Abstract
Endothelium-derived nitric oxide (NO) is an important regulator of vascular function. NO is produced by endothelial NO synthase (eNOS) whose function is modulated, in part, by specific protein interactions. By coimmunoprecipitation experiments followed by MS analyses, we identified a human voltage-dependent anion/cation channel or porin as a binding partner of eNOS. The interaction between porin and eNOS was demonstrated by coimmunoprecipitation studies in nontransfected human endothelial cells and Cos-7 cells transiently transfected with eNOS and porin cDNAs. In vitro binding studies with glutathione S-transferase–porin indicated that porin binds directly to eNOS and that this interaction augmented eNOS activity. The calcium ionophore, A23187, and bradykinin, which are known to activate eNOS, markedly increased porin–eNOS interaction, suggesting a potential role of intracellular Ca2+ in mediating this interaction. Theses results indicate that the interaction between a voltage-dependent membrane channel and eNOS may be important for regulating eNOS activity.
Nitric oxide (NO) is a potent cell-signaling molecule that plays important and diverse roles in biological processes such as neurotransmission, inflammatory response, and vascular homeostasis (1, 2). Because of its important biological effects, NO production by NO synthases (NOS) is under complex and tight control. Three NOS isoforms have been identified to date, namely: neuronal NOS (nNOS or NOS1), inducible NOS (iNOS or NOS2), and endothelial NOS (eNOS or NOS3). nNOS and eNOS are constitutively expressed and activated in response to calcium-calmodulin signaling. In addition, each of the NOS isoforms is regulated at multiple levels, including transcription, and at posttranslational mechanisms. Among the NOS isoforms, the eNOS seems to possess some unique features. For example, eNOS has been shown to undergo myristoylation and palmitoylation, processes that direct eNOS to the caveolae (3, 4). Indeed, localization of eNOS within plasma membrane caveolae seems to be necessary for the efficient release of NO in response to external stimuli (5).
Recent studies indicate that protein–protein interactions are critical to the regulation of eNOS activity. For example, direct interaction of the oxygenase or reductase domains of eNOS with the scaffolding domain of caveolin-1 in endothelial cells or caveolin-3 in cardiac myocytes inhibits eNOS activity (9–11). Interaction of caveolin-1 with eNOS seems to be mutually exclusive, suggesting dynamic regulation of the enzyme by intracellular calcium levels. The carboxyl terminus of the bradykinin B2 receptor has been shown to interact with eNOS and inhibit its activity (12). In contrast, the heat shock protein 90, a molecular chaperone, serves as allosterical activator of eNOS (13).
To determine whether other proteins could be important in regulating eNOS function, we used a MS–protein-sequencing approach to identify proteins that copurify with eNOS from Brij 96 lysates of bovine endothelial cells. From these studies, we have found a voltage-dependent anion channel 1 (VDAC1 or porin) that interacts with eNOS and modulates NO production.
Experimental Procedures
Materials.
All standard culture reagents were obtained from JRH Biosciences (Lenexa, KS). Transfection reagent Fugene6 was obtained from Roche Molecular Biochemicals. eNOS mAb and pAb were purchased from Transduction Laboratories (Lexington, KY). The porin mAb 31HL was obtained from Calbiochem. Calcium ionophore, A23187, was obtained from Sigma. NOS assay kit was obtained from Cayman Biochemicals (Ann Arbor, MI). L-[3H]arginine was supplied by NEN Life Science.
Cell Culture.
Endothelial cells were harvested from human saphenous veins and bovine aortas as described (14, 15). The human endothelial cells were harvested by using type II collagenase (Worthington) and cultured in Medium 199/20 mM Hepes/50 μg/ml endothelial cell growth serum (Collaborative Research), 100 μg/ml heparin sulfate/5 mM L-glutamine (Life Technologies, Grand Island, NY), 5% FCS (HyClone), and an antibiotic mixture of penicillin (100 units/ml)/streptomycin (100 μg/ml)/Fungizone (1.25 μg/ml). The bovine endothelial cells were cultured at 37°C in a growth medium containing DMEM, supplemented with 5 mM L-glutamine (Life Technologies), 10% FCS (HyClone), and an antibiotic mixture of penicillin (100 units/ml), streptomycin (100 mg/ml), and fungizone (250 ng/ml). Relatively pure endothelial cell cultures were confirmed by Nomarski optical microscopy (Olympus IX70, ×40 objective) and by immunofluorescence staining with anti-factor VIII antibodies. All passages were performed with a disposable cell scraper (Costar), and only endothelial cells of fewer than six passages were used.
Immunoprecipitation and Immunoblotting.
After being washed three times with cold PBS, cells were lysed by scraping into 1% Brij 96 (polyoxyethylene 10-oleyl ether; Fluka) in 20 mM Hepes (pH 7.5)/150 mM NaCl/5 mM MgCl2 (HBSM) with 2 mM PMSF/20 μg/ml aprotinin/10 μg/ml leupeptin. After a 1-h extraction at 4°C with rocking, insoluble material was removed by centrifugation, and lysates were precleared for 1 h at 4°C with protein G-Sepharose (Amersham Pharmacia). Specific antibodies were added along with protein G-Sepharose, and immune complexes were collected overnight at 4°C. After being rinsed four times with lysis buffer, immune complexes were eluted by boiling in sample buffer, resolved by SDS/PAGE, and transferred to nitrocellulose. Blots were blocked with 5% nonfat milk in PBS with 0.1% Tween 20 (PBST). Blots were developed with diluted antibodies for eNOS (1:500) and porin (1:1,000), followed by horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse antiserum (diluted 1:8,000 and 1:10,000, respectively) and enhanced chemiluminescence (NEN Life Science) according to the manufacturer's instruction. Band intensities of the autoluminographs were quantitated by densitometry.
Purification of eNOS-Associated Proteins.
Bovine endothelial cells (≈5 × 108) were lysed in a total of 10 ml of 1% Brij 96 lysis buffer and precleared as described above. Anti-eNOS mAb was added at 5 μg/ml along with 150 μl (settled volume) of protein G-Sepharose, and eNOS complexes were collected overnight at 4°C. After extensive rinsing with lysis buffer, eNOS-associated proteins were eluted by incubation with SDS/PAGE sample buffer. The eluate was concentrated with a Microcon-10 microconcentrator (Amicon) and resolved by SDS/PAGE on an 11% minigel. eNOS proteins were stained with 0.1% Coomassie brilliant blue 250 in 40% methanol and 10% acetic acid and destained in 50% methanol. Specific bands presented in eNOS immunocomplex in comparison with immunocomplex obtained by irrelevant mouse antibody were excised, rinsed with 50% HPLC-grade acetonitrile, and stored at −20°C until analysis.
Protein Identification by Capillary Chromatography and MS.
Protein bands were excised, chopped into 1- to 2-mm pieces, and subjected to in-gel digestion with trypsin (16). Tryptic peptides were analyzed by microcapillary liquid chromatography (LC)-MS with automated switching to MS/MS mode for peptide fragmentation. In brief, peptides eluting from the capillary column were automatically fragmented by collision-induced dissociation in a triple quadrupole mass spectrometer (TSQ 7000, Finnigan-MAT, San Jose, CA), and the resulting sequence information was recorded in a tandem (MS/MS) or collision-induced dissociation mass spectrum. Collision-induced dissociation spectra were then computer-searched by using the SEQUEST algorithm (17) against the OWL nonredundant database as well as the EST database (www.ncbi.nlm.nih.gov/dbEST/index.html). Protein identification was unambiguous because multiple peptides from each protein were matched by the database searches.
Expression of GST–Porin Fusion Protein.
The porin cDNA fragment was amplified by PCR and inserted into a bacterial expression vector pGEX-2T (Amersham Pharmacia). Expression of GST–porin fusion proteins was induced by adding isopropyl-D-thiogalactopyranoside. The expressed GST–porin was purified through glutathione-Sepharose beads.
Binding of eNOS and Porin.
Equal amounts of purified baculovirus-expressed bovine eNOS, and either purified GST–porin or the negative control GST alone, were incubated in TBS (10 mM Tris⋅HCl, pH 7.5/150 mM NaCl) for 2 h at 4°C. GST-bind beads, washed in TBS with 0.1% BSA, were added to precipitate the porin, and the beads were then washed extensively in buffer containing 10 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 10 mM mercaptoethanol, 0.5% Tween 20, 0.1% BSA, and 100 mM imidazole. The washed beads were boiled in SDS sample buffer, and bound proteins were resolved by SDS/PAGE and then transferred to nitrocellulose filters. Filters were blocked in TBS containing 0.1% Triton X-100, 5% dry milk, and 0.5% BSA and then incubated with anti-eNOS antibody. Filter was incubated with horseradish peroxidase-conjugated antibody to mouse IgG, followed by detection by using a Renaissance chemiluminescence reagent Plus (Perkin–Elmer Life Sciences).
Transfection.
Plasmids (4 μg each) were transfected into COS-7 cells by using Lipofectamine Plus reagent (Life Technologies). After 48 h of transfection, cells were harvested. The cells were then washed twice with cold PBS and harvested in the extraction buffer (25 mM Tris, pH 7.4/1% Brij 96/1 μg/ml leupeptin/1 μg/ml aprotinin/1 μg/ml pepstatin/1 mM PMSF). After incubation on ice for 15 min, the extract was centrifuged at 14,000 rpm for 15 min to remove cell debris. The protein concentration in supernatant was determined by Pierce bicinchoninic acid protein assay.
eNOS Activity Assay.
eNOS activity was detected by measuring the conversion of L-[3H]arginine to L-[3H]citrulline at 37°C for 30 min with the eNOS assay kit (Calbiochem-Nova Biochem) as described (19). Unlabeled L-arginine was added to L-[3H]arginine (specific activity, 60 Ci/mmol) at a ratio of 3:1. Rat cerebellum extracts, containing elevated amounts of neuronal NOS, were used as positive controls, whereas samples incubated in the presence of the competitive NOS inhibitor, NG-nitro-L-arginine methyl ester (L-NAME) (1 mM), were used to determine nonspecific activity. Nonspecific activity accounted for 20–35% of total activity.
Statistical Analysis.
All data are given as the mean ± SEM. Data were analyzed by using paired and unpaired Student's t test and one-way ANOVA. A P value of <0.05 was taken as significant difference between data sets.
Results
Identification of Porin as an eNOS-Associated Protein.
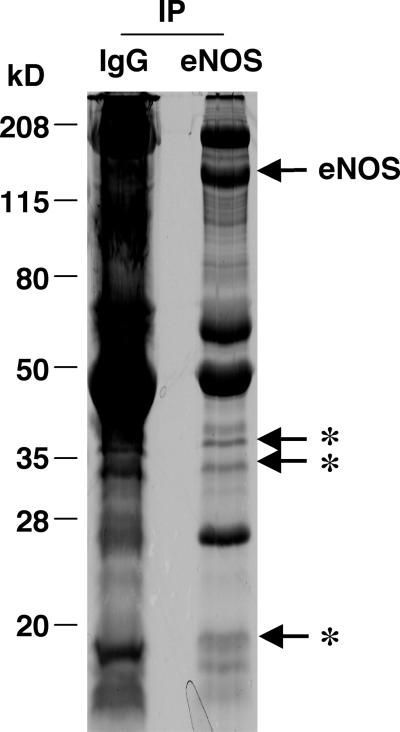
To identify eNOS-associated proteins, eNOS complexes were affinity-purified from a Brij 96 lysate of bovine endothelial cells precleared three times by using normal mouse IgG and fractionated by SDS/PAGE and stained with Coomassie blue. In addition to antibody-derived bands, three bands at 18, 34, and 38 kDa were uniquely present in eNOS complexes in comparison with control proteins resulting from the last preclearing step with mouse IgG (Fig. 1). The 18- and 34-kDa bands were excised and digested with trypsin. Tryptic peptides were separated by capillary chromatography and sequenced by ion-trap tandem MS. LC-MS/MS analysis of the 34-kDa band identified three peptides (VTQSNFAVGYK, LTFDSSFSPNTGK, and SENGLEFTSSGSANTETTK) that correspond to peptides deduced from voltage-dependent anion channel 1 (VDAC1 or porin). A few peptides obtained (from actin, keratin) were generally observed in sensitive mass spectrometric sequencing protocols. The three peptides (DGNGYISAAELR, VFDKDGNGYISAAELR, and EAFSLFDKDGDGTITTK) obtained from the 18-kDa band represented sequence fragments of calmodulin, which has already been shown to associate with eNOS (20).
Figure 1.
Immunoaffinity purification of eNOS complexes. Bovine endothelial cells (≈1 × 108) were lysed in 15 ml of moderately stringent lysis buffer Brij 96. The resultant lysates were precleared three times with mouse nonspecific IgG. Anti-eNOS mAb was then added at 5 μg/ml along with 150 μl (settled volume) of protein G-Sepharose, and eNOS complexes were collected overnight at 4°C. After extensive rinsing with lysis buffer, eNOS-associated proteins were eluted by incubation with SDS-polyacrylamide gel sample buffer, concentrated, and resolved by a SDS-11% polyacrylamide gel. Precipitated proteins were stained with 0.1% Coomassie brilliant blue 250. The immunoprecipitated proteins recovered by using nonspecific IgG were used as control. Asterisks represent some specific proteins present in eNOS immunoprecipitates. IP, immunoprecipitation.
Interaction of eNOS with Porin.
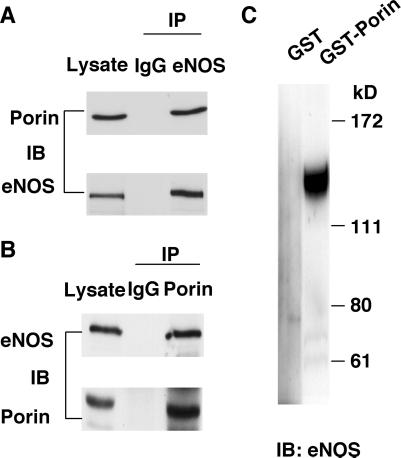
To confirm that eNOS associates with porin in human endothelial cells, we performed coimmunoprecipitations by using the polyclonal anti-eNOS antibody and monoclonal anti-porin antibody, from Brij 96 lysates of nontransfected human endothelial cells (14). Porin was found to coimmunoprecipitate with the eNOS antibody but was not detected in samples in which eNOS antibody was replaced with an irrelevant antibody (Fig. 2A). Similarly, eNOS was found in porin immunoprecipitate (Fig. 2B). These findings indicate that porin associates with eNOS in the intact human endothelial cell. Furthermore, it is estimated that about 6% of total cell porin interacted with eNOS because the amount of porin immunoprecipitated by anti-eNOS antibody from 500 μg of Brij96 cell lysates is similar to the amount of porin that was present in 30-μg cell lysates.
Figure 2.
Direct interaction between eNOS and porin. (A) Western blot analysis of eNOS immunoprecipitates. In each lane, 500 μg Brij 96 lysates from human endothelial cells were immunoprecipitated with mouse preimmune IgG or anti-eNOS polyclonal antibody. For the lysate lane, 30 μg Brij 96 lysates were used. Immunoprecipitated proteins were subjected to immunoblotting with anti-porin (Upper) and anti-eNOS (Lower) antibodies. (B) Western blot analysis of porin immunoprecipitates. Brij 96 lysates (500 μg) from human endothelial cells were immunoprecipitated with mouse preimmune IgG or anti-porin mAb, and 30 μg Brij 96 lysates were used in lysate lane. Immunoprecipitated proteins were subjected to immunoblotting with anti-eNOS (Upper) and anti-porin (Lower) antibodies. (C) Recombinant eNOS incubated with either GST or GST-porin, and proteins were precipitated with glutathione-Sepharose 4B beads. The precipitated proteins were separated through SDS/PAGE, and Western blot analysis was done with anti-eNOS monoclonal antibody. IP, immunoprecipitation; IB, immunoblotting.
A direct interaction between eNOS and porin was further comfirmed by an in vitro binding assay with purified eNOS and GST–porin fusion protein produced in bacteria. Recombinant eNOS was incubated with either GST or GST–porin that is bound to glutathione-Sepharose beads. Proteins that remained bound to the beads after extensive washing were then separated by SDS/PAGE and visualized with Western blot against eNOS. GST alone did not bind to eNOS, but a clear interaction occurred between eNOS and GST–porin (Fig. 2C).
Regulation of eNOS and Porin Interaction.
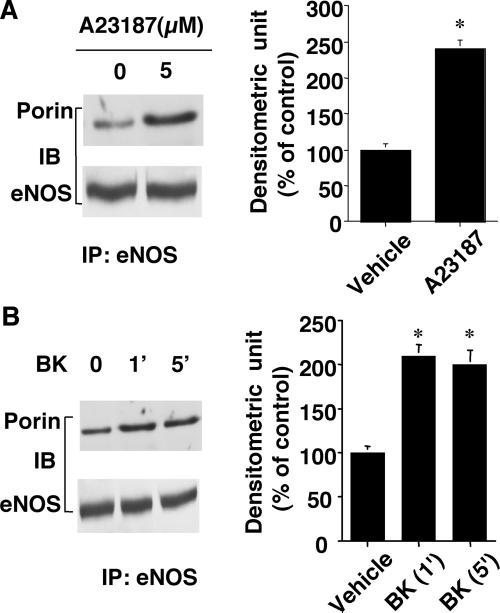
To investigate whether the interaction between eNOS and porin within endothelial cells is regulated by intracellular signals, we stimulated human endothelial cells with the calcium ionophore A23187 (5 μM), which stimulates NO release (21). Compared with untreated cells, endothelial cells stimulated with A23187 for 5 min showed increased amount of porin immunoprecipitated by eNOS antibody (Fig. 3A). Densitometric analysis revealed a 2.5-fold increase in porin bound to eNOS (P < 0.05, n = 3). Bradykinin (1 μM), which has been shown to promote rapid and transient Ca2+-dependent activation of the eNOS (22), also augmented the amount of porin bound to eNOS (Fig. 3B). Taken together, these data suggest that intracellular calcium levels may regulate the interaction between eNOS and porin.
Figure 3.
Calcium ionophore A23187 and bradykinin (BK) augment the binding of porin to eNOS. (A Left) Brij 96 lysates (500 μg) from human endothelial cells treated with either vehicle or A23187 (5 μM) for 2 min were immunoprecipitated with 2.5 μg anti-eNOS polyclonal antibody, and immunoprecipitated proteins were subjected to SDS/PAGE. Western blotting was performed by using anti-porin (Upper) and anti-eNOS (Lower) mAbs. (A Right) Densitometric analysis of porin present in eNOS immunoprecipitates. (B Left) Lysates (500 μg) from human endothelial cells treated with either vehicle or bradykinin (1 μM) were immunoprecipitated with anti-eNOS polyclonal antibody, and precipitated proteins were subjected to SDS/PAGE. Western blotting was performed by using anti-porin (Upper) and anti-eNOS (Lower) antibodies. (B Right) Densitometric analysis of porin in eNOS immunoprecipitates. IP, immunoprecipitation; IB, immunoblotting.
Functional Interaction Between eNOS and Porin.
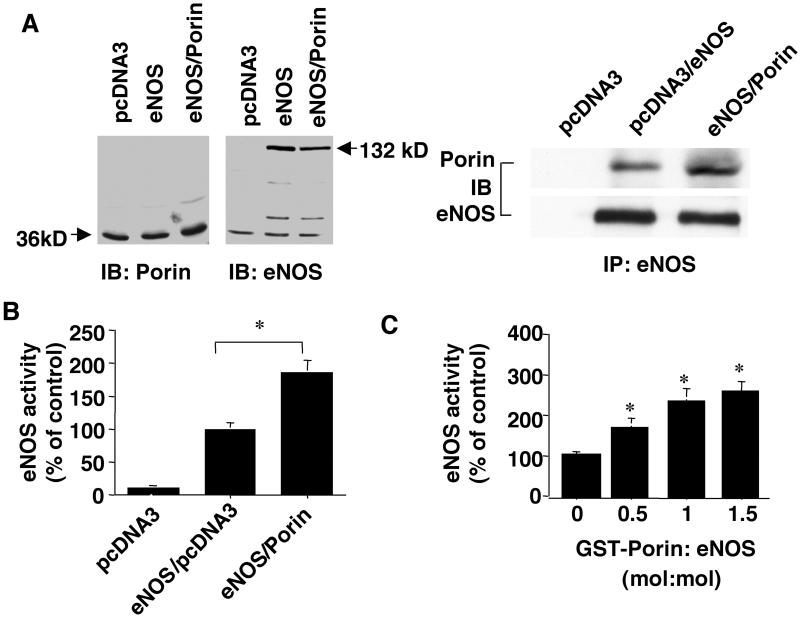
To investigate whether the interaction of eNOS with porin modulates eNOS activation, COS-7 cells were transfected with cDNAs encoding eNOS and porin. Approximately 48 h after transfection, immunocomplexes were examined and eNOS activity was assessed by measuring the conversion of radiolabeled L-arginine to L-citrulline. Under nontransfected conditions, COS-7 cells express porin, but not eNOS (Fig. 4A). Transfection of COS-7 cells with eNOS cDNA alone resulted in its expression and coimmunoprecipitation of eNOS with endogenous porin. Cotransfection of eNOS with porin increased the total amount of porin in total lysates and augmented the amount of porin bound to eNOS.
Figure 4.
Porin enhances eNOS activity in vitro and within cells. (A) Coimmunoprecipitation of eNOS and porin in cotransfected COS-7 cells. COS-7 cells were transfected with 8 μg of empty vector (pcDNA3.1), 4 μg of empty vector plus 4 μg of eNOS DNA, or 4 μg of eNOS DNA plus 4 μg of porin. After 48 h, cell lysates were collected for Western blot analysis with antibodies against eNOS (Left) or porin (Center). Additionally, 300 μg cell lysates were immunoprecipitated with eNOS antibody, and Western blotting was performed with anti-porin antibody (Right). (B) Coexpression of porin with eNOS in COS-7 cells potentiates eNOS activity. COS-7 cells were transfected with different plasmid combinations as described above. After 48 h, cell lysates were prepared for eNOS activity assay as assessed by L-NAME-inhibitable conversion of radiolabeled L-arginine to L-citrulline (*, P < 0.05 vs. eNOS plus empty vector). (C) Recombinant eNOS was incubated with increasing molar concentrations of GST–porin, and then the enzymatic activity of recombinant eNOS was examined by the L-NAME-inhibitable conversion of radiolabeled L-arginine to L-citrulline (*, P < 0.05 vs. GST + eNOS, n = 4). IP, immunoprecipitation; IB, immunoblotting.
Cotransfection of eNOS with porin markedly enhanced eNOS activity in cellular lysates compared with Cos-7 cells transfected with eNOS alone (Fig. 4B). To determine whether porin could directly potentiate eNOS activity, purified recombinant eNOS was incubated with GST alone or with increasing amounts of GST–porin. Addition of porin caused a significant increase in eNOS activity, which was blocked by the NOS inhibitor L-NAME (Fig. 4C). Taken together, these findings indicate a functional association between eNOS and porin.
Discussion
The regulation of eNOS occurs at multiple levels, including gene transcription, posttranscriptional and posttranslational mechanisms, and protein–protein interactions. Most of the previous studies have focused on transcriptional and posttranscriptional mechanisms of eNOS regulation. However, evidence that protein–protein interactions may also contribute to important dynamic regulations of eNOS is growing. To date, eNOS has been shown to associate directly with at least five proteins: heat shock protein 90 (13), calmodulin (20), Dynamin-2 (23), caveolin 1 and caveolin 3 (9–11), and the intracellular domains of certain G protein-coupled receptors (12). Interaction of eNOS with heat shock protein 90, calmodulin, and Dynamin-2 enhances eNOS activity, whereas association with caveolin 1 and 3 causes inhibition of activity. In this study, we have identified a voltage-dependent membrane channel called porin as an eNOS-associated protein. The interaction of eNOS and porin is direct and specific, enhanced by stimulators of eNOS, and ultimately leads to increased eNOS activity.
Porin is a 35-kDa membrane protein that forms a large channel allowing the transport of anions, cations, and various metabolites including substrates and nucleotides (24–27). Through a N-terminal leader sequence, porin localizes to the outer membrane of mitochondria and the plasma membrane (28). The presence of porin has also been detected in the specialized cell surface signal-transducing domains of plasmalemmal caveolae (29, 30). The function of porin in caveolae, however, is not known. Nevertheless, eNOS has also been shown to localize to the caveolae where it interacts with a variety of proteins such as the structural protein caveolin-1 (9–11), the bradykinin receptor B2 (12), and the L-arginine transporter CAT-1 (31). Therefore, it is quite likely that eNOS interacts with porin, especially in the caveolae.
Our findings indicate that eNOS interacts directly with porin. Furthermore, treatment of the human endothelial cells with either a calcium inophore or bradykinin markedly enhanced the association of porin with eNOS, suggesting a potential role of intracellular calcium in regulating this interaction. Indeed, porin contains multiple Ca2+-binding sites, which allows it to participate in intracellular Ca2+ signaling and transport (32). Thus, it is possible that the binding of calcium to porin produces a favorable conformational change that allows it to bind efficiently to eNOS. However, further studies are required to elucidate the precise mechanism of this interaction.
An intriguing possibility is that the interaction between eNOS and porin may play an important role in the trafficking, translocation, and enzymatic regulation of eNOS. For example, the interaction of these proteins may facilitate the localization of eNOS in caveolae. Indeed, localization of eNOS to the caveolae is critical for proper regulation of eNOS activity, because numerous signaling molecules such as G protein-coupled receptors, growth factor receptors, the plasma membrane Ca2+ pump, an inositol 1,4,5-trisphosphate-sensitive Ca2+ channel, and protein kinases such as protein kinase B/Akt are present in caveolae (33). The binding of porin to cholesterol (34, 35) may also play an important role in porin trafficking in caveolae because caveolae are especially rich in cholesterol (36). Cholesterol inhibits eNOS activity and it is possible that competition for porin within the caveolae by cholesterol and eNOS may be the mechanism by which cholesterol regulates the activity of eNOS.
Because porin is a voltage-dependent membrane channel, the interaction of eNOS with porin may also regulate the Ca2+ concentration in the vicinity of eNOS. Recent studies suggest that the subcellular Ca2+ concentration in the caveolae may modulate eNOS activity (18, 37). Porin, by providing Ca2+ transport into and out of mitochondrial and plasma membranes, may play an important role in intracellular Ca2+ signaling (32). Furthermore, the proximity of porin and eNOS may serve to direct substrate and cofactor delivery to eNOS, because the size of porin channels allows for transfer of larger substrates and nucleotides (24–27).
In summary, we have found that eNOS interacts directly with a voltage-dependent membrane channel. This interaction is specific and leads to a functional increase in eNOS activity. Whether the binding of eNOS to porin facilitates trafficking of eNOS to the caveolae and the precise determinants of this interaction, however, remain to be determined.
Acknowledgments
This work was supported by grants from the National Institutes of Health (HL52233, HL48743, and HL70274) and the American Heart Association (Bugher Foundation Award).
Abbreviations
- l-NAME
NG-nitro-l-arginine methyl ester
- NOS
nitric oxide synthase
- eNOS
endothelial NOS
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Moncada S, Palmer R M J, Higgs E. Pharmacol Rev. 1991;43:109–142. [PubMed] [Google Scholar]
- 2.Kubes P, Suzuki M, Granger D N. Proc Natl Acad Sci USA. 1991;88:4651–4655. doi: 10.1073/pnas.88.11.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia-Cardena G, Oh P, Liu J, Schnitzer J E, Sessa W C. Proc Natl Acad Sci USA. 1996;93:6448–6453. doi: 10.1073/pnas.93.13.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson L J, Michel T. Proc Natl Acad Sci USA. 1995;92:11776–11780. doi: 10.1073/pnas.92.25.11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blair A, Shaul P W, Yuhanna I S, Conrad P A, Smart E J. J Biol Chem. 1999;274:32512–32519. doi: 10.1074/jbc.274.45.32512. [DOI] [PubMed] [Google Scholar]
- 6.Fulton D, Gratton J P, McCabe T J, Fontana J, Fujio Y, Walsh K, Franke T F, Papapetropoulos A, Sessa W C. Nature (London) 1999;399:597–601. doi: 10.1038/21218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallis B, Corthals G L, Goodlett D R, Ueba H, Kim F, Presnell S R, Figeys D, Harrison D G, Berk B C, Aebersold R, Corson M A. J Biol Chem. 1999;274:30101–30108. doi: 10.1074/jbc.274.42.30101. [DOI] [PubMed] [Google Scholar]
- 8.Yeh D C, Duncan J A, Yamashita S, Michel T. J Biol Chem. 1999;274:33148–33154. doi: 10.1074/jbc.274.46.33148. [DOI] [PubMed] [Google Scholar]
- 9.Ju H, Zou R, Venema V J, Venema R C. J Biol Chem. 1997;272:18522–18525. doi: 10.1074/jbc.272.30.18522. [DOI] [PubMed] [Google Scholar]
- 10.Michel J B, Feron O, Sase K, Prabhakar P, Michel T. J Biol Chem. 1997;272:25907–25912. doi: 10.1074/jbc.272.41.25907. [DOI] [PubMed] [Google Scholar]
- 11.Michel J B, Feron O, Sacks D, Michel T. J Biol Chem. 1997;272:15583–15586. doi: 10.1074/jbc.272.25.15583. [DOI] [PubMed] [Google Scholar]
- 12.Ju H, Venema V J, Marrero M B, Venema R C. J Biol Chem. 1998;273:24025–24029. doi: 10.1074/jbc.273.37.24025. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Cardena G, Fan R, Shah V, Sorrentino R, Cirino G, Papapetropoulos A, Sessa W C. Nature (London) 1998;392:821–824. doi: 10.1038/33934. [DOI] [PubMed] [Google Scholar]
- 14.Liao J K, Shin W S, Lee W Y, Clark S L. J Biol Chem. 1995;270:3193–3124. doi: 10.1074/jbc.270.1.319. [DOI] [PubMed] [Google Scholar]
- 15.Liao J K, Zulueta J J, Feng-Sheng Y, Peng H, Cote C G, Hassoun P M. J Clin Invest. 1995;96:2661–2666. doi: 10.1172/JCI118332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 17.Eng J, McCormack A, Yates J R. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 18.Paltauf-Doburzynska J, Posch K, Paltauf G, Graier W F. J Physiol (London) 1998;513:369–379. doi: 10.1111/j.1469-7793.1998.369bb.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Endres M, Laufs U, Huang Z, Nakamura T, Huang P, Moskowitz M A, Liao J K. Proc Natl Acad Sci USA. 1998;95:8880–8885. doi: 10.1073/pnas.95.15.8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Busse R, Mulsch A. FEBS Lett. 1990;265:133–136. doi: 10.1016/0014-5793(90)80902-u. [DOI] [PubMed] [Google Scholar]
- 21.Sessa W C, Garcia-Cardena G, Liu J, Keh A, Pollock J S, Bradley J, Thiru S, Braverman I M, Desai K M. J Biol Chem. 1995;270:17641–17644. doi: 10.1074/jbc.270.30.17641. [DOI] [PubMed] [Google Scholar]
- 22.Prabhakar P, Thatte H S, Goetz R M, Cho M R, Golan D E, Michel T. J Biol Chem. 1998;273:27383–27388. doi: 10.1074/jbc.273.42.27383. [DOI] [PubMed] [Google Scholar]
- 23.Cao S, Yao J, McCabe T J, Yao Q, Katusic Z S, Sessa W C, Shah V. J Biol Chem. 2001;276:14249–14256. doi: 10.1074/jbc.M006258200. [DOI] [PubMed] [Google Scholar]
- 24.Benz R. Biochim Biophys Acta. 1994;1197:167–196. doi: 10.1016/0304-4157(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 25.Mannella C A. J Bioenerg Biomembr. 1997;29:525–531. doi: 10.1023/a:1022489832594. [DOI] [PubMed] [Google Scholar]
- 26.Colombini M. Curr Top Membr Transp. 1994;42:73–101. [Google Scholar]
- 27.Hodge T, Colombini M. J Membr Biol. 1997;157:271–279. doi: 10.1007/s002329900235. [DOI] [PubMed] [Google Scholar]
- 28.Buettner R, Papoutsoglou G, Scemes E, Spray D C, Dermietzel R. Proc Natl Acad Sci USA. 2000;97:3201–3206. doi: 10.1073/pnas.060242297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lisanti M P, Scherer P E, Vidugiriene J, Tang Z, Hermanowski-Vosatka A, Tu Y H, Cook R F, Sargiacomo M. J Cell Biol. 1994;126:111–126. doi: 10.1083/jcb.126.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bathori G, Parolini I, Tombola F, Szabo I, Messina A, Oliva M, De Pinto V, Lisanti M, Sargiacomo M, Zoratti M. J Biol Chem. 1999;274:29607–29612. doi: 10.1074/jbc.274.42.29607. [DOI] [PubMed] [Google Scholar]
- 31.McDonald K K, Zharikov S, Block E R, Kilberg M S. J Biol Chem. 1997;272:31213–31216. doi: 10.1074/jbc.272.50.31213. [DOI] [PubMed] [Google Scholar]
- 32.Gincel D, Zaid H, Shoshan-Barmatz V. Biochem J. 2001;358:147–155. doi: 10.1042/0264-6021:3580147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richard G W. Annu Rev Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- 34.Freitag H, Neupert W, Benz R. Eur J Biochem. 1982;123:629–636. doi: 10.1111/j.1432-1033.1982.tb06578.x. [DOI] [PubMed] [Google Scholar]
- 35.Popp B, Schmid A, Benz R. Biochemistry. 1995;34:3352–3361. doi: 10.1021/bi00010a026. [DOI] [PubMed] [Google Scholar]
- 36.Uittenbogaard A, Ying Y, Smart E J. J Biol Chem. 1998;273:6525–6232. doi: 10.1074/jbc.273.11.6525. [DOI] [PubMed] [Google Scholar]
- 37.Teubl M, Groschner K, Kohlwein S D, Mayer B, Schmidt K. J Biol Chem. 1999;274:29529–29535. doi: 10.1074/jbc.274.41.29529. [DOI] [PubMed] [Google Scholar]