Ivanova, T. Doniger, H. Wang, D. Guenther, D. Pinto-Gonzalez, D. Genetti, A. Dominguez, U. Duru, J. Lemus, S. Backus, and C. Hansen. We thank A. Beavis for expert flow cytometry. We thank Drs. Christa Muller-Sieberg and Margaret Baron for critical reviews of the manuscript. This work was supported by National Institutes of Health Grant HL58739 (to K.A.M.), and by Association pour la Recherche sur le Cancer Grant 5837 and Association Française Contre les Myopathies/Institut National de la Santé et de la Recherche Médicale Grant 4CS02F (to P.C.). Additional support for sequencing and microarray development was provided by ImClone Systems, Inc., New York. Supplementary information is available at http://stromalcell.princeton.edu.
Abbreviations
- dlk
delta-like
- pref-1
preadipocyte factor-1
- LTC
long-term cultures
- BM
bone marrow
- LT-CAFC
long term-cobblestone area forming cell
- LT-CRSC
long term-competitive repopulating stem cell
- StroCDB
Stromal Cell Database
Footnotes
References
- 1.Lagasse E, Shizuru J A, Uchida N, Tsukamoto A, Weissman I L. Immunity. 2001;14:425–436. doi: 10.1016/s1074-7613(01)00123-6. [DOI] [PubMed] [Google Scholar]
- 2.Morrison S J, Shah N M, Anderson D J. Cell. 1997;88:287–298. doi: 10.1016/s0092-8674(00)81867-x. [DOI] [PubMed] [Google Scholar]
- 3.Anderson D J, Gage F H, Weissman I L. Nat Med. 2001;7:393–395. doi: 10.1038/86439. [DOI] [PubMed] [Google Scholar]
- 4.Blau H M, Brazelton T R, Weimann J M. Cell. 2001;105:829–841. doi: 10.1016/s0092-8674(01)00409-3. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs E, Segre J A. Cell. 2000;100:143–155. doi: 10.1016/s0092-8674(00)81691-8. [DOI] [PubMed] [Google Scholar]
- 6.Ema H, Nakauchi H. Blood. 2000;95:2284–2288. [PubMed] [Google Scholar]
- 7.Moore K A, Ema H, Lemischka I R. Blood. 1997;89:4337–4347. [PubMed] [Google Scholar]
- 8.Wineman J, Moore K, Lemischka I, Muller-Sieburg C. Blood. 1996;87:4082–4090. [PubMed] [Google Scholar]
- 9.Miller J S, McCullar V, Punzel M, Lemischka I R, Moore K A. Blood. 1999;93:96–106. [PubMed] [Google Scholar]
- 10.Punzel M, Wissink S D, Miller J S, Moore K A, Lemischka I R, Verfaillie C M. Blood. 1999;93:3750–3756. [PubMed] [Google Scholar]
- 11.Thiemann F T, Moore K A, Smogorzewska E M, Lemischka I R, Crooks G M. Exp Hematol. 1998;26:612–619. [PubMed] [Google Scholar]
- 12.Laborda J, Sausville E A, Hoffman T, Notario V. J Biol Chem. 1993;268:3817–3820. [PubMed] [Google Scholar]
- 13.Smas C M, Sul H S. Cell. 1993;73:725–734. doi: 10.1016/0092-8674(93)90252-l. [DOI] [PubMed] [Google Scholar]
- 14.Moore K A, Pytowski B, Witte L, Hicklin D, Lemischka I R. Proc Natl Acad Sci USA. 1997;94:4011–4016. doi: 10.1073/pnas.94.8.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spangrude G J, Brooks D M, Tumas D B. Blood. 1995;85:1006–1016. [PubMed] [Google Scholar]
- 16.Ploemacher R E, van der Sluijs J P, Voerman J S, Brons N H. Blood. 1989;74:2755–2763. [PubMed] [Google Scholar]
- 17.Taswell C. J Immunol Methods. 1984;72:29–40. doi: 10.1016/0022-1759(84)90430-7. [DOI] [PubMed] [Google Scholar]
- 18.Phillips R L, Ernst R E, Brunk B, Ivanova N, Mahan M A, Deanehan J K, Moore K A, Overton G C, Lemischka I R. Science. 2000;288:1635–1640. doi: 10.1126/science.288.5471.1635. [DOI] [PubMed] [Google Scholar]
- 19.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uchida N, Aguila H L, Fleming W H, Jerabek L, Weissman I L. Blood. 1994;83:3758–3779. [PubMed] [Google Scholar]
- 21.Ploemacher R E, van der Sluijs J P, van Beurden C A, Baert M R, Chan P L. Blood. 1991;78:2527–2533. [PubMed] [Google Scholar]
- 22.Weilbaecher K, Weissman I, Blume K, Heimfeld S. Blood. 1991;78:945–952. [PubMed] [Google Scholar]
- 23.Austin T W, Solar G P, Ziegler F C, Liem L, Matthews W. Blood. 1997;89:3624–3635. [PubMed] [Google Scholar]
- 24.Van Den Berg D J, Sharma A K, Bruno E, Hoffman R. Blood. 1998;92:3189–3202. [PubMed] [Google Scholar]
- 25.Mahoney P A, Weber U, Onofrechuk P, Biessmann H, Bryant P J, Goodman C S. Cell. 1991;67:853–868. doi: 10.1016/0092-8674(91)90359-7. [DOI] [PubMed] [Google Scholar]
- 26.Moloney D J, Panin V M, Johnston S H, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine K D, Haltiwanger R S, Vogt T F. Nature (London) 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 27.Kidd T, Bland K S, Goodman C S. Cell. 1999;96:785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 28.Wu J Y, Feng L, Park H T, Havlioglu N, Wen L, Tang H, Bacon K B, Jiang Z, Zhang X, Rao Y. Nature (London) 2001;410:948–952. doi: 10.1038/35073616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsui M, Mizuseki K, Nakatani J, Nakanishi S, Sasai Y. Proc Natl Acad Sci USA. 2000;97:5291–5296. doi: 10.1073/pnas.090020497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terskikh A V, Easterday M C, Li L, Hood L, Kornblum H I, Geschwind D H, Weissman I L. Proc Natl Acad Sci USA. 2001;98:7934–7939. doi: 10.1073/pnas.131200898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bjornson C R, Rietze R L, Reynolds B A, Magli M C, Vescovi A L. Science. 1999;283:534–537. doi: 10.1126/science.283.5401.534. [DOI] [PubMed] [Google Scholar]
- 32.Brazelton T R, Rossi F M, Keshet G I, Blau H M. Science. 2000;290:1775–1779. doi: 10.1126/science.290.5497.1775. [DOI] [PubMed] [Google Scholar]
- 33.Mezey E, Chandross K J, Harta G, Maki R A, McKercher S R. Science. 2000;290:1779–1782. doi: 10.1126/science.290.5497.1779. [DOI] [PubMed] [Google Scholar]
- 34.Tavazoie S, Hughes J D, Campbell M J, Cho R J, Church G M. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 35.Karanu F N, Murdoch B, Gallacher L, Wu D M, Koremoto M, Sakano S, Bhatia M. J Exp Med. 2000;192:1365–1372. doi: 10.1084/jem.192.9.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Temple S. Nature (London) 2001;414:112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- 37.Alon U, Surette M G, Barkai N, Leibler S. Nature (London) 1999;397:168–171. doi: 10.1038/16483. [DOI] [PubMed] [Google Scholar]
- 38.von Dassow G, Meir E, Munro E M, Odell G M. Nature (London) 2000;406:188–192. doi: 10.1038/35018085. [DOI] [PubMed] [Google Scholar]
- 39.Grakoui A, Bromley S K, Sumen C, Davis M M, Shaw A S, Allen P M, Dustin M L. Science. 1999;285:221–227. [PubMed] [Google Scholar]