Abstract
Our discovery of dominant-negative inhibition of prion formation in cultured cells provided an explanation for the resistance of some sheep to scrapie and humans to Creutzfeldt–Jakob disease. To determine whether dominant-negative inhibition occurs in vivo, we produced transgenic (Tg) mice expressing prion protein (PrP) with either the Q167R or Q218K mutation alone or in combination with wild-type (wt) PrP. Tg(MoPrP,Q167R)Prnp0/0 mice expressing mutant PrP at levels equal to non-Tg mice remained healthy for >550 days, indicating that inoculation with prions did not cause disease. Immunoblots of brain homogenates and histologic analysis did not reveal abnormalities. Tg(MoPrP,Q167R)Prnp+/+ mice expressing both mutant and wt PrP did not exhibit neurologic dysfunction, but their brains revealed low levels of the PrP pathogenic isoform (PrPSc), and sections showed numerous vacuoles and severe astrocytic gliosis at 300 days after inoculation. Both Tg(MoPrP,Q218K)Prnp0/0 and Tg(MoPrP,Q218K)Prnp+/+ mice expressing high levels of the transgene product remained healthy for >300 days after inoculation. Neither PrPSc nor neuropathologic changes were found. Our studies demonstrate that although dominant-negative inhibition of wt PrPSc formation occurs, expression of the dominant-negative PrP at the same level as wt PrP does not prevent prion formation completely. However, expression of dominant-negative PrP alone had no deleterious effects on the mice and did not support prion propagation.
In prion diseases, the aberrantly folded isoform (PrPSc) of the normal, cellular prion protein (PrPC) stimulates the conversion of PrPC into nascent PrPSc. The accumulation of PrPSc leads to CNS dysfunction and neuronal degeneration (1). At present, there is no accepted therapy for prion diseases, and whether the drug quinacrine will prove to be effective in treating these diseases remains to be established (2). In addition to quinacrine (3), other compounds that block prion replication as well as stimulate the clearance of existing prions include branched polyamines (4), phthalocyanines and porphyrin derivatives (5), Congo red (6), compound 60 (7), β-breaker peptides (8), and anti-PrP antibodies (9, 10). Although many of the foregoing compounds are able to clear prions in scrapie-infected neuroblastoma cells, none have been shown to be effective in animals or humans to date (11). It is noteworthy that both vaccination and passive immunization have effectively decreased Aβ amyloid deposits in the brains of transgenic (Tg) mice expressing mutant amyloid precursor protein (12–14). Unfortunately, some attempts to vaccinate humans with Aβ have resulted in an allergic meningoencephalitis, which halted a clinical trial (15).
One compound noted above was designed to mimic dominant-negative inhibition of prion replication (7). Dominant-negative inhibition occurs when the product of the mutant or variant allele interferes with a function of the wild-type (wt) allelic protein. Naturally occurring polymorphic variants of PrP, Q171R and E219K, known to render sheep and humans resistant to scrapie and Creutzfeldt–Jakob disease, respectively (16–18), were found to act as dominant negatives in scrapie-infected neuroblastoma cells (19, 20).
Based on these findings, we undertook studies on dominant-negative PrP. Tg mice expressing mutant PrP with either Q167R or Q218K or coexpressing mutant and wt PrP were inoculated with Rocky Mountain Laboratory (RML) prions, and incubation times were determined. We found that expression of dominant-negative PrP at the same level as wt PrP dramatically slowed PrPSc formation. Moreover, dominant-negative PrP was not converted into PrPSc, and its expression, even at high levels, had no deleterious effects on the mice.
Materials and Methods
Nomenclature.
Residue 171 in sheep PrP corresponds to codon 167 in mouse PrP (MoPrP) and codon 168 in human PrP (HuPrP). Residue 219 in HuPrP corresponds to codon 218 in MoPrP.
Laboratory Animals and Inoculum.
We obtained wt FVB mice from Charles River Breeding Laboratories. Tg(MoPrP-A)4053 mice have been described (21). MoPrP,Q167R and MoPrP,Q218K mutant genes were subcloned into the cosTet vector for microinjection (19). Tg(MoPrP,Q167R)Prnp0/0 and Tg(MoPrP,Q218K)Prnp0/0 founder mice were identified by PCR screening for transgene integration by using a Beckman robotic workstation.
Tg(MoPrP,Q167R)Prnp+/+ and Tg(MoPrP,Q218K)Prnp+/+ mice were produced by repeated back-crossing of Tg(MoPrP,Q167R)Prnp0/0 and Tg(MoPrP,Q218K)Prnp0/0 mice, respectively, with FVB mice until we obtained the third (F3) generation. In these mice, both the transgene and wt Prnp genes were identified by PCR screening.
The RML prion inoculum was as described (22).
Determination of Incubation Periods.
Control and Tg mice were inoculated intracerebrally with 30 μl of a 1% RML preparation or 10% brain homogenate prepared in PBS. Beginning 50 days after inoculation, the mice were monitored daily, and the neurologic status was assessed semiweekly as described (23). Mice scored positively for prion disease when two or three signs of neurologic dysfunction were present and progressive deterioration was apparent according to 16 diagnostic criteria as described (24, 25).
Antibodies.
Two chimeric human–mouse (HuM) recombinant antibody fragments (recFab), D13 and R1, that recognize PrP(97–106) and PrP(225–231), respectively, were used for immunoblot analysis (9, 26).
Preparation of Brain Homogenates and Immunoblot Analysis.
We prepared 10% brain homogenates in PBS by using a 5-ml syringe coupled to gauge needles of decreasing diameters and by repeated suctions and extrusions of the solution. After the solution was centrifuged for 5 min at 2,000 rpm in a Beckman centrifuge, the supernatant was collected. Protein concentration was measured with the bicinchoninic acid reagent (BCA, Pierce) and corresponds to 10 mg/ml. Volumes (500 μl) of 1% homogenates were prepared in PBS and 2% Sarkosyl and digested with 20 μg/ml of proteinase K (PK) at a ratio of 1:50 (PK/protein) for 1 h at 37°C. The digestion was stopped with 5 mM phenylmethylsulfonyl fluoride. PK-treated samples were mixed with an equal volume of SDS-loading buffer and boiled for 5 min, and 30 μl were loaded on 12% SDS/PAGE precast gels (Bio-Rad). Undigested samples were prepared by mixing an aliquot of 10% homogenate with an equal volume of SDS-loading buffer and then boiled for 5 min. Prepared samples (10 μl) were loaded on 12% SDS/PAGE precast gels. Immunoblot analysis was performed according to a protocol described previously (28).
Quantification of PrPSc by Conformation-Dependent Immunoassay (CDI).
To detect levels of PrPSc, an immunoassay was performed on Syrian hamster PrP according to a technique described previously (29).
Preparation of the Calibration Curve.
Brains from normal or RML-inoculated FVB mice were resuspended to 5% solution, in PBS and 2% Sarkosyl, as described above. Fivefold serial dilutions were performed by diluting the 5% RML-FVB brain homogenate with 5% normal FVB brain homogenate. Ten-point dilutions were completed starting at 1% and continuing with 0.2, 0.04, 0.008%, etc. Then, 1-ml aliquots were digested with 25 μg/ml PK at a 1:200 ratio (PK/protein) for 1 h at 37°C on a shaker. The reaction was stopped by a mixture of protease inhibitors; samples were precipitated with sodium phosphotungstate and processed as described in ref. 29. The samples were quantified by CDI using time-resolved fluorescence spectroscopy (29). The europium-labeled HuM-D13 Fab was used to detect mutant and wt MoPrP.
Preparation of Modified MoPrP.
The 10% brain homogenates previously analyzed by immunoblots were tested also by CDI. Homogenates were diluted to 5% solution in PBS/2% Sarkosyl and rehomogenized with a syringe to break aggregates. Aliquots of 1 ml were digested with 25 μg/ml PK for 1 h at 37°C on a shaker. The samples were processed as described above.
Neuropathology.
Brain tissue was immersion-fixed in 10% buffered formalin immediately after the mice were killed. Histological sections were prepared and stained with hematoxylin/eosin as described (30).
Histoblots.
Histoblots were performed as described in ref. 31.
Results
RML Prions in Tg(MoPrP,Q167R) Mice.
To study the effect of Q167R on prion formation, we constructed Tg mice expressing mutated MoPrP(Q167R) on the PrP-deficient (Prnp0/0) background. These mice, designated Tg(MoPrP,Q167R)Prnp0/0, express PrP at the same levels as FVB mice and were inoculated with RML prions. None of the 12 inoculated mice showed signs of disease after 550 days (Table 1). Uninoculated Tg(MoPrP,Q167R)Prnp0/0 mice showed no signs of spontaneous neurodegeneration. FVB control mice developed signs of CNS dysfunction at 127 ± 1 days after inoculation. Prnp0/0 mice inoculated with RML prions did not show signs of disease after >550 days (Table 1).
Table 1.
Susceptibility of Tg(MoPrP,Q167R) mice to RML prions
| Host | PrP expression level*
|
Inoculum | Incubation period, days ± SEM | n/n0† | |
|---|---|---|---|---|---|
| Mutant | wt | ||||
| FVB | 0 | 1× | RML | 127 ± 1 | 50/50 |
| Tg(MoPrP,Q167R)Prnp0/0 | 1× | 0 | RML | >550 | 0/8 |
| Tg(MoPrP,Q167R)Prnp0/0 | 1× | 0 | RML | >557 | 0/4 |
| Tg(MoPrP,Q167R)Prnp0/0 | 1× | 0 | None | >557 | 0/10 |
| FVB/Prnp0/0 | 0 | 0 | RML | >557 | 0/7 |
| Tg(MoPrP,Q167R)Prnp+/+ | 1× | 1× | RML | 447 ± 11 | 7/10‡ |
| Tg(MoPrP,Q167R)Prnp+/+ | 1× | 1× | None | >256 | 0/6 |
Expression levels were determined by comparing serial dilutions of Tg mouse brain homogenates to that of normal FVB mice (1 × PrP level) by immunoblot.
n, number of sick mice; n0, number of inoculated mice.
Seven animals died atypically of prolapsus at the mean incubation time indicated; three healthy animals were killed at 300 days for Western blot and histoblot analyses.
Tg(MoPrP,Q167R)Prnp+/+ mice express the same levels of both wt MoPrPC and mutant MoPrP(Q167R). Seven of the 10 Tg(MoPrP,Q167R)Prnp+/+ mice showed signs of CNS dysfunction at more than 400 days after inoculation with prions.
Immunoblot Analysis of Tg(MoPrP,Q167R) Mice.
Tg(MoPrP,Q167R)-Prnp0/0 mice that remained healthy were killed for immunoblot analysis to determine the level of PrP transgene expression (Fig. 1a). Equal amounts of protein were loaded on the gel as judged by immunoblotting of glyceraldehyde 3-phosphate dehydrogenase.
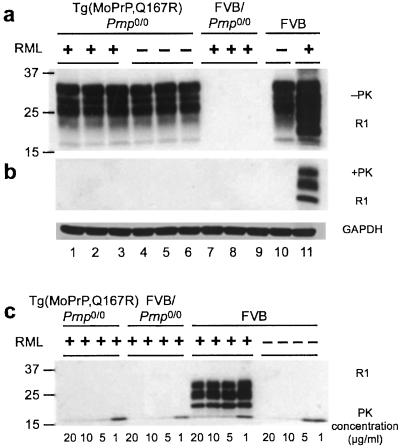
Figure 1.
Immunoblots of Tg(MoPrP,Q167R)Prnp0/0 mice. (a and b) Brain homogenates were analyzed before (a) and after (b) PK digestion. The plus (+) and minus (−) signs indicate that the mice were and were not inoculated, respectively, with RML prions. Lanes 1–6, six independent Tg(MoPrP,Q167R)Prnp0/0 mice killed after 550 (lanes 1–3) and 420 (lanes 4–6) days; lanes 7–9, three FVB/Prnp0/0 mice killed after 420 days; lane 10, normal, wt FVB mouse; lane 11, wt FVB mouse inoculated with prions and sick after ≈120 days. (a) In each lane, 50 μg of 10% brain homogenate was loaded. GAPDH, glyceraldehyde 3-phosphate dehydrogenase. (c) PK digestion of brain homogenates from Tg(MoPrP,Q167R)Prnp0/0 and wt FVB mice. The PK concentrations used are indicated. All membranes were probed with HuM-R1 Fab at a final concentration of 1 μg/ml. A secondary antibody coupled with horseradish peroxidase was also diluted to a final concentration of 1 μg/ml. Blots were developed with the enhanced chemiluminescence (ECL) kit (Amersham Pharmacia). The numbers to the left of each blot indicate the molecular mass of protein standards in kDa.
Brain homogenates from FVB mice inoculated with RML prions were subjected to limited proteolysis; these samples gave a strong protease-resistant PrPSc signal (Fig. 1b, lane 11). PK-digested samples from inoculated Tg(MoPrP,Q167R)Prnp0/0 (Fig. 1b, lanes 1–3), uninoculated Tg(MoPrP,Q167R)Prnp0/0 (Fig. 1b, lanes 4–6), inoculated Prnp0/0 (Fig. 1b, lanes 7–9), and uninoculated FVB mice (Fig. 1b, lane 10) showed no detectable PrPSc. Prolonged exposure of the film, up to 15 min, did not reveal a PrPSc signal. A second Western blot was probed with HuM-D13 Fab and showed no PrPSc in Tg(MoPrP,Q167R)Prnp0/0 mice inoculated with prions (data not shown).
Because the Q167R mutation might render PrPSc more sensitive to PK-catalyzed hydrolysis, we digested the samples with PK concentrations ranging from 1 to 20 μg/ml (Fig. 1c). Brains from FVB mice inoculated with RML prions showed an increased PrPSc signal with lower PK concentrations; however, no PrPSc band was present in the brain extracts of both inoculated and uninoculated Tg(MoPrP,Q167R)Prnp0/0 mice even at 1 μg/ml PK. Based on these immunoblots and the lack of neuropathology in Tg(MoPrP,Q167R)Prnp0/0 mice, we conclude that MoPrP(Q167R) is unable to support PrPSc formation.
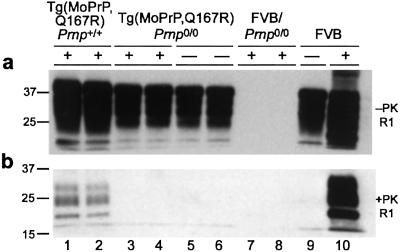
None of the 10 Tg(MoPrP,Q167R)Prnp+/+ mice, which express same levels of both wt and mutant MoPrP, showed signs of CNS dysfunction more than 400 days after inoculation with prions (Table 1). To determine whether wt MoPrPC in Tg(MoPrP,Q167R)-Prnp+/+ mice supported prion replication, we killed two apparently healthy Tg(MoPrP,Q167R)Prnp+/+ mice 300 days after inoculation and performed immunoblot analysis (Fig. 2). PK-digested homogenates revealed the presence of low amounts of protease-resistant PrP, with similar intensity (Fig. 2b, lanes 1 and 2). The intensity of these bands corresponds to ≈10% of the signal obtained with FVB mice inoculated with prions (Fig. 2b, lane 10), arguing for a diminished rate of PrPSc formation. The remaining seven inoculated Tg(MoPrP,Q167R)Prnp+/+ mice presented atypical neurologic signs with a mean incubation time of 447 ± 11 days. Because we detected some wt PrPSc replication in the brains of these mice at 300 days, it seems likely that further prion replication occurred over the subsequent 150 days. The additional presumed replication of prions and neuropathology described below argue that these mice died of prion disease, but the cause of death remains to be established.
Figure 2.
Immunoblots of Tg(MoPrP,Q167R) mice. (a and b) Brain homogenates were analyzed by immunoblot before (a) and after (b) PK digestion. The plus (+) and minus (−) signs indicate that the mice were and were not inoculated, respectively, with RML prions. Lanes 1 and 2, two independent Tg(MoPrP,Q167R)Prnp+/+ mice killed after 300 days; lanes 3–6, Tg(MoPrP,Q167R)Prnp0/0 mice killed after 550 (lanes 3 and 4) and 420 (lanes 5 and 6) days; lanes 7 and 8, FVB/Prnp0/0 mice killed after 420 days; lane 9, normal, wt FVB mouse; lane 10, wt FVB mouse that became ill after ≈120 days. Membranes were probed with HuM-R1 Fab at a final concentration of 1 μg/ml.
Neuropathology of Tg(MoPrP,Q167R) Mice.
Pathological studies were performed on the midbrains of the Tg(MoPrP,Q167R) mice analyzed by immunoblotting (Figs. 1 and 2). Both inoculated and uninoculated Tg(MoPrP,Q167R)Prnp0/0 mice showed no vacuolation in the hippocampus (Fig. 5 e and g, which is published as supporting information on the PNAS web site, www.pnas.org) but showed minimal astrocytic gliosis, which is consistent with aging (compare with Fig. 5 f and h).
In contrast, hippocampal sections from Tg(MoPrP,Q167R)-Prnp+/+ mice revealed widespread vacuolation associated with severe astrocytic gliosis at 300 days after inoculation (Fig. 5 i and j). The severity of the neuropathologic changes resembled that of prion-inoculated FVB mice (Fig. 5 a and b). However, neurodegeneration was localized in the ventral hippocampus in Tg(MoPrP,Q167R)Prnp+/+ mice, whereas both the ventral and dorsal hippocampus of FVB mice were affected, with the most intense changes in the dorsal area. Histopathologic analyses of uninoculated Tg(MoPrP,Q167R)Prnp+/+ mice (Fig. 5 k and l) resembled those of uninoculated FVB mice (Fig. 5 c and d). Our findings show an excellent correlation between Western blot and neuropathological analysis; the presence of PrPSc was accompanied by vacuolation and astrocytic gliosis.
Localization of PrPSc in the Brains of Tg(MoPrP,Q167R) Mice.
Histoblotting demonstrated widespread intense staining of the brains of RML-inoculated FVB mice with clinical signs of neurologic dysfunction (Fig. 6a, which is published as supporting information on the PNAS web site). Histoblots of the brains of Tg(MoPrP,Q167R)Prnp0/0 mice inoculated with prions (Fig. 6b) were indistinguishable from uninoculated controls (Fig. 6c) and inoculated FVB/Prnp0/0 mice (Fig. 6d). Minimal PrPSc deposits, localized primarily to the ventral hippocampus, were found in the brains of inoculated Tg(MoPrP,Q167R)Prnp+/+ mice (Fig. 6e).
CDI of Tg(MoPrP,Q167R) Brain Homogenates.
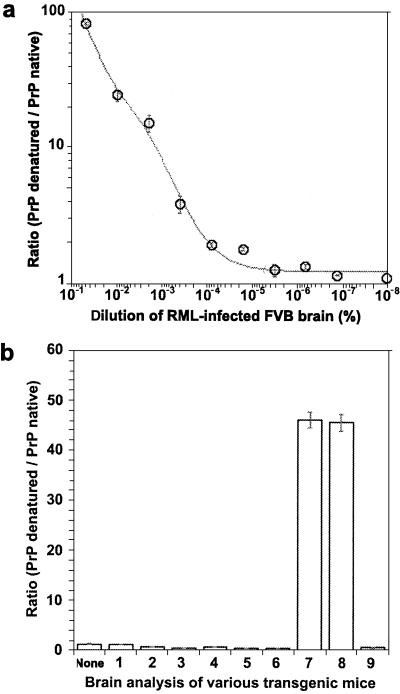
To quantify the PrPSc detected in Western blots, we used the CDI to measure both protease-resistant and protease-sensitive PrPSc (29). The CDI uses time-resolved fluorescence to measure the binding of antibodies to PrP before and after denaturation. The epitope for antibody binding is typically exposed in native (N) PrPC, denatured (D) PrPC, and denatured PrPSc but buried in native, infectious PrPSc. To establish a calibration curve for MoPrPSc, brain homogenates were prepared from FVB mice infected with RML prions (Fig. 3a). A linear relationship between PrPSc level and a brain dilution up to 3.2 × 10−5 was demonstrated. Whether MoPrPSc was detected after a dilution of 10−6, which corresponds to a D/N ratio of 1.25 ± 0.04, is unclear. Control brain homogenates from uninoculated FVB mice gave a D/N ratio of 1.1 ± 0.04 (Fig. 3b) in contrast to a value of 1.67 for Syrian hamster brains (29).
Figure 3.
CDI analysis of Tg(MoPrP,Q167R) mice. (a) Calibration curve performed on brain extracts of inoculated FVB mice. The europium-labeled HuM-D13 Fab was used for the detection of PrP. (b) CDI analysis of various brain homogenates. None, uninoculated wt FVB mice; 1 and 2, two prion-inoculated Tg(MoPrP,Q167R)Prnp0/0 mice killed after 550 days; 3 and 4, two uninoculated Tg(MoPrP,Q167R)Prnp0/0 mice killed after 420 days; 5 and 6, prion-inoculated FVB/Prnp0/0 mice killed after 420 days; 7 and 8, prion-inoculated Tg(MoPrP,Q167R)Prnp+/+ mice killed after 300 days; 9, an uninoculated Tg(MoPrP,Q167R)Prnp+/+ mouse killed after 150 days. Data points and bars are average ± SEM obtained from three independent measurements.
Brain homogenates (1%, wt/vol) prepared from RML-inoculated FVB mice gave a D/N ratio of 82 ± 2.3. We prepared 5% brain homogenates from Tg mice to detect potential traces of PrPSc. Using the CDI, we detected PrPSc only in homogenates from RML-inoculated Tg(MoPrP,Q167R)Prnp+/+ mice, with samples from two animals giving D/N ratios of 46 ± 1.5 and 45 ± 1.6 (Fig. 3b), which correspond to 0.05 μg/ml PrPSc. These D/N values for Tg(MoPrP,Q167R)Prnp+/+ mice were measured in homogenates that were five times more concentrated than those from FVB mice, therefore equivalent 1% homogenates would give values of 9.2 ± 0.3 and 9.0 ± 0.32. Comparing D/N values, we have ≈9 times less PrPSc in Tg(MoPrP,Q167R)Prnp+/+ than in normal FVB mice, a finding that correlates with Western blot analysis (Fig. 2b, lanes 1 and 2) and histoblots (Fig. 6e). In contrast, uninoculated Tg(MoPrP,Q167R)Prnp+/+ mice as well as inoculated Tg(MoPrP,Q167R)Prnp0/0 and Prnp0/0 mice gave D/N ratios between 0.58 ± 0.04 and 1.2 ± 0.01, indicating the absence of PrPSc.
RML Prions in Tg(MoPrP,Q218K) Mice.
Because dominant-negative inhibition of prion replication in Tg(MoPrP,Q167R)Prnp+/+ mice was incomplete, we produced mice expressing high levels of MoPrP(Q218K). Two lines were established, Tg(MoPrP,Q218K)-22500/Prnp0/0 and Tg(MoPrP,Q218K)21603/Prnp0/0, that express mutant PrP at levels 32 and 16 times that of FVB mice, respectively. Additionally, we produced Tg(MoPrP,Q218K)22500/Prnp+/+ and Tg(MoPrP,Q218K)21603/Prnp+/+ mice, which overexpress mutant PrP and express wt PrP at 1× level. We inoculated these mice with RML prions.
Of 16 inoculated Tg(MoPrP,Q218K)22500/Prnp0/0 mice, 12 showed signs of disease with a mean incubation time of 476 ± 31 days. It is likely that these animals developed a spontaneous disease because 6 of 10 uninoculated mice also died with a mean survival time of 442 ± 49 days (Table 2). This result is not surprising because of the high expression level (32×) of the transgene; high levels of PrP expression have been reported to cause disease (32).
Table 2.
Susceptibility of Tg(MoPrP,Q218K) mice to RML prions
| Host | PrP expression level*
|
Inoculum | Incubation period, days ± SEM | n/n0† | |
|---|---|---|---|---|---|
| Mutant | wt | ||||
| Tg(MoPrP-A)4053/Prnp0/0 | 0 | 8× | RML | 50 ± 2 | 16/16 |
| FVB/Prnp0/0 | 0 | 0 | RML | >550 | 0/6 |
| Tg(MoPrP,Q218K)22500/Prnp0/0 | 32× | 0 | RML | 476 ± 31 | 12/16 |
| Tg(MoPrP,Q218K)22500/Prnp0/0 | 32× | 0 | None | 442 ± 49 | 6/10 |
| Tg(MoPrP,Q218K)22500/Prnp+/+ | 32× | 1× | RML | 374 ± 24 | 5/8 |
| Tg(MoPrP,Q218K)22500/Prnp+/+ | 32× | 1× | None | >376 | 1/6‡ |
| Tg(MoPrP,Q218K)21603/Prnp0/0 | 16× | 0 | RML | >550 | 0/8 |
| Tg(MoPrP,Q218K)21603/Prnp0/0 | 16× | 0 | RML | >468 | 0/10 |
| Tg(MoPrP,Q218K)21603/Prnp0/0 | 16× | 0 | None | >481 | 0/10 |
| Tg(MoPrP,Q218K)21603/Prnp+/+ | 16× | 1× | RML | >508 | 5/9§ |
| Tg(MoPrP,Q218K)21603/Prnp+/+ | 16× | 1× | None | >438 | 0/10 |
Expression levels were determined as described for Table 1.
n, number of sick mice; n0, number of inoculated mice.
One animal got sick at 319 days, and two animals presented signs of ataxia at 370 days.
The incubation period of the five sick mice was 412 ± 18 days.
Similar results were obtained with Tg(MoPrP,Q218K)20250/Prnp0/0 mice, which express mutant PrP at 32× level. All inoculated mice developed CNS dysfunction after 315 ± 33 days, whereas uninoculated mice showed clinical signs after 322 ± 30 days. Neuropathological examination revealed numerous vacuoles and pronounced astrocytic gliosis in both inoculated and uninoculated animals, and neither group had PrPSc based on immunoblots (data not shown). These findings argue that Tg(MoPrP,Q218K)22500/Prnp0/0 mice develop spontaneous neurologic disease but are resistant to infection by RML prions.
Five of eight Tg(MoPrP,Q218K)22500/Prnp+/+ mice inoculated with prions became ill, with a mean incubation time of 374 ± 24 days. It is possible that these mice also developed spontaneous neurodegeneration because noninoculated controls became ill, with one animal dying at 319 days and two mice still living but presenting signs of ataxia at 370 days (Table 2).
In contrast, Tg(MoPrP,Q218K)21603/Prnp0/0 mice that express lower levels of PrP did not develop spontaneous disease. None of the 18 inoculated Tg(MoPrP,Q218K)21603/Prnp0/0 mice showed signs of disease after more than 468 and 550 days, which is remarkable because of the 16× expression level. Five of nine Tg(MoPrP,Q218K)21603/Prnp+/+ mice inoculated with prions developed disease with a mean incubation time of 412 ± 18 days. By comparison with Tg(MoPrP-A)4053 mice, Tg(MoPrP,Q218K)-21603/Prnp+/+ mice exhibited a prolonged incubation time by a factor of approximately 8. Tg(MoPrP-A)4053 mice, expressing wt PrP at 8×, developed disease at 50 days after inoculation with RML prions (Table 2). Currently, two Tg(MoPrP,Q218K)21603/Prnp+/+ mice remain healthy after 508 days, as is true for all uninoculated mice from this Tg line (Table 2).
Immunoblots of Tg(MoPrP,Q218K) Mice.
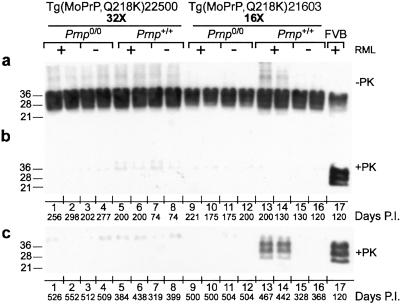
To distinguish between neurodegeneration caused by prions and that caused by misprocessing of the transgene product, we performed immunoblotting on brain homogenates of Tg(MoPrP,Q218K)22500/Prnp0/0 and Tg(MoPrP,Q218K)21603/Prnp0/0 mice. We were interested in whether Tg(MoPrP,Q218K)22500/Prnp+/+ and Tg(MoPrP,Q218K)21603/Prnp+/+ mice were able to form nascent wt PrPSc as seen in Tg(MoPrP,Q167R)Prnp+/+ mice. For each experimental group in Table 2, two animals were killed when they were young, and immunoblots were performed on their brain homogenates before and after PK digestion (Fig. 4 a and b). None of the samples from young mice with the Prnp0/0 or Prnp+/+ background showed detectable levels of PrPSc regardless of whether the animals were inoculated with RML prions (Fig. 4b). These samples were tested by CDI, and resulting D/N ratios were always below the 1.67 cut-off value for PrPSc (data not shown). Additional samples from Tg mice were taken between 319 and 552 days after inoculation for analysis. Protease-digested brain homogenates from ill Tg(MoPrP,Q218K)21603/Prnp+/+ mice revealed low amounts of PrPSc (Fig. 4c, lanes 13 and 14), whereas PK-digested samples from ill Tg(MoPrP,Q218K)22500/Prnp+/+ mice did not (Fig. 4c, lanes 5 and 6).
Figure 4.
Immunoblots of inoculated Tg(MoPrP,Q218K) mice. Brain homogenates were analyzed before (a) and after (b and c) limited PK digestion. Plus (+) and minus (−) signs indicate that mice were and were not inoculated, respectively, with RML prions. Membranes were probed with HuM-D13 Fab at a final concentration of 1 μg/ml. The numbers to the left of each blot indicate the molecular mass of protein standards in kDa. The number of days elapsed before the animal became ill or was killed is indicated as Days P.I. (a and b) Brain homogenates of mice at ≈200 days. Lanes 1–4, Tg(MoPrP,Q218K)22500/Prnp0/0 mice that fell ill (lanes 1 and 3) or remained healthy and were killed (lanes 2 and 4); lanes 5–8, healthy Tg(MoPrP,Q218K)22500/Prnp+/+ mice killed at the days indicated; lanes 9–12, healthy Tg(MoPrP,Q218K)21603/Prnp0/0 mice killed at the days indicated; lanes 13–16, healthy Tg(MoPrP,Q218K)21603/Prnp+/+ mice killed at the days indicated; lane 17, inoculated FVB mouse that died after ≈120 days. (c) Brain homogenates of Tg(MoPrP,Q218K) mice at ≈500 days. Lanes 1–4, ill Tg(MoPrP,Q218K)22500/Prnp0/0 mice; lanes 5–8, ill Tg(MoPrP,Q218K)22500/Prnp+/+ mice; lanes 9–12, healthy Tg(MoPrP,Q218K)21603/Prnp0/0 mice killed at the days indicated; lanes 13–16, Tg(MoPrP,Q218K)21603/Prnp+/+ mice that either fell ill (lanes 13 and 14) or were killed while still healthy (lanes 15 and 16); lane 17, inoculated FVB mouse that died after ≈120 days.
From these findings, we conclude that inoculated Tg(MoPrP,Q218K)22500/Prnp+/+ mice developed CNS dysfunction not from prion accumulation but from overexpression of the transgene product. The presence of low but readily detectable amounts of PrPSc in inoculated, ill Tg(MoPrP,Q218K)21603/Prnp+/+ mice raises the possibility that these mice developed prion disease. Presumably the lower level of MoPrP(Q218K) expression in Tg(MoPrP,Q218K)21603/Prnp+/+ mice compared with Tg(MoPrP,Q218K)22500/Prnp+/+ mice permitted prion replication to occur. Why the levels of PrPSc are not higher in the ill Tg(MoPrP,Q218K)21603/Prnp+/+ mice is unknown. One argument is that these mice died of brain degeneration caused by the accumulation of the transgene product, but none of the other Tg(MoPrP,Q218K)21603 mice developed CNS illness (Table 2). Alternatively, relatively low levels of PrPSc might have caused illness or the combined presence of PrPSc and mutant MoPrP(Q218K) produced disease.
Discussion
The discovery of dominant-negative inhibition of prion replication began with the finding that chimeric HuM PrP transgenes rendered mice susceptible to human prions (33, 34). Using scrapie-infected neuroblastoma cells, we mapped the residues on PrPC that are critical for dominant-negative inhibition of prion replication (19). The side chains of these residues on the surface of PrPC form a discontinuous epitope near the C terminus, which is thought to bind to a macromolecule provisionally designated protein X. The avid binding of dominant-negative PrP to protein X is presumed to result in the sequestering of the protein, which in turn prevents conversion of wt PrPC into PrPSc.
Scrapie-infected neuroblastoma-cell studies and investigations of naturally occurring PrP polymorphisms in sheep and humans that protect them from prion diseases argued for dominant-negative inhibition of prion replication. Although these polymorphisms may be more effective in the context of the ovine or human sequence, we postulated that experimental testing of this hypothesis in Tg mice would allow better characterization of the protective effects of dominant-negative variants. We produced mice expressing mutant MoPrP transgenes that carry either polymorphic residue protecting sheep or humans. The substitution of arginine at codon 167 or lysine at 218 rendered MoPrPC unconvertible into PrPSc. When the transgene product and wt MoPrPC were coexpressed, prion formation was retarded.
Dominant-Negative Inhibition of Scrapie.
At codon 171, both heterozygous Q/R and homozygous R/R Suffolk sheep were found to be resistant to natural scrapie (17). In studies of experimental scrapie, Cheviot sheep carrying Q/R or R/R at residue 171 were found to be resistant to prion infection (16, 35–37). Similar findings have been reported for flocks of Texel sheep with natural scrapie. Sheep expressing ARR/ARR or ARR/AHQ at positions 136, 154, and 171, respectively, were the most resistant to scrapie, with 89% of animals surviving; sheep expressing VRQ/VRQ, ARQ/VRQ, or ARH/VRQ were the most susceptible to disease, with only 5% unaffected (38, 39). These findings are readily explained by our results with Tg(MoPrP,Q167R)Prnp+/+ mice; position 167 in MoPrP corresponds to 171 in sheep PrP. Therefore, we conclude that protection of sheep expressing Q/R at position 171 from scrapie is caused by dominant-negative inhibition. Sheep expressing R/R at 171 were protected also because MoPrP(Q167R) was not converted into PrPSc even after 500 days after prion inoculation.
Dominant-Negative Inhibition of Prion Disease in Humans.
Among the Japanese population, 12% carry the E/K polymorphism at position 219, whereas remaining individuals are E/E. None of 85 autopsied sporadic Creutzfeldt–Jakob disease cases were found to be E/K, suggesting that heterozygosity at 219 protects humans from this disease (40, 41). These findings are readily explained by our results with Tg(MoPrP,Q218K)Prnp+/+ mice; position 218 in MoPrP corresponds to 219 in HuPrP. Thus, protection of humans expressing E/K at codon 219 from Creutzfeldt–Jakob disease is caused by dominant-negative inhibition.
Transgene Expressions Levels and Dominant-Negative Inhibition.
In planning the studies described here, we sought to produce one mouse line with relatively low levels of mutant PrP expression and at least one line with high levels of mutant PrP expression. Tg(MoPrP,Q167R)Prnp0/0 mice express MoPrP(Q167R) at about the same level that FVB mice express wt MoPrP. Tg(MoPrP,Q218K)21603/Prnp0/0 and Tg(MoPrP,Q218K)22500/Prnp0/0 mice express MoPrP(Q218K) at 16 and 32×, respectively. Although none of the uninoculated Tg(MoPrP,Q167R)Prnp0/0 and Tg(MoPrP,Q218K)21603/Prnp0/0 mice developed spontaneous neurologic disease, 18 of 26 Tg(MoPrP,Q218K)22500/Prnp0/0 mice developed what appears to be spontaneous neurodegeneration (Table 2).
Although Tg(MoPrP,Q167R)Prnp+/+ mice were asymptomatic at 300 days after inoculation with RML prions, their brains contained detectable levels of PrPSc (Figs. 2 and 3). Presumably, PrPSc was derived from wt PrPC, but we have no antibodies that distinguish wt PrPSc from putative PrPSc(Q167R). However, our results clearly show the inhibitory effect of MoPrP(Q167R) on the conversion of wt PrPC into PrPSc as reflected by the prolonged incubation times in Tg(MoPrP,Q167R)Prnp+/+ mice compared with FVB mice (Table 1).
Tg(MoPrP,Q218K)21603/Prnp+/+ and Tg(MoPrP,Q218K)-22500/Prnp+/+ mice reacted differently to inoculation with prions. After incubation times as long as 500 days, Tg(MoPrP,Q218K)22500/Prnp+/+ mice were unable to form nascent wt PrPSc, whereas Tg(MoPrP,Q218K)21603/Prnp+/+ mice developed CNS dysfunction at 412 ± 18 days and exhibited low levels of PrPSc (Fig. 4c). This result is most readily explained by the different levels of transgene expression. Whether Tg(MoPrP,Q218K)21603/Prnp+/+ mice became ill because of prion disease remains uncertain. One possibility is that the neurologic disease seen in these mice is caused by combined effects of PrPSc accumulation and the high level of MoPrP(Q218K) expression. Certainly MoPrP(Q218K) expression alone in Tg(MoPrP,Q218K)21603/Prnp0/0 mice does not seem sufficient to cause disease (Table 2), and low levels of wt MoPrPSc are generally not associated with clinical signs or neuropathologic changes (Fig. 4c).
Prevention of Prion Disease.
The inability of MoPrP(Q167R) and MoPrP(Q218K) to support prion replication raises the possibility of producing prion-resistant livestock that express PrP with a single amino acid substitution. Because sheep homozygous for R at position 171 already exist, breeding populations of resistant sheep is a reasonable undertaking. Presumably, this was the genetic basis of Parry's scrapie eradication program in Great Britain 40 years ago (42, 43). However, this natural protection might be restricted to certain prion strains. Recently, experimental transmission of bovine spongiform encephalopathy prions to sheep showed that animals carrying ARQ/ARQ at codon 136, 154, and 171, respectively, died after 672 days, whereas sheep harboring ARR/ARR were resistant to bovine spongiform encephalopathy prions (44). The ARR/ARR sheep showed neither neurologic signs nor PrPSc deposits in their brains. It would seem prudent to explore the utility of such an approach by inoculating Tg(ShePrP,Q171R)Prnp0/0 mice with many different prion strains. A similar approach, inoculating Tg(BoPrP,Q179R)Prnp0/0 or Tg(BoPrP,Q230K)Prnp0/0 mice with many different prion strains, might be useful in evaluating the utility of producing prion-resistant cattle. Although the introduction of a point mutation may not produce complete resistance to prion infection as disruption of the Prnp gene does (45, 46), it may prove to be more desirable. Whether disruption of the Prnp gene produces some subtle but important aberration in cattle, sheep, or pigs is unknown; moreover, such livestock may be considered unacceptable to consumers. In contrast, the introduction of a naturally occurring point mutation may be less detrimental biologically and accepted more readily by consumers. Both germ-line and somatic cell gene therapy strategies could be applied in principle.
Should single point mutations in sheep or bovine PrP transgenes prove to be ineffective with some strains, it may be prudent to construct a transgene carrying both substitutions. Whether Tg mice expressing BoPrP(Q179R,Q230K) will be healthy or develop spontaneous neurodegeneration is unknown (32). Moreover, whether such mice will be more resistant to prion disease remains to be established.
Supplementary Material
Acknowledgments
We thank Jeff Monaghan for his excellent technical support with the Tg mice screening. This work was supported by grants from the National Institutes of Health as well as a gift from the G. Harold and Leila Y. Mathers Foundation. V.P. received fellowships from the Fondation pour la Recherche Médicale and the French Alzheimer's Foundation.
Abbreviations
- PrP
prion protein
- PrPSc
PrP pathogenic isoform
- PrPC
PrP normal cellular isoform
- Tg
transgenic
- wt
wild type
- RML
Rocky Mountain Laboratory
- MoPrP
mouse PrP
- HuPrP
human PrP
- HuM
human–mouse
- PK
proteinase K
- CDI
conformation-dependent immunoassay
- N
native
- D
denatured
References
- 1.DeArmond S J, Mobley W C, DeMott D L, Barry R A, Beckstead J H, Prusiner S B. Neurology. 1987;37:1271–1280. doi: 10.1212/wnl.37.8.1271. [DOI] [PubMed] [Google Scholar]
- 2.Korth C, May B C H, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 2001;98:9836–9841. doi: 10.1073/pnas.161274798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doh-ura K, Iwaki T, Caughey B. J Virol. 2000;74:4894–4897. doi: 10.1128/jvi.74.10.4894-4897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Supattapone S, Nguyen H-O B, Cohen F E, Prusiner S B, Scott M R. Proc Natl Acad Sci USA. 1999;96:14529–14534. doi: 10.1073/pnas.96.25.14529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caughey W S, Raymond L D, Horiuchi M, Caughey B. Proc Natl Acad Sci USA. 1998;95:12117–12122. doi: 10.1073/pnas.95.21.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demaimay R, Harper J, Gordon H, Weaver D, Chesebro B, Caughey B. J Neurochem. 1998;71:2534–2541. doi: 10.1046/j.1471-4159.1998.71062534.x. [DOI] [PubMed] [Google Scholar]
- 7.Perrier V, Wallace A C, Kaneko K, Safar J, Prusiner S B, Cohen F E. Proc Natl Acad Sci USA. 2000;97:6073–6078. doi: 10.1073/pnas.97.11.6073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soto C, Kascsak R J, Saborío G P, Aucouturier P, Wisniewski T, Prelli F, Kascsak R, Mendez E, Harris D A, Ironside J, et al. Lancet. 2000;355:192–197. doi: 10.1016/s0140-6736(99)11419-3. [DOI] [PubMed] [Google Scholar]
- 9.Peretz D, Williamson R A, Kaneko K, Vergara J, Leclerc E, Schmitt-Ulms G, Mehlhorn I R, Legname G, Wormald M R, et al. Nature (London) 2001;412:739–743. doi: 10.1038/35089090. [DOI] [PubMed] [Google Scholar]
- 10.Enari M, Flechsig E, Weissmann C. Proc Natl Acad Sci USA. 2001;98:9295–9299. doi: 10.1073/pnas.151242598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Priola S A, Raines A, Caughey W S. Science. 2000;287:1503–1506. doi: 10.1126/science.287.5457.1503. [DOI] [PubMed] [Google Scholar]
- 12.Schenk D, Barbour R, Dunn W, Gordon G, Grajeda H, Guido T, Hu K, Huang J, Johnson-Wood K, Khan K, et al. Nature (London) 1999;400:173–177. doi: 10.1038/22124. [DOI] [PubMed] [Google Scholar]
- 13. Bard, F., Cannon, C., Barbour, R., Burke, R.-L., Games, D., Grajeda, H., Guido, T., Hu, K., Huang, J., Johnson-Wood, K., et al. (2000) Nat. Med. 916–919. [DOI] [PubMed]
- 14.DeMattos R B, Bales K R, Cummins D J, Dodart J C, Paul S M, Holtzman D M. Proc Natl Acad Sci USA. 2001;98:8850–8855. doi: 10.1073/pnas.151261398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birmingham K, Frantz S. Nat Med. 2002;8:199–200. doi: 10.1038/nm0302-199b. [DOI] [PubMed] [Google Scholar]
- 16.Goldmann W, Hunter N, Smith G, Foster J, Hope J. J Gen Virol. 1994;75:989–995. doi: 10.1099/0022-1317-75-5-989. [DOI] [PubMed] [Google Scholar]
- 17.Westaway D, Zuliani V, Cooper C M, Da Costa M, Neuman S, Jenny A L, Detwiler L, Prusiner S B. Genes Dev. 1994;8:959–969. doi: 10.1101/gad.8.8.959. [DOI] [PubMed] [Google Scholar]
- 18.Shibuya S, Higuchi J, Shin R-W, Tateishi J, Kitamoto T. Ann Neurol. 1998;43:826–828. doi: 10.1002/ana.410430618. [DOI] [PubMed] [Google Scholar]
- 19.Kaneko K, Zulianello L, Scott M, Cooper C M, Wallace A C, James T L, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1997;94:10069–10074. doi: 10.1073/pnas.94.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zulianello L, Kaneko K, Scott M, Erpel S, Han D, Cohen F E, Prusiner S B. J Virol. 2000;74:4351–4360. doi: 10.1128/jvi.74.9.4351-4360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Telling G C, Haga T, Torchia M, Tremblay P, DeArmond S J, Prusiner S B. Genes Dev. 1996;10:1736–1750. doi: 10.1101/gad.10.14.1736. [DOI] [PubMed] [Google Scholar]
- 22.Chandler R L. Lancet. 1961;1:1378–1379. doi: 10.1016/s0140-6736(61)92008-6. [DOI] [PubMed] [Google Scholar]
- 23.Carlson G A, Goodman P A, Lovett M, Taylor B A, Marshall S T, Peterson-Torchia M, Westaway D, Prusiner S B. Mol Cell Biol. 1988;8:5528–5540. doi: 10.1128/mcb.8.12.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlson G A, Kingsbury D T, Goodman P A, Coleman S, Marshall S T, DeArmond S, Westaway D, Prusiner S B. Cell. 1986;46:503–511. doi: 10.1016/0092-8674(86)90875-5. [DOI] [PubMed] [Google Scholar]
- 25.Scott M, Foster D, Mirenda C, Serban D, Coufal F, Wälchli M, Torchia M, Groth D, Carlson G, DeArmond S J, et al. Cell. 1989;59:847–857. doi: 10.1016/0092-8674(89)90608-9. [DOI] [PubMed] [Google Scholar]
- 26.Supattapone S, Muramoto T, Legname G, Mehlhorn I, Cohen F E, DeArmond S J, Prusiner S B, Scott M R. J Virol. 2001;75:1408–1413. doi: 10.1128/JVI.75.3.1408-1413.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serban D, Taraboulos A, DeArmond S J, Prusiner S B. Neurology. 1990;40:110–117. doi: 10.1212/wnl.40.1.110. [DOI] [PubMed] [Google Scholar]
- 28.Scott M R, Köhler R, Foster D, Prusiner S B. Protein Sci. 1992;1:986–997. doi: 10.1002/pro.5560010804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen F E, Prusiner S B. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 30.Muramoto T, Kitamoto T, Tateishi J, Goto I. Am J Pathol. 1992;140:1411–1420. [PMC free article] [PubMed] [Google Scholar]
- 31.Taraboulos A, Jendroska K, Serban D, Yang S-L, DeArmond S J, Prusiner S B. Proc Natl Acad Sci USA. 1992;89:7620–7624. doi: 10.1073/pnas.89.16.7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westaway D, DeArmond S J, Cayetano-Canlas J, Groth D, Foster D, Yang S-L, Torchia M, Carlson G A, Prusiner S B. Cell. 1994;76:117–129. doi: 10.1016/0092-8674(94)90177-5. [DOI] [PubMed] [Google Scholar]
- 33.Telling G C, Scott M, Hsiao K K, Foster D, Yang S-L, Torchia M, Sidle K C L, Collinge J, DeArmond S J, Prusiner S B. Proc Natl Acad Sci USA. 1994;91:9936–9940. doi: 10.1073/pnas.91.21.9936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Telling G C, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen F E, DeArmond S J, Prusiner S B. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 35.Clousard C, Beaudry P, Elsen J M, Milan D, Dussaucy M, Bounneau C, Schelcher F, Chatelain J, Launay J-M, Laplanche J-L. J Gen Virol. 1995;76:2097–2101. doi: 10.1099/0022-1317-76-8-2097. [DOI] [PubMed] [Google Scholar]
- 36.Bossers A, Schreuder B E C, Muileman I H, Belt P B G M, Smits M A. J Gen Virol. 1996;77:2669–2673. doi: 10.1099/0022-1317-77-10-2669. [DOI] [PubMed] [Google Scholar]
- 37.Hunter N, Cairns D, Foster J D, Smith G, Goldmann W, Donnelly K. Nature (London) 1997;386:137. doi: 10.1038/386137a0. [DOI] [PubMed] [Google Scholar]
- 38.Belt P B G M, Muileman I H, Schreuder B E C, Ruijter J B, Gielkens A L J, Smits M A. J Gen Virol. 1995;76:509–517. doi: 10.1099/0022-1317-76-3-509. [DOI] [PubMed] [Google Scholar]
- 39.Baylis M, Houston F, Goldmann W, McClean A. International Symposium on Prion Diseases and Related Processes. Les Pensières, Veyrier-du-Lac, France: Fondation Marcel Mérieux; 2000. [Google Scholar]
- 40.Kitamoto T, Tateishi J. Philos Trans R Soc London B. 1994;343:391–398. doi: 10.1098/rstb.1994.0034. [DOI] [PubMed] [Google Scholar]
- 41.Shibuya S, Higuchi J, Shin R-W, Tateishi J, Kitamoto T. Lancet. 1998;351:419. doi: 10.1016/S0140-6736(05)78358-6. [DOI] [PubMed] [Google Scholar]
- 42.Parry H B. Heredity. 1962;17:75–105. doi: 10.1038/hdy.1962.4. [DOI] [PubMed] [Google Scholar]
- 43.Parry H B. In: Scrapie Disease in Sheep. Oppenheimer D R, editor. New York: Academic; 1983. pp. 31–59. [Google Scholar]
- 44.Baron T G, Madec J Y, Calavas D, Richard Y, Barillet F. Neurosci Lett. 2000;284:175–178. doi: 10.1016/s0304-3940(00)01047-8. [DOI] [PubMed] [Google Scholar]
- 45.Büeler H, Aguzzi A, Sailer A, Greiner R-A, Autenried P, Aguet M, Weissmann C. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 46.Prusiner S B, Groth D, Serban A, Koehler R, Foster D, Torchia M, Burton D, Yang S-L, DeArmond S J. Proc Natl Acad Sci USA. 1993;90:10608–10612. doi: 10.1073/pnas.90.22.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.