Abstract
The p300 and closely related cAMP response element binding protein (CREB)-binding protein (CBP) acetyltransferases function as global transcriptional coactivators and play important roles in a broad spectrum of biological processes, including cell proliferation and differentiation. A role of p300/CBP in tumor suppression has been proposed from the fact that these coactivators are targeted by viral oncoproteins and that biallelic mutations of p300 have been identified in carcinomas. Here, we show that transcriptional response to the transforming growth factor β (TGF-β), an inhibitor of epithelial cell growth, was severely impaired in human carcinoma cell lines carrying p300 mutations accompanied by inactivation of the second allele, and that wild-type expression restored TGF-β-dependent transcriptional activity. Furthermore, reintroduction of wild-type p300 suppressed the growth of p300-deficient carcinoma cells, whereas p300 did not inhibit the growth of carcinoma cells examined, which have no detectable alterations in p300 protein and retain the TGF-β-dependent transcriptional response. In addition, tumor-derived mutants missing the bromodomain or glutamine-rich region, which are respectively important for chromatin interaction and coactivator activities, lost the suppressive activity. In contrast, CBP exhibited no or reduced ability to suppress the growth of p300-deficient carcinoma cells. These results provide experimental evidence to show that p300 acts as a suppressor of tumor cell growth and suggest a distinct role of p300 in suppression of epithelial tumors.
The p300 (1) and the closely related cAMP response element binding protein (CREB)-binding protein (CBP) (2), function as molecular scaffolds bridging a variety of sequence-specific transcription factors, coactivators, and the basal transcriptional machinery, including the RNA polymerase II holoenzyme (3) through multiple domains that serve as docking sites for these associated proteins, such as Cys/His rich regions (CH1, CH2, and CH3), and a glutamine (Q)-rich region (Fig. 1A). CBP/p300 also possess chromatin remodeling activity through histone acetyltransferase (HAT) activity intrinsic to p300 and CBP (4, 5) and/or to other associated coactivators, including P/CAF and the SRC/p160 family coactivators (6–8), and acetylate lysine residues of histones and nonhistone proteins, such as p53 (9). In addition, p300/CBP have a bromodomain, which is conserved among chromatin-associated proteins, including nuclear HATs, and which interacts specifically with acetyl-lysines in the N-terminal tails of histones (10, 11).
Figure 1.
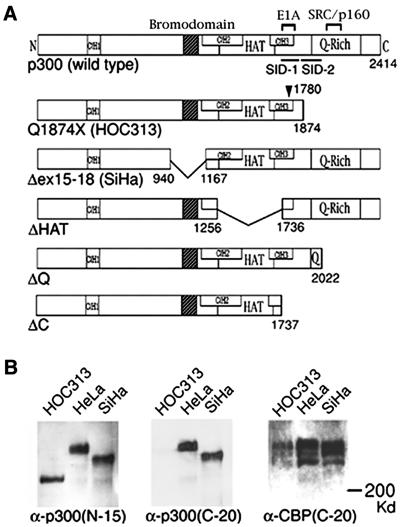
Mutations of p300 in HOC313 and SiHa cells. (A) Schematic diagram of wild type and p300 mutants. Functional domains of p300 protein are indicated: Cys/His-rich regions (CH1, CH2, and CH3), bromodomain, HAT domain (HAT), E1A-binding region (E1A), SRC/p160-binding region, Q-Rich, and Smad interaction domains (SID-1 and SID-2). Numbers indicate amino acids at boundaries. Position of mutation at codon 1780 is indicated by an arrowhead. (B) Alterations of p300 protein in HOC313 and SiHa cells. Western blotting of HeLa, HOC313, and SiHa cells. The blot was probed with α-p300 (N-15; Left), then reprobed for α-p300 (C-20; Center), and α-CBP (C-20; Right).
Both p300 and CBP participate in the regulation of a wide range of biological processes, such as cell proliferation and differentiation (3), and both are essential for normal development (12). In addition, the fact that p300/CBP double heterozygous mice are embryonic lethal indicates that the total gene dosage of p300 and CBP is important in development (13). However, although p300 and CBP share many functional properties, recent evidence indicates that these coactivators have distinct functions in certain types of cells (13–16).
A role for p300/CBP in tumor suppression has been proposed by the fact that disturbance of p300/CBP function by viral oncoproteins, the adenovirus E1A and SV40 large T antigen, is essential for the transformation of rodent primary cells. Consistent with this hypothesis, biallelic mutations of p300 have been identified in certain types of human cancers of epithelial origin (17–19). In contrast, CBP-, but not p300-, heterozygous mice develop tumors in hematopoietic cells, accompanied by a loss of heterozygosity at the CBP locus (15), suggesting cell type-specific roles for p300 and CBP in tumorigenesis. Nevertheless, little is known about the basis for the differences between p300 and CBP in this process. Furthermore, although Rb, which is inactivated by E1A and large T antigen, suppresses tumor cell growth, no direct evidence has yet shown that p300 and CBP can suppress the growth of tumor cells lacking normal counterparts. We therefore examined the roles of p300 in the suppression of tumor cell growth by using human carcinoma cell lines lacking normal p300. We found that p300 plays critical roles in growth suppression and transcriptional regulation of epithelial cells and that p300 and CBP perform nonoverlapping functions in these processes.
Materials and Methods
Cell Culture.
HOC313, HSC-7 (human oral squamous cell carcinoma), SiHa, HeLa (human cervical carcinoma), COS-7 (obtained from Riken Cell Bank), Mv1Lu (mink lung epithelial cells), and HaCaT (human keratinocytes) cells were cultured at 37°C in DMEM supplemented with 10% FCS.
Western Blotting and Antibodies.
Whole cell extracts were prepared and Western blotted as described (19, 20). The antibodies were as follows: rabbit polyclonal α-p300 (N-15), α-p300 (C-20), and α-CBP (C-20) antibodies (Santa Cruz Biotechnology); and mouse monoclonal α-Flag M2 and α-Myc antibodies (Sigma).
Plasmids.
pcDEFp300Δex15–18 was described elsewhere (19). To create a plasmid expressing Flag-tagged p300Q1874X, cDNA from HOC313 cells was amplified with C2 primers as described (19). pcDEFp300Q1874X was generated by inserting the PCR fragment into the appropriate restriction sites in the full-length Flag-tagged p300 cDNA of pcDEFp300. The antisense construct pcDEFp300AS was created by religation of the fragment encoding Flag-tagged p300 to pcDEF3 in the opposite direction and used for controls. p300ΔHAT and ΔQ were created by removing the BglII-Aor51HI and HincII (nucleotide 7266)-EcoRV fragments from pcDEFp300, respectively. pcDEFCBP was generated by inserting the CBP cDNA (a gift from R. H. Goodman, Oregon Health and Science University, Portland, OR) into pcDEF3 vector. The constructions were confirmed by DNA sequencing. The retinoic acid receptor (RAR) reporter vector (pTK-βREX2-Luc) was a gift from K. Umezono, Kyoto University, Kyoto.
Transfection and Luciferase Assays.
Cells were transfected by using FuGENE 6 Transfection Reagent (Roche Diagnostics) according to the manufacturer's instructions. pTK-RL (Toyo Ink, Tokyo) was used as an internal control. Luciferase and Renilla luciferase activities were measured 48 h after transfection. Luciferase assays were performed as described (19). Luciferase activity was normalized to Renilla-luciferase activity.
Colony Formation Assay and 5-Bromo-2′-deoxyuridine (BrdUrd) Incorporation.
Colony formation was assayed as described (21). Cells were transfected with Flag-tagged p300 or CBP expression plasmids, and selected with G418 (450 μg⋅ml−1 for HSC-7, 500 μg⋅ml−1 for SiHa and HaCaT, 550 μg⋅ml−1 for HeLa, and 600 μg⋅ml−1 for HOC313 cells) for 3 wk. In some experiments, SiHa and HaCaT cells were grown in the presence of 2 μg⋅ml−1 of transforming growth factor (TGF)-β1 during G418 selection. To monitor proliferation, cells were labeled with BrdUrd for 24 h before fixing. BrdUrd incorporation was detected by using the BrdUrd Labeling and Detection Kit I (Roche Diagnostic). To detect the expression of transfected Flag-tagged cDNAs, cells were stained with α-Flag M2 antibody. Immunostaining was visualized by using HistoMark BLACK horseradish peroxidase reporter reagent (Kirkegaard & Perry Laboratories). The number of BrdUrd-positive cells was counted before Flag staining. Data represent average of three independent experiments.
Results
Mutations of p300 in HOC313 Oral Carcinoma Cells.
We recently identified a homozygous internal deletion of exons 15–18 of the p300 gene in the SiHa cervical carcinoma cell line, which results in a loss of the bromodomain (p300Δex15–18) (ref. 19; Fig. 1A). To further examine alterations of p300 in human cancers, we performed reverse transcription–PCR-single strand conformation polymorphism (SSCP) analysis of p300 in human oral squamous cell carcinoma cell lines. We detected mobility-shifted bands in two PCR products derived from HOC313 cells. Direct sequencing of these PCR products revealed two point mutations in HOC313 cells; a missense mutation (CCC→GCC; Pro→Ala) at codon 1780 within the E1A binding region, and a nonsense mutation (CAG→TAG; Gln→Stop) at codon 1874 (see Fig. 6 and supporting Materials and Methods, which are published as supporting information on the PNAS web site, www.pnas.org). In addition, neither of the PCR products contained any normal sequences. The nonsense mutation leads to a 541-aa truncation of the C-terminal glutamine-rich (Q-rich) region (named p300Q1874X; Fig. 1A). Coimmunoprecipitation assays revealed that the missense mutation of the p300Q1874X protein in the E1A binding region did not affect binding to E1A and P/CAF, which bind this region (refs. 1 and 6; data not shown). Western blotting revealed that HOC313 cells expressed a truncated protein that was detected only by the p300 (N-15) antibody specific for the N terminus of the protein, whereas the size of CBP did not differ in all three cell lines examined (Fig. 1B). Consistent with the RT-PCR-SSCP and sequencing results, wild-type p300 protein was not detectable in either HOC313 or SiHa cells (19), indicating that the second allele of the gene is not expressed in HOC313 cells. Nevertheless, Southern blotting detected no gross rearrangements or deletions of either allele of the p300 gene in HOC313 cells (data not shown). Thus, the basis for the absence of p300 expression is unknown.
Critical Role of p300 in TGF-β-Dependent Transcriptional Activation.
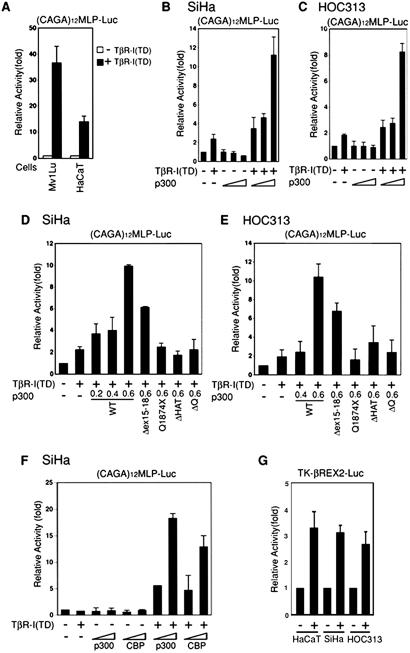
TGF-β inhibits the proliferation of epithelial cells, and the TGF-β response is frequently lost in carcinomas. The p300 and CBP mediate the TGF-β signaling pathway through interaction with the transcription factor Smads (22), and our recent study has demonstrated that p300 is critical for TGF-β-dependent activation of the p21CIP1 promoter in SiHa carcinoma cells (19). We further examined the role of p300 in transcriptional responses to TGF-β signaling in carcinoma cells lacking normal p300. SiHa and HOC313 cells were transiently transfected with a reporter construct driven by multimerized Smad-binding sites [(CAGA)12 MLP-Luc; ref. 23], together with a plasmid encoding a constitutively active form of the TGF-β type I receptor, TβR-I (TD) (24). We compared the TGF-β response in control Mv1Lu and HaCaT epithelial cells, both of which respond to TGF-β signaling (25). TβR-I (TD) expression activated the reporter about 37- and 14-fold in Mv1Lu and HaCaT cells, respectively (Fig. 2A), whereas it only weakly enhanced (≈2-fold) reporter activity in both SiHa and HOC313 cells (Fig. 2 B and C). Transfection with increasing amounts of p300 in SiHa and HOC313 cells led to an 8- to 11-fold increase in TGF-β-dependent activation of the reporter activity (Fig. 2 B and C). The results obtained by using 3TP-Lux, a well-known TGF-β responsive reporter construct (26), were similar (data not shown).
Figure 2.
TGF-β-dependent transcriptional activity is restored by p300 in SiHa and HOC313 cells. (A) Mv1Lu and HaCaT cells were transfected with 0.3 μg of (CAGA)12MLP-Luc reporter, together with 0.1 μg of pTβR-I (TD). (B and C) SiHa (B) and HOC313 (C) cells were transfected with (CAGA)12MLP-Luc, together with pTβR-I (TD) as in A, and increasing amounts (0.2, 0.4, and 0.8 μg in B or 0.4, 0.6, and 0.8 μg in C) of p300. (D and E) Experiments were performed as described in B and C except that the indicated amounts (in μg) of wild-type or mutant p300 plasmids were used. (F) Experiments were performed as described in B except that 0.4 and 0.6 μg of p300 or CBP plasmids were used. (G) Response of SiHa and HOC313 cells to RAR activity. SiHa, HOC313, and HaCaT cells were transfected with 0.25 μg of pTK-βREX2-Luc reporter, together with a plasmid expressing RARα and then stimulated with 9cis-RA (10−6 M) 24 h before harvest. Results are shown as relative luciferase levels. Data represent the average of three independent experiments, each performed in duplicate.
We also examined the response to retinoic acid in SiHa and HOC313 cells by using an RAR-responsive luciferase reporter construct (pTK-βREX2-Luc), because previous experiments using fibroblasts derived from p300–null mice have shown that p300 is critical for RAR function (13). However, retinoic acid treatment significantly increased the RAR-responsive reporter activity in both SiHa and HOC313 cells to the levels similar to that in control HaCaT cells (Fig. 2G), indicating that normal p300 appears not to be essential for this activity in these cell lines. Taken together, these results indicate that loss of normal p300 functions causes critical effects on TGF-β, but not RAR signaling in p300-deficient carcinoma cell lines.
We next examined the effects of p300 mutations on TGF-β-dependent transcriptional activation. The nonsense mutation of p300Q1874X is located in the middle of the Smad interaction domain (SID), resulting in a loss of the C-terminal half of SID, SID-2 (Fig. 1A; ref. 27). The ability of the p300Q1874X mutant to enhance transcription of the TGF-β-responsive reporter was severely impaired in both SiHa and HOC313 cells (Fig. 2 D and E). We next determined whether the loss of SID-2 affects Smad binding by using coimmunoprecipitation assays in vivo. Flag-tagged p300Q1874X protein expressed in COS-7 cells was coimmunoprecipitated with 6Myc-tagged Smad2 in a manner that depended on TGF-β signaling, although to a lesser extent than to wild type, whereas no interaction was detected p300ΔC lacking the whole SID domain (Fig. 7A, which is published as supporting information on the PNAS web site; ref. 27). The results for Smad3 were similar (data not shown). To determine whether the deletion of the Q-rich region in p300Q1874X protein may account for the loss of its ability to mediate TGF-β signaling, we tested an additional mutant that lacks the Q-rich region but has an intact SID domain (p300ΔQ, Fig. 1A). As shown in Fig. 2 D and E, p300ΔQ severely compromised TGF-β-dependent transcription, indicating that deletion of the Q-rich region including the SRC/p160 binding region critically affects the ability of p300 to mediate TGF-β signaling.
We next examined the effect of the deletion of the bromodomain in p300Δex15–18 on TGF-β signaling. As shown in Fig. 2 D and E, the p300Δex15–18 mutant reproducibly showed reduced activity but retained some coactivator function in the transient reporter assay. We next examined the ability of p300Δex15–18 to acetylate histones by using the immunoprecipitation (IP)-HAT assay (see supporting Materials and Methods). Flag-tagged p300 proteins expressed in COS-7 cells were immunoprecipitated with the α-Flag antibody, and HAT activity in the immunoprecipitates was then measured. The HAT activity of p300Δex15–18 was reduced (70%), compared with that of p300Q1874X and the wild-type control (Fig. 7B). Endogenous p300 immunoprecipitated from SiHa cell lysates also exhibited less HAT activity than control HeLa cells (data not shown). We next examined whether the HAT activity of p300 is required for TGF-β-dependent transcription. Fig. 2 D and E shows that the ability of p300ΔHAT lacking the HAT domain to activate the TGF-β-dependent reporter was severely impaired, suggesting that HAT activity of p300 is essential for mediating TGF-β signaling. However, given the severely impaired TGF-β response in SiHa cells, the residual activity of exogenous p300Δex15–18 is probably due to high-level expression of the protein in the transient reporter assay. In addition, CBP expression showed moderate coactivator activity in TGF-β-dependent transcription as reported (Fig. 2F). These observations imply that transient reporter assays may not precisely determine whether the total level of these coactivators or distinct functions of p300 and its functional domains, such as the bromodomain, are critical for TGF-β signaling.
p300 Suppresses Tumor Cell Growth.
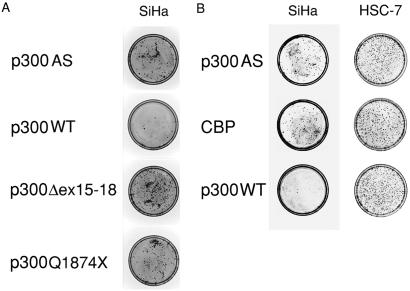
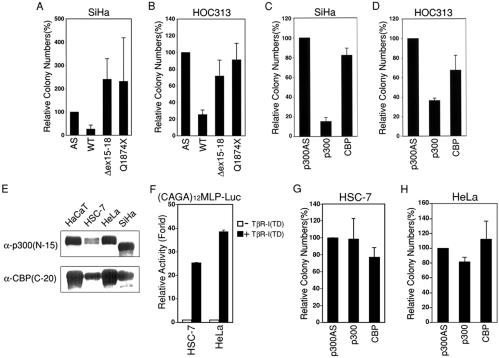
To further examine the role of p300 and the biological relevance of p300 mutations in epithelial tumorigenesis, we next tested whether the reintroduction of wild-type p300 suppresses the proliferation of carcinoma cells lacking normal p300 in colony formation assay. SiHa and HOC313 cells were transfected with either Flag-tagged wild-type or mutant p300 expression constructs that also express the neomycin resistance gene and counted G418-resistant colonies 3 wk later. For comparison, we also tested a Flag-tagged CBP expression construct. Both SiHa and HOC313 cells transfected with wild-type p300 formed 4-fold fewer colonies than those transfected with a control plasmid (p300AS). Notably, cells transfected with both of the tumor-derived mutants formed colonies comparable to the control in HOC313 cells and even more in SiHa cells (Fig. 3A and Fig. 4 A and B). Furthermore, compared with p300, CBP did not exhibit apparent suppression activity in SiHa cells (Fig. 3B and Fig. 4C), and CBP was moderately (50%) suppressive in HOC313 cells (Fig. 4D), even though Flag-tagged p300 and CBP were expressed at comparable levels in COS-7 cells (data not shown).
Figure 3.
Colony formation assay with SiHa and HSC-7 cells. Cells were stably transfected with 2.5 μg of control pDEFp300AS or indicated Flag-tagged p300 or CBP expression plasmids in a 35-mm plate, split into two 100-mm plates, and then cultured in media containing G418 for 3 wk. Cells were stained with Giemsa solution. (A and B) Photographs of plates showing colony formation assay of SiHa and HSC-7 cells.
Figure 4.
Suppression of carcinoma cell growth by p300. Colony formation assays proceeded as in Fig. 3, and then the number of colonies was counted. Data represent the average of the number of colonies obtained from at least three independent experiments. (A and B) Suppression of colony formation by wild-type, but not mutant, p300 in SiHa (A) and HOC313 (B) cells. The mark at 100% represents 780 and 114 colonies per plate of SiHa and HOC313 cells, respectively. (C and D) Suppression of colony formation by wild-type p300, but not CBP, in SiHa (C) and HOC313 (D) cells. The mark at 100% represents 141 and 340 colonies per plate of SiHa and HOC313 cells, respectively. (E) Expression of p300 and CBP measured by Western blotting using α-p300 (N-15) (Upper) and α-CBP (C-20; Lower) in the indicated cells. (F) The TGF-β response in HSC-7 and HeLa cells. Transient reporter assays using (CAGA)12MLP-Luc proceeded as in Fig. 2A. (G and H) Colony formation assay with HSC-7 and HeLa cells. The mark at 100% represents 150 and 87 colonies per plate of HSC-7 and HeLa cells, respectively.
We next examined whether p300 overexpression might have any growth inhibitory effect on cells expressing wild-type p300. To this end, we have used the HSC-7 oral squamous cell carcinoma and HeLa cervical carcinoma cell lines that represent the same types of carcinoma as HOC313 and SiHa cell lines, respectively, and had no apparent alterations in p300 and CBP proteins as determined by Western blotting (Fig. 4E). Furthermore, TβR-I (TD) expression activated the (CAGA)12 MLP-Luc reporter about 25- and 38.5-fold in HSC-7 and HeLa cells, respectively, in transient reporter assay (Fig. 4F), which are comparable levels in control Mv1Lu and HaCaT cells (Fig. 2A), supporting the notion that these cells express wild-type p300. However, because TGF-β did not induce growth arrest of HSC-7 and HeLa cells (data not shown), these cells might have some defect other than p300 in the TGF-β signaling pathway. In contrast to the results obtained with the p300-deficient carcinoma cells, transfection of neither p300 nor CBP significantly suppressed the colony formation of HSC-7 and HeLa cells (Fig. 3B, and Fig. 4 G and H).
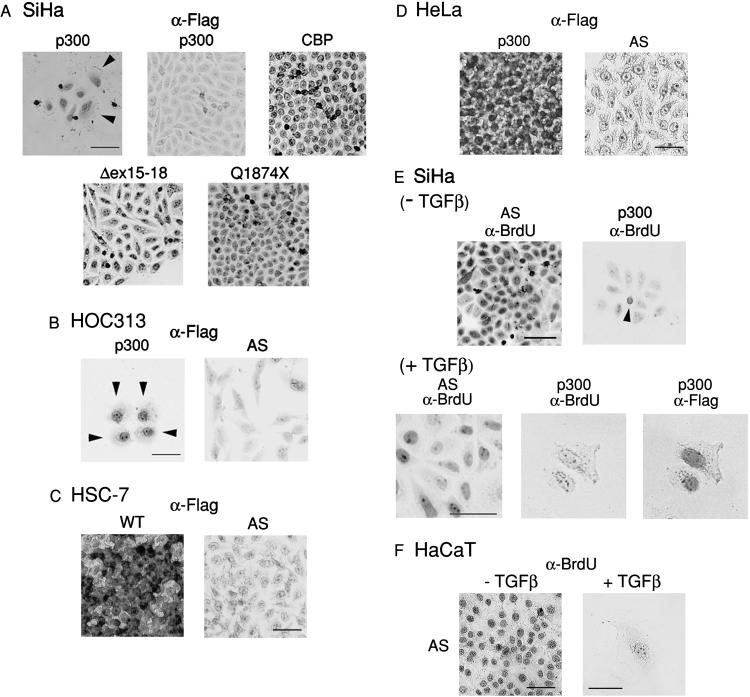
To confirm the expression of transfected Flag-tagged p300 and CBP proteins, colonies of SiHa cells were stained with α-Flag antibody. Nuclei of cells in colonies formed by cells transfected with CBP (Fig. 5A, top right) and the p300 mutants (Fig. 5A, bottom) were clearly stained with the antibody. In contrast, even though wild-type p300 transfection produced some visible G418-resistant colonies, none of such colonies were stained with α-Flag antibody, indicating that the cells in these colonies did not express wild-type p300 (Fig. 5A, top middle). Flag-positive cells transfected with wild-type p300 were retained in small populations of fewer than 20 cells (Fig. 5A, top left). The results were similar with HOC313 cells (data not shown), except that cells expressing wild-type p300 were retained in groups of fewer than 5 cells, the morphology of which was enlarged and flat compared with that of Flag-negative cells (Fig. 5B, arrowhead). In SiHa cells, Flag-positive colonies formed by wild-type p300 transfection consisted of large and flat cells similar to those seen among HOC313 cells as well as those that remained unchanged, with a morphology that resembled that of the parent cells, whereas the cells in colonies formed by CBP transfection showed no apparent morphological changes.
Figure 5.
Expression of p300 and CBP proteins and BrdUrd incorporation in stably transfected carcinoma cells. (A–D) Expression of Flag-tagged p300 or CBP proteins in stably transfected SiHa (A), HOC313 (B), HSC-7 (C), and HeLa (D). SiHa, HSC-7, HeLa (3 wk after transfection), and HOC313 cells (10 days after transfection) were stained with α-Flag antibody. Flag-positive cells were visualized by DAB staining. Arrowheads indicate large, flat cells. (E and F) BrdUrd incorporation of SiHa (E) and HaCaT (F) cells stably transfected with the indicated plasmids. Cells were cultured in the absence or presence of TGF-β1 for 3 wk (the last 24 h in the presence of BrdUrd). Colonies were fixed and stained with α-BrdUrd, followed by α-Flag antibodies. Immunostaining was visualized by using DAB. Arrowhead indicates BrdUrd-positive cell. (Bar = 100 μm.)
In HSC-7 and HeLa cells, the majority of cells (80.3 ± 5.0% for HSC-7 and 78.9 ± 11.4% for HeLa cells) in colonies formed by wild-type p300 transfection were stained with α-Flag antibody (Fig. 5 C and D). Flag-tagged p300 was localized both in the nucleus and cytoplasm in these cell lines. In addition, these colonies consisted of heterogeneous populations of cells, about 20% of which had no or very weak Flag expression, suggesting the possibility that p300 overexpression might have some growth inhibitory effect on HSC-7 and HeLa cells. However, p300 transfection did not generate any large, flat cells in HSC-7 and HeLa cells as observed in SiHa and HOC313 cells. These results indicate that, even though HeLa and HSC-7 carcinoma cells derive from the same tissues as the SiHa and HOC313 cells, the effect of p300 expression differs depending on cell lines. Thus, the growth of p300-deficient SiHa and HOC313 cells was probably not suppressed by nonspecific toxic effects of p300 expression on the transfected cells.
Taken together, we conclude that the reintroduction of p300 suppresses the colony formation of the p300-deficient carcinoma cells and that the bromodomain and Q-rich region are critical for the suppressive activity. Furthermore, the limited growth suppressive activity of CBP in the p300-deficient cells suggests that p300 plays a role distinct from CBP in the growth suppression of carcinoma cells.
Effect of TGF-β on the Proliferation of SiHa Cells Stably Transfected with p300.
To examine the effect of TGF-β on the proliferative rate of SiHa cells stably transfected with p300 in the colony formation assay, the transfected SiHa and control HaCaT cells were cultured in the presence or absence of TGF-β1 for 3 wk, and then labeled with BrdUrd for 24 h before fixing. In the presence of TGF-β1, we obtained colony numbers similar to those in the absence of TGF-β1 in SiHa cells, whereas no large colonies were formed in HaCaT cells transfected with the control vector in the presence of TGF-β1 (data not shown). In SiHa cells, most of the cell populations (84.6%) formed by p300, which were confirmed by subsequent staining with α-Flag antibody (not shown), contained at least one BrdUrd-positive cell in the absence of TGF-β1 (Fig. 5E, right panel, arrowhead), indicating that expression of p300 is not sufficient to cause complete growth arrest in SiHa cells. Addition of TGF-β1 reduced the percentage of BrdUrd-positive populations to 15.4% in p300-transfected SiHa cells, whereas all of the colonies transfected with the control vector incorporated BrdUrd even in the presence of TGF-β1 (Fig. 5E). Moreover, under these conditions, the morphology of BrdUrd-negative cells transfected with p300 was enlarged and flat similar to HaCaT cells treated with TGF-β (Fig. 5 E and F). No morphological changes were evident in Flag-negative cells or in those transfected with the control vector, p300 mutants, or CBP, which continued to incorporate BrdUrd in the presence of TGF-β1 (Fig. 5E; data not shown). These results demonstrated that p300 can suppress the proliferation of p300-deficient carcinoma cells in the absence of TGF-β and that p300 potentiates the growth-suppressing effects of TGF-β. However, BrdUrd incorporation assays in transient transfected SiHa cells revealed that transfection of p300 moderately inhibited BrdUrd incorporation (45%) and that addition of TGF-β had only a slight effect on the inhibition (2 days after transfection; data not shown), indicating that this effect become apparent in stably transfected cells (3 wk after transfection). SiHa cells contain integrated human papillomavirus type 16 DNA and express transcripts encoding E6 and E7 oncoproteins, inhibition of which has been shown to cause antiproliferative effects on SiHa cells (28, 29). Because E7 leads to the abrogation of a variety of growth inhibitory signals, including TGF-β-mediated growth suppression (30), expression of this protein may contribute to the delay in antiproliferative response to TGF-β in p300-transfected SiHa cells.
Discussion
The present study demonstrates that reintroduction of p300 leads to growth suppression of p300-deficient carcinoma cell lines, providing experimental evidence that p300 acts as a suppressor of tumor cell growth. Given the fact that total levels of p300 and CBP in the cell are critical for embryogenesis, one might argue that the growth suppression of carcinoma cells by p300 overexpression might be caused by nonspecific cytostatic or cytotoxic effect on the transfected cells. However, we show that introduction of p300 did not suppress the growth of the same types of carcinoma cell lines that appear to express functional p300 (Fig. 4 E and F). Therefore, it seems unlikely that the growth arrest was attributed to such nonspecific effects of p300 overexpression. Rather, our results indicate that reintroduction of normal p300 into p300-deficient cells compensates for functional defects resulting from lack of normal p300. In agreement with this, CBP is incapable of suppressing the growth of carcinoma cell lines that have no apparent alterations in CBP, except for HOC313 cells, in which CBP moderately suppressed the growth. It is possible that inappropriate amounts of CBP might partially compensate p300 functions that are defective in this cell line.
We also show that distinct functional domains of p300, the bromodomain and Q-rich region, are critical for tumor suppression and TGF-β signaling. The importance of the Q-rich region in tumorigenesis is also supported by the recent findings of similar C-terminal truncating mutants in human carcinomas (18). On the other hand, critical effects of the bromodomain deletion on p300 functions were apparent in stably (colony formation assays), but not transiently (reporter assays), transfected cells. The precise role of the bromodomain in p300 functions is unknown in vivo, but recent studies have shown that the p300 bromodomain is important for nucleosomal histone acetylation and chromatin binding activities in vitro (31, 32). Thus, the bromodomain may play a critical role in the p300-dependent transcription of endogenous genes residing in their normal chromosomal sites, which may not be precisely reflected in transient transfection assays using reporter plasmids that are not integrated in chromatin structure.
Given the multiple interactions of p300 with various transcription factors and coactivators, mutations of p300 may affect a wide range of transcriptional events. The present study provides evidence of a mechanism by which the loss of normal p300 functions contributes to epithelial tumorigenesis. Although both p300 and CBP mediate the TGF-β/Smads signaling pathway, our results indicate that p300 is essential for TGF-β transcriptional activation in epithelial cells. The TGF-β/Smads pathway is important for the negative growth regulation of epithelial cells, and it is also targeted for mutations in carcinomas. Thus, TGF-β signaling impaired by p300 mutation results in deregulated expression of TGF-β target genes important for growth suppression, such as p21CIP1 (19).
The fact that p300 can suppress tumor cell growth without TGF-β treatment suggests that p300-transfected SiHa and HOC313 cells may respond to TGF-β or TGF-β-like factors contained in the serum used to culture these cells. It is also possible that TGF-β-independent mechanisms may contribute to p300-mediated growth suppression. Despite important roles of p300/CBP in the regulation of p53 activity, the inactivation of p53 in both SiHa and HOC313 cells (28, 29, 33) indicates that the growth suppression activity of p300 is independent of p53 action. On the other hand, p300/CBP have been shown to play an important role in biological processes involved in differentiation. We show that p300 causes the morphological changes in p300-deficient cells (Fig. 5 A and B), which is similar to Rb that causes flat cell formation and induces differentiation markers in certain Rb-deficient cells (21, 34). Therefore, it is possible that p300 expression may trigger a pathway leading to the differentiation of epithelial cells, and thereby resulting in the growth suppression of p300-deficient cells. Additionally, recent studies have indicated roles of p300 in TGF-β-independent growth suppression, although the effects of p300 mutation remain to be investigated. For instance, the inhibition of p300 expression by antisense-mediated depletion of p300 causes up-regulation of c-myc and S phase induction in quiescent cells (35). In addition, p300, but not CBP, forms a complex with the SYT protooncoprotein to mediate contact inhibition (16).
Finally, accumulating evidence demonstrates that p300 and CBP have distinct as well as overlapping functions (13–16). The present study shows that CBP has limited growth-suppressive activity in p300-deficient carcinoma cells. In this regard, cell type-specific roles of p300 and CBP in tumor suppression suggest that genes critical for proliferation and differentiation differ depending on cell type. The evidence presented here, together with the identification of biallelic p300 mutations in human carcinomas (18, 19), indicates that p300 plays distinct roles in epithelial tumor suppression.
Supplementary Material
Acknowledgments
We thank R. Eckner, D. M. Livingston, R. H. Goodman, and K. Umezono for materials; K. Eto for support; K. Ohtani for useful discussion; and K. Miyazono for critical reading of the manuscript. This work was supported in part by Grants-in-Aid for Cancer Research (08264105 to M.I.) and Scientific Research (10470400 and 10877295 to M.I., and 10307043 and 08557097 to K. Eto) from the Ministry of Education, Science, Sports, and Culture of Japan and the Japan Society for the Promotion of Science, and by part of the “Ground Research for Space Utilization” promoted by National Space Development Agency of Japan and Japan Space Forum (K. Eto).
Abbreviations
- CBP
CREB-binding protein
- HAT
histone acetyltransferase
- TGF-β
transforming growth factor β
- RAR
retinoic acid receptor
- SID
Smad interaction domain
- Q-rich
glutamine-rich
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 2.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Nature (London) 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 3.Goodman R H, Smolik S. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 4.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 5.Bannister A J, Kouzarides T. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 6.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 7.Yao T P, Ku G, Zhou N, Scully R, Livingston D M. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, et al. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 9.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 10.Dhalluin C, Carlson J E, Zeng L, He C, Aggarwal A K, Zhou M M. Nature (London) 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 11.Jacobson R H, Ladurner A G, King D S, Tjian R. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 12.Oike Y, Takakura N, Hata A, Kaname T, Akizuki M, Yamaguchi Y, Yasue H, Araki K, Yamamura K, Suda T. Blood. 1999;93:2771–2779. [PubMed] [Google Scholar]
- 13.Yao T P, Oh S P, Fuchs M, Zhou N D, Ch'ng L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 14.Kawasaki H, Eckner R, Yao T P, Taira K, Chiu R, Livingston D M, Yokoyama K K. Nature (London) 1998;393:284–289. doi: 10.1038/30538. [DOI] [PubMed] [Google Scholar]
- 15.Kung A L, Rebel V I, Bronson R T, Ch'ng L E, Sieff C A, Livingston D M, Yao T P. Genes Dev. 2000;14:272–277. [PMC free article] [PubMed] [Google Scholar]
- 16.Eid J E, Kung A L, Scully R, Livingston D M. Cell. 2000;102:839–848. doi: 10.1016/s0092-8674(00)00072-6. [DOI] [PubMed] [Google Scholar]
- 17.Muraoka M, Konishi M, Kikuchi-Yanoshita R, Tanaka K, Shitara N, Chong J M, Iwama T, Miyaki M. Oncogene. 1996;12:1565–1569. [PubMed] [Google Scholar]
- 18.Gayther S A, Batley S J, Linger L, Bannister A, Thorpe K, Chin S F, Daigo Y, Russell P, Wilson A, Sowter H M, et al. Nat Genet. 2000;24:300–303. doi: 10.1038/73536. [DOI] [PubMed] [Google Scholar]
- 19.Ohshima T, Suganuma T, Ikeda M A. Biochem Biophys Res Commun. 2001;281:569–575. doi: 10.1006/bbrc.2001.4389. [DOI] [PubMed] [Google Scholar]
- 20.Ikeda M A, Jakoi L, Nevins J R. Proc Natl Acad Sci USA. 1996;93:3215–3220. doi: 10.1073/pnas.93.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin X Q, Chittenden T, Livingston D M, Kaelin W G., Jr Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 22.Massagué J, Blain S W, Lo R S. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 23.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier J M. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wieser R, Wrana J L, Massagué J. EMBO J. 1995;14:2199–2208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pouponnot C, Jayaraman L, Massagué J. J Biol Chem. 1998;273:22865–22868. doi: 10.1074/jbc.273.36.22865. [DOI] [PubMed] [Google Scholar]
- 26.Cárcamo J, Zentella A, Massagué J. Mol Cell Biol. 1995;15:1573–1581. doi: 10.1128/mcb.15.3.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishihara A, Hanai J, Imamura T, Miyazono K, Kawabata M. J Biol Chem. 1999;274:28716–28723. doi: 10.1074/jbc.274.40.28716. [DOI] [PubMed] [Google Scholar]
- 28.Hamada K, Sakaue M, Alemany R, Zhang W W, Horio Y, Roth J A, Mitchell M F. Gynecol Oncol. 1996;63:219–227. doi: 10.1006/gyno.1996.0310. [DOI] [PubMed] [Google Scholar]
- 29.Venturini F, Braspenning J, Homann M, Gissmann L, Sczakiel G. Nucleic Acids Res. 1999;27:1585–1592. doi: 10.1093/nar/27.7.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demers G W, Espling E, Harry J B, Etscheid B G, Galloway D A. J Virol. 1996;70:6862–6869. doi: 10.1128/jvi.70.10.6862-6869.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraus W L, Manning E T, Kadonaga J T. Mol Cell Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manning E T, Ikehara T, Ito T, Kadonaga J T, Kraus W L. Mol Cell Biol. 2001;21:3876–3887. doi: 10.1128/MCB.21.12.3876-3887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakai E, Tsuchida N. Oncogene. 1992;7:927–933. [PubMed] [Google Scholar]
- 34.Sellers W R, Novitch B G, Miyake S, Heith A, Otterson G A, Kaye F J, Lassar A B, Kaelin W G. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolli S, Buchmann A M, Williams J, Weitzman S, Thimmapaya B. Proc Natl Acad Sci USA. 2001;98:4646–4651. doi: 10.1073/pnas.081141998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.