Abstract
With genomics well established in modern molecular biology, recent studies have sought to further the discipline by integrating complementary methodologies into a holistic depiction of the molecular mechanisms underpinning cell function. This genomic subdiscipline, loosely termed “systems biology,” presents the biology educator with both opportunities and obstacles: The benefit of exposing students to this cutting-edge scientific methodology is manifest, yet how does one convey the breadth and advantage of systems biology while still engaging the student? Here, I describe an active-learning approach to the presentation of systems biology. In graduate classes at the University of Michigan, Ann Arbor, I divided students into small groups and asked each group to interpret a sample data set (e.g., microarray data, two-hybrid data, homology-search results) describing a hypothetical signaling pathway. Mimicking realistic experimental results, each data set revealed a portion of this pathway; however, students were only able to reconstruct the full pathway by integrating all data sets, thereby exemplifying the utility in a systems biology approach. Student response to this cooperative exercise was extremely positive. In total, this approach provides an effective introduction to systems biology appropriate for students at both the undergraduate and graduate levels.
Keywords: systems biology, functional genomics, bioinformatics, active learning, cooperative learning, yeast, graduate, undergraduate
INTRODUCTION
As a discipline, biology is constantly evolving. Even in recent years, newly emerging technologies have fueled fundamental shifts in the nature and implementation of biological studies, with the concomitant promise of novel and useful data sets to follow. For example, advances in DNA sequencing technology were critical to the sequencing of entire genomes during the early 1990s. These large-scale efforts have resulted in complete DNA sequences for the human genome as well as for most major eukaryotic model organisms (International Human Genome Sequencing Consortium, 2004; Koonin, 1997; Olsen and Woese, 1997). To effectively utilize this resource of sequence data, researchers have developed innovative experimental approaches to define and assess gene function on a global, genomewide scale. At present, a variety of such approaches are in place. DNA microarray technologies, for one, provide a means by which we can measure expression levels for tens of thousands of genes in parallel (DeRisi et al., 1997; Schena et al., 1995). Genomewide collections of informative mutant alleles (e.g., precise gene deletions, transposable element insertions, regulated promoter constructs) are now available in many organisms (Coelho et al., 2000). Libraries of constructs facilitating RNA interference are being developed in metazoans such as Caenorhabditis elegans and Drosophila melanogaster (Boutros et al., 2004; Kamath et al., 2003). In addition, techniques are now in place by which proteome-scale applications of the yeast two-hybrid method and mass spectrometry are feasible (Kumar and Snyder, 2001).
With these technologies drawing a strong foothold in modern molecular biology, a growing subdiscipline of genomics is seeking to integrate large-scale data sets in order to achieve a more complete representation of the cell as a whole. This integrative approach to genomic biology is one branch of the subdiscipline termed “systems biology” (Ideker et al., 2001; Kitano, 2002). The rationale behind this integrative approach is simple: While no one data set can comprehensively define a cellular pathway or response, several complementary data sets may be integrated in order to reveal such pathways — or provide insight into a response or process unobtainable from the consideration of a single given data set. For example, DNA microarray analysis may be used to identify genes differentially regulated at the level of transcription; however, it cannot be used to identify genes regulated posttranscriptionally (e.g., by protein phosphorylation). In contrast, mass spectrometry is extremely useful as a means of identifying proteins modified by phosphorylation but is not applicable as a means of directly identifying differentially transcribed genes. Considered in union, however, data sets derived by microarray analysis and mass spectrometry may provide an indication of both transcriptional and posttranscriptional regulatory events and, therefore, a more comprehensive understanding of the genes and regulatory mechanisms driving a given cell response.
Systems biology is rapidly increasing in popularity. While it is difficult, if not impossible, to conclusively trace the lineage of this field, most studies in systems biology reference back to the work of Ideker et al. (2001), in which the authors implemented an integrative genomics strategy to define pathway components involved in galactose utilization in yeast. In the years since, the term systems biology has been used in over 110 published articles, and several institutes are now dedicated to this genomic discipline. The widespread application of systems biology is unlikely to diminish in the immediate future, with the increasing numbers of integrative genomic studies under way.
As promising research paradigms are developed, it is both our responsibility and privilege as educators to communicate this work. The advent of systems biology provides us with just such an opportunity; however, many obstacles do exist in accurately presenting this material to graduate and upper-level undergraduate students. In particular, it is difficult to convey effectively the unique advantage provided by systems biology: namely, that by integrating the data from multiple approaches, we can potentially generate novel findings beyond those that can be derived from the examination of any one individual data set. Furthermore, in order to appreciate such studies, students must possess a basic understanding of the approaches in use; typically, an introduction to these approaches is not provided in lower-division biology courses.
Recently, a number of educators have reported the benefits of “active learning” as applied to the teaching of undergraduate and graduate biology (Klionsky, 2002; Lord, 1994; Malacinski and Zell, 1996; Wyckoff, 2001). Briefly, active learning refers to the application of any teaching strategy in which students actively participate in academic exercises or projects rather than passively listen to an instructor's lecture (Baines et al., 2004; Harwood, 2003; Malacinski, 2003). Defined as such, active learning draws from a wide range of teaching practices; a comprehensive overview of these teaching strategies is provided in Paulson and Faust (2002). Active-learning techniques encompass individual exercises designed to foster effective listening practices, written exercises designed to promote student retention of lecture material, and group exercises in which students may learn from one another. In particular, the latter approach represents a subset of active learning termed “cooperative learning” (Sharan, 1994). In cooperative learning, students typically work in groups of three or more to complete fairly complex tasks, such as multistep exercises, research projects, and presentations (McKinney and Graham-Buxton, 1993). By performing these tasks in groups, the students may utilize the specific expertise of their respective classmates — a particular advantage in considering material that is interdisciplinary in nature.
Here, I present a cooperative-learning–based approach suitable for the introductory overview of systems biology at the graduate or upper-division undergraduate level. As an introduction to systems biology, I asked my graduate class at the University of Michigan, Ann Arbor, to reconstruct a cellular pathway using a modification of the “jigsaw” learning method. The class was broken into small groups, and each group was asked to interpret a different sample data set. Each group's independent findings were subsequently presented before the class as a whole to be ultimately integrated into a single comprehensive interpretation of the pathway. The message in this exercise became clear very quickly: The sum conclusions drawn by integrating the various data sets outweighed the individual conclusions drawn from any one data set in isolation. In total, this exercise proved very successful in actively engaging students as well as in disseminating relevant information. Furthermore, the approach provides a readily modifiable template for the instruction of many genomic disciplines at a variety of student levels.
CLASS FORMAT/STRUCTURE
This cooperative-learning approach was implemented in a graduate level class in Bioinformatics with a student enrollment of 16; however, this approach may be implemented easily in larger classes of 20–30 students without significant modification. Although students were of diverse educational backgrounds, most students possessed bachelor's degree–level experience in computer programming and an understanding of biology commensurate with that of a sophomore majoring in biology. Class sessions were 90 min in length, meeting twice a week. The systems biology module described herein encompassed one class session; note that this module may be expanded easily into a larger course component as discussed.
MODIFIED JIGSAW GROUP PROJECTS
As an introduction to the large-scale technologies encompassed in systems biology, I presented a 30-min lecture at the start of class describing relevant experimental methods in functional genomics and proteomics (e.g., DNA microarray analysis, yeast two-hybrid methods, co-immunoprecipitation/mass spectrometry–based approaches). This lecture encompassed background information regarding the methods, practical protocols as to the implementation of each approach, and enumeration of the potential limitations inherent in a given technique. Alternatively, much of this information may be presented as written notes, with the lecture serving principally as a brief review.
To instill within my students an appreciation of the potential benefit in a systems biology–based approach, I utilized a teaching strategy featuring a modified form of the jigsaw approach (Clarke, 1994). In this learning method, the class is divided into small groups; each group is asked to complete a discrete part of a total project. Once all groups have completed their assigned tasks, the findings from each group are presented before the class as a whole to be integrated into a finished project. By analogy, the individual group findings may be thought of as pieces in a jigsaw puzzle; the jigsaw puzzle can only be solved by properly integrating the various component findings.
In my class, after the introductory lecture, students were divided randomly into small groups of five or six. I presented each group with a different sample data set generated by one of the approaches discussed in my lecture; each data set related to a single hypothetical eukaryotic signaling pathway. I instructed each group to interpret the data regarding this pathway as completely as possible. The data sets were realistic and are discussed in greater detail below. As each group analyzed its respective data set, I circulated around the room to answer questions and provide assistance as necessary. After 40 min, one representative from each group was asked to present the group's findings; this student would draw or otherwise indicate the group's representation of the pathway on a whiteboard at the front of our classroom. During the final 20 min of class, the students and I integrated each group's findings into one comprehensive representation of the pathway. By integrating the results from all the data sets, we were able to construct a truer representation of the pathway than could be determined from any one data set alone, thereby exemplifying the utility of systems biology.
LARGE-SCALE DATA SETS
For the jigsaw exercise presented here, I devised three sample data sets of DNA microarray results, yeast two-hybrid analyses, and homology-search outputs from which students were able to construct a hypothetical eukaryotic signaling pathway. Specifically, the data sets describe a simplified “genome” of 26 genes (labeled genes A–Z). Despite the fact that the pathway itself was hypothetical (unrelated to bacterial genes), each data set was realistic in format and, in some cases, represented genuine experimental results. Each data set is described in greater detail below.
DNA Microarray Analysis
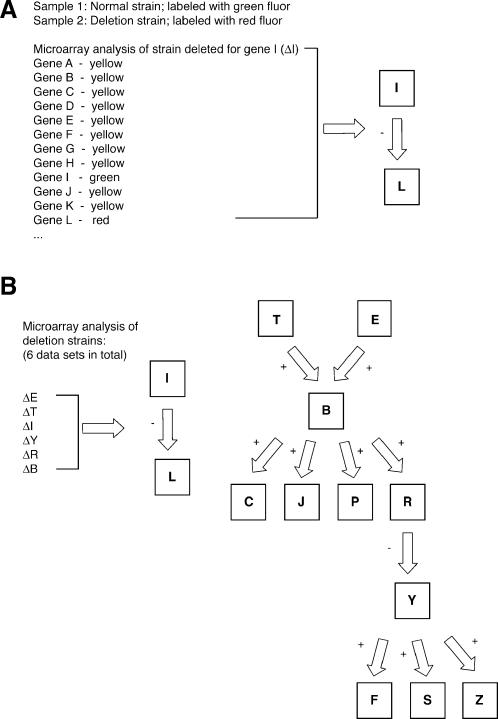
Students were presented with sample results from a comparative hybridization of RNA extracted from a strain deleted for a given gene and RNA extracted from a wild-type strain (Figure 1A). Students were told that complementary DNA (cDNA) prepared from the wild-type RNA had been labeled with a green fluor; cDNA prepared from the deletion strain RNA had been labeled with a red fluor. In the data set presented to the students, hybridization results for each gene (corresponding to a “spot” on the array) were listed with an accompanying color: red, green, or yellow. According to the indicated labeling scheme, a red spot indicates an increased concentration of deletion strain RNA and, therefore, a gene whose expression is induced in the deletion strain. Similarly, a green spot indicates a gene repressed in the deletion strain, while a yellow spot indicates a gene whose expression level is unchanged between the two strains. These microarray data are intended to reflect a classic approach in which polymerase chain reaction products are spotted onto a glass microscope slide (DeRisi et al., 1997).
Figure 1. Microarray-based data for student analysis. (A) Partial subset of microarray data describing comparative hybridization of RNA extracted from a strain deleted for gene I with RNA extracted from its wild-type counterpart. The data are presented as a simplified color representation to ease student analysis. The complete microarray data set is provided online as supplementary material. The conclusions drawn from the brief subset of microarray data shown here are indicated to the right. (B) The transcriptional network drawn from the complete microarray data sets. In total, six microarray data sets (indicated on the left) were used to define the signaling modules illustrated on the right. Multiple representations of this transcriptional network can be drawn from the data; these differing interpretations provide ground for further in-class discussion if desired.
By analyzing the relative expression pattern of each gene in the various deletion strains, students can formulate a transcriptional network of pathway components. The network drawn from the complete set of gene deletion/microarray analysis is shown in Figure 1B.
Yeast Two-Hybrid/Mass Spectrometric Analysis
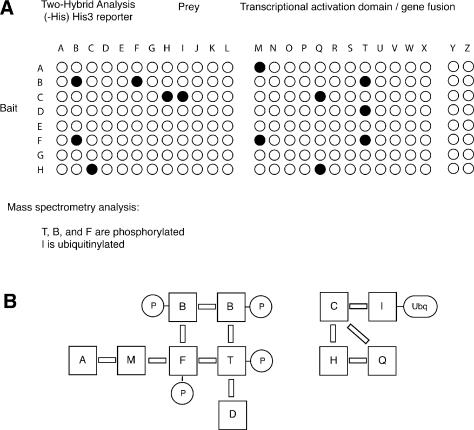
A second group of students was presented with sample results from a large-scale two-hybrid analysis of all pathway components in pairwise combination (Figure 2A). The two-hybrid system described here utilizes a simple HIS3 reporter, such that growth on medium lacking histidine may serve as an indicator of two-hybrid reporter activity and a corresponding protein–protein interaction. The hypothetical analysis presented to my students encompassed all possible pairwise combinations between the 26 genes included in this pathway; results were depicted as a grid of 676 assays, wherein colony growth indicates a protein–protein interaction.
Figure 2. Data sets describing protein–protein interaction and posttranslational modifications. (A) Partial subset of two-hybrid data describing comprehensive pairwise analysis of all pathway components. Data are presented in grid format to simplify student analysis. The complete data set (available online) also presents the results of simple mass spectrometric analysis in which proteins T, B, and F have been found to be phosphorylated, and protein I is ubiquitinylated. (B) Conclusions drawn from the data set in full; multiple interpretations are possible. Solid bars are used to represent protein–protein interactions. Abbreviations: P, phosphoryl group; Ubq, ubiquitin.
In addition, students were given a small set of data from a mass spectrometric analysis of pathway components identifying three phosphorylated proteins (T, B, and F) and one ubiquitinylated protein (I). Conclusions drawn from these two-hybrid and mass spectrometric analyses are indicated in Figure 2B.
Homology Searching
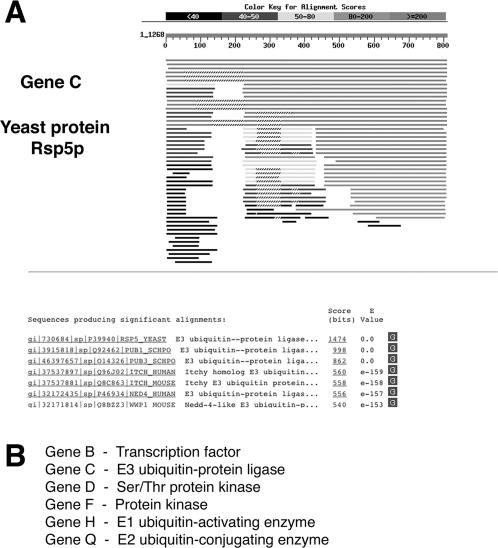
Finally, a third group of students was presented with output from a series of homology searches using the basic local alignment search tool (BLAST; Altschul et al., 1990). Specifically, BLAST protein (BLASTP) alignments were generated for five genes: B, C, F, H, and Q; the other genes were said to possess no orthologs. Complete amino acid sequences were available for each gene. A typical BLASTP alignment output is shown in Figure 3A. The alignments were created using actual yeast genes of a type identical to the hypothetical gene in the student data set. For example, in the pathway constructed here, gene C is intended to be an E3 ubiquitin-protein ligase; therefore, I downloaded the amino acid sequence of the yeast E3 ligase Rsp5p and used this sequence to query the nonredundant SwissProt database by BLAST. By interpreting the resulting sequence alignment output, students can identify the function of gene C as an E3 ubiquitin ligase. The conclusions drawn from these BLAST alignments are listed in Figure 3B.
Figure 3. Sample data set presenting the output format of a typical BLAST search. (A) Partial BLAST report for gene C (yeast protein Rsp5p). In total, BLAST search results were provided for six hypothetical genes in the sample pathway; again, full data sets are available as supplementary material. (B) The conclusions drawn from these sequence alignments are listed.
Data Integration
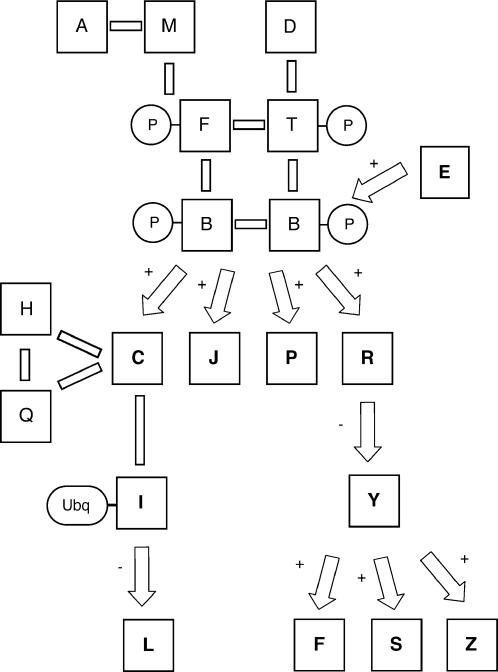
As indicated in Figures 1B, 2B, and 3B, each data set reveals useful and complementary insight into the structure of the pathway; however, no one data set is sufficient to define comprehensively all pathway components and connections. Necessarily, microarray analysis is effective only as a means of identifying genes regulated at the level of transcription, while the yeast two-hybrid method and mass spectrometric analysis identify protein–protein interactions and posttranslational modifications, respectively. Considered collectively with the homology-search results, the integrated data sets may be used to derive the pathway indicated in Figure 4.
Figure 4. Complete signaling pathway as determined by the integration of each individual genomic data set. Solid bars indicate protein–protein interactions; arrows indicate regulatory interactions.
STUDENT RESPONSE
The class lessons outlined here were specifically designed to address two objectives: 1) introduce students to the field of systems biology and 2) actively engage students in the learning process. In reality, both objectives are linked, as student participation is generally correlated with increased retention and understanding of class material (Wyckoff, 2001). As assessed through written evaluations as well as through individual discussions with students outside class, this cooperative-learning approach was very effective in communicating the benefits and rationale behind systems biology while actively encouraging student participation.
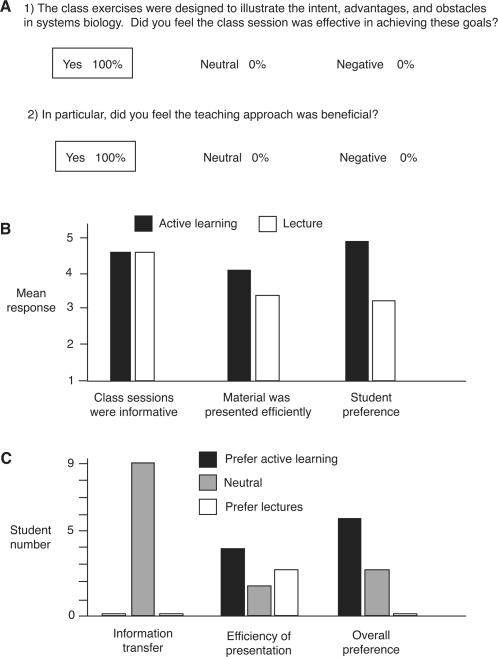
Of the 16 students enrolled in Bioinformatics 526, 13 were full-time students in the Bioinformatics Program at the University of Michigan. These 13 students were asked to assess critically the active-learning sessions after completion of this course. One student was absent during the systems biology classes; three students could not be reached for comment. The remaining nine students completed an anonymous survey assessing the utility and effectiveness of this teaching strategy. Results from this survey are presented in Figure 5.
Figure 5. Student response to the active-learning approach. (A) Surveyed graduate students were asked to respond to the two questions listed here. We received nine responses from 13 bioinformatics graduate students in total. As indicated, all student responses were uniformly positive in regard to the group-learning strategy. (B) Students were asked to assess specific aspects of the cooperative-learning and lecture-based strategies presented in Bioinformatics 526 using a numeric scale (5 = strongly agree and 1 = strongly disagree). The mean response is indicated. Students also used this numeric scale in response to the statements “I preferred the lecture classes” and “I preferred the active-learning approach”; mean responses to these statements are indicated as “student preference.” (C) In corollary to the mean numeric scores, a summary of individual student responses to the three questions/statements presented in Figure 5B is indicated. No single student in the class preferred a lecture format to the active-learning sessions.
As indicated, respondents were unanimously positive in regards to the active-learning–based approach, affirming that the group strategy was both effective and beneficial (Figure 5A). One student wrote that the active-learning approach “allowed an individual group to focus on its problem, brainstorm, collaborate but then come back and hear about the other groups, which allowed for depth and breadth.” Another student stated that an understanding of methods for data interpretation “can best be learned through a hands-on approach.” One student suggested that it would be helpful to distribute the data sets prior to class — a valid suggestion that can be easily incorporated into future sessions.
To enable quantitative assessment, students were asked to utilize a numeric scale in comparing active-learning and lecture-based classes in Bioinformatics 526 (Figures 5B, C). Students did not draw a distinction between the two teaching strategies in regards to information transfer, finding both approaches to be equally informative. Opinion was mildly split in regards to the efficiency of each approach; the class generally felt that the active-learning sessions were a more efficient use of time, although three students offered contrasting viewpoints. Opinion was decidedly uniform, however, when students were asked to select the teaching approach they preferred. Not one student preferred a lecture-based approach, and 75% of the class (six of nine respondents) strongly favored a cooperative-learning format. In interpreting these results, it should be noted that my class sessions were taught using active-learning approaches; the lecture-based sessions in Bioinformatics 526 were presented by other instructors, and the comparisons indicated here must be viewed in this light.
To assess retention of course material, each surveyed student was asked to define systems biology and list an advantage and disadvantage of this genomics approach. Each student was able to define systems biology at a level consistent with the expectations of this course. Nearly all respondents emphasized the integrative nature of the discipline and were equally accurate in identifying advantages (e.g., identification of whole-pathway effects and interpathway cross-talk) and disadvantages (e.g., expense, computational requirements, the preponderance of false-positive results in genomic data sets).
By other metrics as well, student response was extremely positive. Students undertook the jigsaw exercise with genuine enthusiasm; in fact, two groups asked for additional time to continue their respective analyses. After 30 min, student groups remained unanimously focused on their assigned data sets. I overheard two students commenting to each other: “It's amazing how time flies when you're really working.” Furthermore, each group was accurate in interpreting the data, suggesting that the students were indeed gaining valuable experience in analyzing realistic experimental results. By the end of class, several students were able to point out a minor error I had made in integrating the various data sets. It is possible that the success of this exercise results, in part, from the fact that the audience consisted of a small group of bioinformatics students; it will be interesting to consider the success of these class exercises when used on a larger group of undergraduate students.
To gain additional insight into the effectiveness of my cooperative-learning–based strategy, I met individually with several members from this class. From even a cursory conversation, it was evident that these students had grasped a basic understanding of systems biology. One student went so far as to discuss with me methods by which a systems biology–based approach might be incorporated into his own doctoral research. Another student commented upon the limitations of bioinformatics within systems biology and on the need for computational methods by which the process of data integration can be automated. This level of understanding far exceeded my expectations for an introductory overview of such a challenging field.
A FLEXIBLE TEMPLATE FOR ACTIVE LEARNING
The class paradigm presented here provides a readily modifiable template for both graduate and undergraduate courses. In undergraduate courses, the data sets selected for use may be pared down to emphasize simpler techniques. Plus, additional lecture time may be needed to communicate the necessary experimental methodology. Most importantly, the class sessions described above may be easily expanded; in fact, brief sessions may not be as effective in a larger class of undergraduate students. Even in a graduate-level class, several sessions may be spent presenting in detail a single experimental methodology, such as DNA microarray analysis. Subsequent to this discussion, students may consider a less-processed data set of raw expression values from microarray analysis — an exercise particularly well suited for a bioinformatics course. Similar class lessons may be designed for other experimental approaches (e.g., two-hybrid studies, co-immunoprecipitation/mass spectrometric studies, homology searching, comparative genomics) with a systems biology module taught in summation. In addition, a wide diversity of systems biology–based research papers may be addressed in cooperative groups in order to emphasize the breadth of this field. For example, the work of Davidson et al. (2002) may be well suited for these studies. Collectively, this template alone can be expanded to encompass the majority of a quarter-based course or approximately one-half of a semester course.
Obviously, the data sets used for this exercise need not be identical to the ones I have selected — they are included in this manuscript strictly as a suggested model. In fact, for future classes, I intend to utilize data sets related to a genuine eukaryotic pathway; I expect the added authenticity of the data will enhance student interest and enthusiasm (Campbell, 2003). Furthermore, additional data sets may be incorporated into this exercise; for example, large-scale data sets describing protein localization and abundance may complement the data sets described here.
CONCLUSIONS
In summary, I suggest that cooperative-learning strategies are ideally suited to the effective instruction of systems biology. Students, almost necessarily, enter discussions of systems biology with some background in a given research method or two; however, very few are well versed in all the technologies encompassed within a typical systems biology study. Individually, many, if not most, students would be unfamiliar with at least one of the techniques employed in such studies; however, collectively, the students in a group can complement each other's individual unfamiliarity, such that the group as a whole can function with an appropriate knowledge base. As an additional advantage, cooperative groups pose relatively little potential for student embarrassment, facilitating a more open and interactive classroom setting.
Acknowledgments
I would like to thank the University of Michigan Active Learning2 Community (supported by a National Science Foundation Director's Award for Distinguished Teaching Scholars to D.J. Klionsky) for guidance and support during the development of this exercise.
REFERENCES
- Altschul S.F, Gish W, Miller W, Meyers E.W, Lipman D.J. Basic local alignment search tool. J. Mol. Biol. (1990);215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baines A.T, McVey M, Rybarczyk B, Thompson J.T, Wilkins H.R. Mystery of the toxic flea dip: an interactive approach to teaching aerobic cellular respiration. Cell Biol. Educ. (2004);3:62–68. doi: 10.1187/cbe.03-06-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M, Kiger A.A, Armknecht S, Kerr K, Hild M, Koch B, Haas S.A, Consortium H.F, Paro R, Perrimon N. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. (2004);303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- Campbell A.M. Public access for teaching genomics, proteomics, and bioinformatics. Cell Biol. Educ. (2003);2:98–111. doi: 10.1187/cbe.03-02-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J. Pieces of the puzzle: the jigsaw method. In: Sharan S, editor. Handbook of Cooperative Learning Methods. Westport, CT: Greenwood Press; (1994). pp. 34–50. In: [Google Scholar]
- Coelho P.S, Kumar A, Snyder M. Genome-wide mutant collections: toolboxes for functional genomics. Curr. Opin. Microbiol. (2000);3:309–315. doi: 10.1016/s1369-5274(00)00095-3. [DOI] [PubMed] [Google Scholar]
- Davidson E.H, Rast J.P, Oliveri P, Ransick A, Calestani C, Yuh C.H, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown C.T, Livi C.B, Lee P.Y, Revilla R, Rust A.G, Pan Z, Schilstra M.J, Clarke P.J, Arnone M.I, Rowen L, Cameron R.A, McClay D.R, Hood L, Bolouri H. A genomic regulatory network for development. Science. (2002);295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- DeRisi J.L, Iyer V.R, Brown P.O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. (1997);278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Harwood W.S. Course enhancement: a road map for devising active-learning and inquiry-based science courses. Int. J. Dev. Biol. (2003);47:213–221. [PubMed] [Google Scholar]
- Ideker T, Thorsson V, Ranish J.A, Christmas R, Buhler J, Eng J.K, Bumgarner R, Goodlett D.R, Aebersold R, Hood L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. (2001);292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2004). Finishing the euchromatic sequence of the human genome. Nature. pp. 931–945. [DOI] [PubMed]
- Kamath R, Fraser A.G, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman D.P, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. (2003);421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kitano H. Systems biology: a brief overview. Science. (2002);295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- Klionsky D.J. Constructing knowledge in the lecture hall: a quiz-based group-learning approach to introductory biology. J. Coll. Sci. Teach. (2002);31:246–251. [Google Scholar]
- Koonin E.V. Big time for small genomes. Genome Res. (1997);7:418–421. doi: 10.1101/gr.7.5.418. [DOI] [PubMed] [Google Scholar]
- Kumar A, Snyder M. Emerging technologies in yeast genomics. Nat. Rev. Genet. (2001);2:302–312. doi: 10.1038/35066084. [DOI] [PubMed] [Google Scholar]
- Lord T.R. Using constructivism to enhance student learning in college biology. J. Coll. Sci. Teach. (1994);23:346–348. [Google Scholar]
- Malacinski G.M. Student-oriented learning: an inquiry-based developmental biology lecture course. Int. J. Dev. Biol. (2003). pp. 135–140. [PubMed]
- Malacinski G.M, Zell P.W. Learning molecular biology means more than memorizing the formula for tryptophan. J. Coll. Sci. Teach. (1996);25:198–202. [Google Scholar]
- McKinney K, Graham-Buxton M. The use of collaborative learning groups in the large class: is it possible? Teach. Sociol. (1993);21:403–408. [Google Scholar]
- Olsen G.J, Woese C.R. Archaeal genomics: an overview. Cell. (1997);89:991–994. doi: 10.1016/s0092-8674(00)80284-6. [DOI] [PubMed] [Google Scholar]
- Paulson D.R, Faust J.L. Active Learning for the College Classroom. (2002). http://www.calstatela.edu/dept/chem/chem2/Active/index.htm (accessed 30 November, 2004).
- Schena M, Shalon D, Davis R.W, Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. (1995);270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Sharan S. Handbook of Cooperative Learning Methods. Westport, CT: Greenwood Press; (1994). [Google Scholar]
- Wyckoff S. Changing the culture of undergraduate science teaching. J. Coll. Sci. Teach. (2001);29:409–414. [Google Scholar]