Abstract
The tumor suppressor p53 is a labile protein whose level is known to be regulated by the Mdm-2–ubiquitin–proteasome degradation pathway. We have found another pathway for p53 proteasomal degradation regulated by NAD(P)H quinone oxidoreductase 1 (NQO1). Inhibition of NQO1 activity by dicoumarol induces p53 and p73 proteasomal degradation. A mutant p53 (p53[22,23]), which is resistant to Mdm-2-mediated degradation, was susceptible to dicoumarol-induced degradation. This finding indicates that the NQO1-regulated proteasomal p53 degradation is Mdm-2-independent. The tumor suppressor p14ARF and the viral oncogenes SV40 LT and adenovirus E1A that are known to stabilize p53 inhibited dicoumarol-induced p53 degradation. Unlike Mdm-2-mediated degradation, the NQO1-regulated p53 degradation pathway was not associated with accumulation of ubiquitin-conjugated p53. In vitro studies indicate that dicoumarol-induced p53 degradation was ubiquitin-independent and ATP-dependent. Inhibition of NQO1 activity in cells with a temperature-sensitive E1 ubiquitin-activating enzyme induced p53 degradation and inhibited apoptosis at the restrictive temperature without ubiquitination. Mdm-2 failed to induce p53 degradation under these conditions. Our results establish a Mdm-2- and ubiquitin-independent mechanism for proteasomal degradation of p53 that is regulated by NQO1. The lack of NQO1 activity that stabilizes a tumor suppressor such as p53 can explain why humans carrying a polymorphic inactive NQO1 are more susceptible to tumor development.
Wild-type p53 is a tumor suppressor that accumulates in response to DNA damage and other types of stress and induces either growth arrest (1–4) or apoptosis (5–8). p53 level is regulated by the rate of its degradation, which is known to be mediated by the Mdm-2–ubiquitin–proteasome degradation pathway (9, 10). Mdm-2 is an E3 ubiquitin ligase that binds to the N terminus of p53 and ubiquitinates it. Stabilization of p53 can be enhanced by disruption of p53–Mdm-2 interaction, which prevents Mdm-2 from ubiquitinating p53. This stabilization occurs by posttranslational modifications of p53 and Mdm-2 (3, 4) or by their interaction with other proteins including simian virus 40 (SV40) large T antigen (LT; refs. 11 and 12), hsp90 (13–15), and p14ARF (16, 17). Ubiquitin-dependent degradation is a multistep process that plays an important role in the degradation of abnormal and short-lived proteins. Ubiquitin is first activated by an E1 ubiquitin-activating enzyme, transferred to one of several E2 ubiquitin-conjugating enzymes, and then ligated to lysine residues of target proteins by specific E3 ubiquitin ligases. Further ubiquitination leads to the formation of polyubiquitinated proteins that are recognized by proteasomes for degradation (18, 19).
We have previously reported that NAD(P)H quinone oxidoreductase 1 (NQO1) regulates p53 stability and that inhibition of NQO1 activity by dicoumarol induces proteasomal degradation of p53 and inhibits p53-mediated apoptosis (20). We have also shown that wild-type NQO1, but not an inactive polymorphic NQO1, stabilizes wild-type p53, and that NQO1 does not inhibit Mdm-2-mediated p53 degradation (15). We have now analyzed the mechanism of p53 degradation in the NQO1-regulated pathway. Our results indicate that NQO1 regulates degradation of p53 in the proteasomes by a mechanism that is independent of both Mdm-2 and ubiquitin.
Materials and Methods
Cells, Cell Culture, and Compounds.
The cell lines used were: HCT116 human colon carcinoma, p53 null HCT116, M1-t-p53 mouse leukemia, and COS 1 (described in refs. 15 and 20), 293 human kidney, A31N and A31N-ts20, a BALB/c mouse cell line that has a temperature-sensitive E1 ubiquitin-activating enzyme (21). A31N and A31N-ts20 cells were grown in DMEM (GIBCO) supplemented with 10% FBS, 100 units/ml penicillin, and 100 mg/ml streptomycin and cultured at 32°C in a humidified incubator with 5.6% CO2. Dicoumarol, MG132, and thapsigargin were from Sigma, calpeptin was from Calbiochem, and tumor necrosis factor α was from PeproTech (Rocky Hill, NJ).
Plasmids.
The plasmids used were: pRc/CMV human p53, pCOC-mouse mdm-2 × 2, pRc/CMV-E6, and pEFIRES-HA-NQO1 (described in ref. 15), pEFIRES-HA-ubiquitin, pRc/CMV human mutant p53 with a Leu to Gln conversion at position 22, and Trp to Ser at position 23 (p53[22,23]) (22), pCDNA3-HA-p14ARF, pCDNA3-E1A, and pSUPER short interfering RNA (siRNA; ref. 23) into which NQO1 (Su-NQO1) or RFX (Su-RFX) were cloned.
Transfection.
Cells were seeded at 60% confluence in six-well plates 16 h before transient transfection with the desired plasmids. Transfections of 293 and HCT116 cells were performed by the calcium phosphate method. In HCT116, cells were treated with 10% glycerol for 30 s, 6 h after transfection. The amount of plasmid used in each experiment is indicated in the corresponding figure legend. Whenever needed, an empty vector was used to maintain a constant amount of total DNA in each transfection mixture. Transfections of A31N-ts20 BALB/c cells were performed with the lipofectamine reagent (Life Technologies, Grand Island, NY) according to the manufacturer's protocol. Cell extracts were generally prepared 24 h after transfection.
Immunoblot Analysis.
Cell extracts from 293 cells were prepared by lysis with 1% SDS/TBS (Tris-buffered saline: 0.05 M Tris, 0.15 M NaCl) followed by two cycles of vortexing and heating at 95°C for 5 min and then addition of an equal volume of 1.5% Triton X-100 in TBS. Cell extracts from all other cells were prepared by lysis with radioimmunoprecipitation assay buffer. Immunoblot analysis of human and mouse p53, Mdm-2, hemagglutinin (HA), β actin, β tubulin, Bcl-2, and IκBα was performed as described (15, 20), and p73 was detected with rabbit anti-human p73 antibody.
Apoptosis Assay.
Both adherent and floating cells of A31N-ts20 were harvested, washed with PBS, fixed in 70% ethanol, and stained with 25 μg/ml propidium iodide. Cell cycle analysis was performed with a fluorescence-activated cell sorter, and the percent of sub-G1 cells was determined.
In Vitro Protein Degradation Assay.
Reticulocyte lysate degradation mixture contained: 40 mM Tris⋅HCl, pH 7.5, 5 mM MgCl2, 2 mM DTT, 0.5 mM ATP, 10 mM phosphocreatine (Sigma), 1.6 mg/ml creatine phosphokinase (Roche Molecular Biochemicals), 200 μg/ml ubiquitin (Sigma), 50,000 cpm of in vitro reticulocyte lysate translated [35S]methionine-labeled p53 (5% of the reaction volume), and the reticulocyte lysate fraction indicated. Degradation mixture was incubated at 37°C for 90 and 180 min and then mixed with Laemmli sample buffer, heated at 95°C for 5 min, and loaded on a SDS/12.5% polyacrylamide gel. After electrophoresis, gels were dried at 70°C for 2 h, exposed to x-ray film (Fuji), and developed. ATP dependence was determined by omitting ATP, phosphocreatine, and creatine phosphokinase and adding 20 mM 2-deoxyglucose and 20 μg/ml hexokinase to the reaction mixture. Ubiquitin dependence was determined by omitting ubiquitin from the reaction mixture and by using fraction II of reticulocyte lysate depleted of ubiquitin as described (24).
Results
Inhibition of NQO1 Activity Induces Proteasomal Degradation of p53 and p73.
Inhibition of NQO1 activity by dicoumarol induces p53 proteasomal degradation (20). Dicoumarol also induces degradation of p73 (20) and this degradation was blocked by the proteasome inhibitor MG132 (Fig. 1A). Similar results were obtained by using the proteasome inhibitor lactacystin. Calcium-activated calpains can cause p53 degradation (25). However, elevation of intracellular Ca2+ by thapsigargin did not cause p53 degradation and the calpain-inhibitor calpeptin did not inhibit dicoumarol-induced p53 degradation (Fig. 1B), so that dicoumarol-induced p53 degradation is not mediated by calpain. Overexpression of wild-type NQO1, but not an inactive polymorphic NQO1, promotes p53 stability (15). In addition, transfection with a plasmid encoding NQO1-specific siRNA (Su-NQO1) reduced the level of p53, whereas the control RFX-specific siRNA (Su-RFX) did not (Fig. 1C). These results indicate a direct role of NQO1 in p53 and p73 stabilization and can explain the recent finding of reduced p53 and p73 levels in NQO1 knockout mice (26).
Figure 1.
Inhibition of NQO1 activity induces p53 and p73 proteasomal degradation. (A) HCT116 cells were incubated for 4 h without (−) or with 200 or 400 μM dicoumarol (Dic.) and without (−) or with (+) 100 μM MG 132. (B) M1-t-p53 cells were preincubated at 32°C without (−) or with (+) 10 nM thapsigargin or 40 μM calpeptin for 1 h followed by culture for an additional 5 h without (−) or with (+) 400 μM dicoumarol. (C) A31N- ts20 cells were transfected with 200 ng of pRc/CMV human wild-type p53 alone or cotransfected with 1.5 μg of pSUPER RFX (Su-RFX) or pSUPER NQO1 (Su-NQO1) with and without 1 μg of pEFIRES-HA-NQO1. The cells were cultured at 32°C. Levels of human p53 were determined 24 h after transfection.
Inhibition of NQO1 Activity Induces Mdm-2-Independent p53 Degradation That Is Blocked by p14ARF.
Ubiquitination is an important step in proteasomal protein degradation (18, 19). Mdm-2, an E3 ubiquitin ligase, ubiquitinates p53 and targets it to proteasomal degradation (9, 10), whereas SV40 LT stabilizes p53 (11). In contrast to p53, p73 is not targeted to degradation by Mdm-2 (27) and is not stabilized by LT (28). Unlike p53, dicoumarol-induced degradation of p73 was also not inhibited in COS-1 cells that overexpress LT (Fig. 2A). The p14ARF tumor suppressor, which inhibits the ability of Mdm-2 to target p53 for degradation (16, 17), also inhibited dicoumarol-induced p53 degradation (Fig. 2B). To study whether degradation of p53 by inhibition of NQO1 activity is mediated by Mdm-2, we used p53[22,23], a mutant p53 that cannot bind to Mdm-2 and is thus resistant to Mdm-2-mediated degradation (22). p53 null HCT116 cells were transfected either with wild-type p53 or p53[22,23] in combination with an Mdm-2-expressing plasmid and were then treated with dicoumarol. As expected, the degradation of wild-type p53 but not p53[22,23] was stimulated by Mdm-2 (Fig. 2C, lanes 1, 3, 4, and 6). However, both wild-type p53 and p53[22,23] were degraded in the presence of dicoumarol (Fig. 2C, lanes 2 and 5), indicating that dicoumarol induces p53 degradation in an Mdm-2-independent manner. The tumor suppressor p14ARF also protected p53[22,23] from dicoumarol-induced degradation (Fig. 2D, lanes 1–3), which indicates that p14ARF can also protect p53 against degradation by an Mdm-2-independent pathway. The adenovirus E1A oncogene that stabilizes p53 by inducing p14ARF (29) also protected p53[22,23] from dicoumarol-induced degradation (Fig. 2D, lanes 4–7).
Figure 2.
Inhibition of NQO1 activity induces Mdm-2-independent degradation of p53 that is inhibited by LT, p14ARF, and E1A. (A) COS 1 cells expressing SV40 LT were γ-irradiated at 6 Gy and incubated for 4 h without (−) or with dicoumarol (Dic.). (B) p53 null HCT116 cells were transfected with 150 ng of pRc/CMV human wild-type p53 alone or cotransfected with 3 μg of pCDNA3-HA-p14ARF. Twenty-four hours after transfection, cells were cultured for 5 h without (−) or with (+) 300 μM dicoumarol. (C) p53 null HCT116 cells were transfected with 150 ng of pRc/CMV human wild-type (wt) p53 or pRc/CMV human mutant p53[22,23] alone or cotransfected with 300 ng of pCOC-mdm2 X2. Twenty-four hours after transfection, cells were cultured for 5 h without (−) or with (+) 300 μM dicoumarol (Dic.). (D) p53 null HCT116 cells were transfected with 150 ng of pRc/CMV human mutant p53[22,23] alone or cotransfected with 3 μg of pCDNA3-HA-p14ARF or pCDNA3-E1A. Twenty-four hours after transfection, cells were cultured for 5 h without (−) or with (+) 300 μM dicoumarol (Dic.).
Inhibition of NQO1 Activity Induces Ubiquitin-Independent Degradation of p53.
Mdm-2-independent degradation of p53 can be induced by the human papilloma virus protein HPV-E6 (30), JNK (31), and adenovirus E4orf6 and E1B55K (32). In all these cases proteasomal degradation of p53 is ubiquitin-dependent and is associated with p53 polyubiquitination. We therefore determined whether dicoumarol-induced p53 degradation is also associated with p53 polyubiquitination. Human kidney 293 cells were cotransfected with wild-type p53 and HA-ubiquitin in the presence or absence of Mdm-2. The results show that transfection with Mdm-2 or treatment with dicoumarol induced a similar level of p53 degradation (Fig. 3, lanes 2 and 3). Whereas p53 degradation by Mdm-2 was associated with formation of high molecular weight p53-polyubiquitin conjugates, no such p53-polyubuiquitin conjugates were detected after treatment with dicoumarol (Fig. 3, lanes 2 and 3, overexposed). This result indicates that dicoumarol-induced proteasomal degradation of p53 is not mediated by formation of polyubiquitin-p53 conjugates. To determine whether ubiquitin is required for dicoumarol-induced p53 degradation, we used a cell-free degradation system. Labeled p53 was added to a reticulocyte lysate-based degradation mixture supplemented with ATP, an ATP-regenerating system, and ubiquitin and incubated at 37°C for 90 and 180 min. As expected, p53 was stable under these conditions (Fig. 4A). However, p53 was efficiently degraded when either HPV-E6 (Fig. 4B) or dicoumarol (Fig. 4C) were added. When ATP and an ATP-regenerating system were omitted from the degradation mixture, dicoumarol-induced p53 degradation was not observed (Fig. 4D). This result indicates that dicoumarol-induced p53 degradation requires ATP, which is typical of a proteasome-mediated degradation system (18, 19). Similar experiments were performed by using a reticulocyte lysate degradation mixture depleted of ubiquitin (fraction II) (24). As expected, the HPV-E6-mediated ubiquitin-dependent degradation of p53 was completely prevented under these conditions (Fig. 4E), whereas dicoumarol still efficiently induced p53 degradation (Fig. 4F).
Figure 3.
Inhibition of NQO1 activity induces degradation of p53 without polyubiquitin-p53 conjugates. Human kidney 293 cells were cotransfected with 200 ng of pRc/CMV human wild-type p53 and 1 μg of pEFIRES-HA-ubiquitin (HA ubiq.) without or with 200 ng of pCOC-mdm2 X2. Twenty-four hours after transfection, cells were cultured for 5 h without (−) or with (+) 300 μM dicoumarol (Dic.).
Figure 4.
Inhibition of NQO1 activity induces ATP-dependent and ubiquitin-independent degradation of p53 in vitro. In vitro translated 35S-labeled wild-type p53 was added to reticulocyte lysate degradation mixture and incubated at 37°C for 90 and 180 min. (A) Without any additions (None). (B) With in vitro translated HPV-E6. (C) With 200 μM dicoumarol (Dic.). (D) In a reticulocyte lysate degradation mixture lacking ATP and an ATP-regenerating system (−ATP) and treated with 200 μM dicoumarol. (E and F) In a fraction II of reticulocyte lysate degradation mixture depleted of ubiquitin (−ubiquitin) and treated with in vitro translated HPV-E6 (E) or with 200 μM dicoumarol (F). Degradation of radiolabeled p53 was followed by autoradiography as described in Materials and Methods.
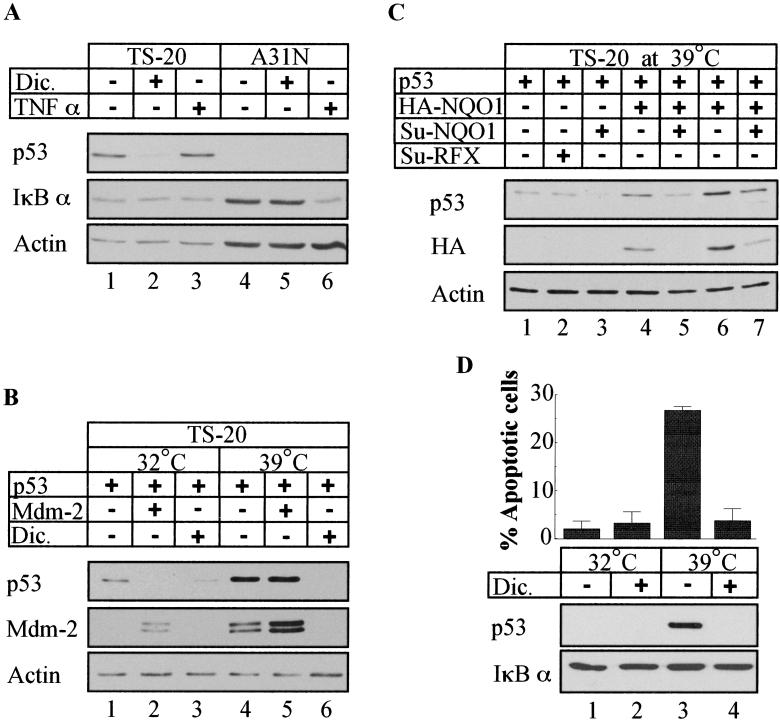
To examine further whether ubiquitin is required for dicoumarol-induced p53 degradation in cells, we used the A31N-ts20 BALB/c mouse cell line that has a temperature-sensitive E1 ubiquitin-activating enzyme (21). Only one E1 enzyme has been found in mammalian cells (33). Therefore, when these cells are cultured at the nonpermissive temperature (39°C), the E1 enzyme is inactivated and protein ubiquitination is repressed, facilitating accumulation of short-lived proteins such as p53 (21, 34). Addition of dicoumarol 24 h after cell transfer to 39°C induced p53 degradation (Fig. 5A, lanes 1 and 2). To rule out the possibility that some residual E1 activity still remains in A31N-ts20 cells at 39°C, we tested for tumor necrosis factor-induced ubiquitin-dependent IκBα degradation and for Mdm-2-mediated p53 degradation in this system. The results show that IκBα degradation was completely prevented in A31N-ts20 cells at 39°C (Fig. 5A, lanes 1 and 3), whereas it was effectively degraded at this temperature in the parental A31N cells (Fig. 5A, lanes 4 and 6). In addition, transfected Mdm-2 caused p53 degradation in A31N-ts20 cells at 32°C but not at 39°C (Fig. 5B, lanes 1, 2, 4, and 5), whereas dicoumarol induced p53 degradation at both temperatures (Fig. 5B, lanes 2 and 6). As expected, when A31N-ts20 cells were cultured at the permissive temperature (32°C), wild-type p53 level was low, but after transfer to 39°C, p53 accumulated (Fig. 5B lanes 1 and 4). Overexpression of HA-NQO1 further increased p53 level in A31N-ts20 cells at 39°C (Fig. 5C, lanes 1, 4, and 6), whereas expression of SuNQO1 reduced the level of p53 accumulated at 39°C (Fig. 5C, lanes 1, 3, 5, and 7). These results indicate that inhibition of NQO1 expression or activity induces ubiquitin-independent p53 degradation.
Figure 5.
Inhibition of NQO1 activity induces ubiquitin-independent proteasomal degradation of p53 and inhibits apoptosis. (A) A31N and A31N-ts20 mouse cells were preincubated for 24 h at the nonpermissive (39°C) and then cultured at 39°C without (−) or with (+) 200 μM dicoumarol (Dic.) for additional 5 h or without (−) or with (+) 2 ng/ml of tumor necrosis factor α for 10 min. (B) A31N-ts20 cells were transfected with 200 ng of pRc/CMV human wild-type p53 without or with 300 ng pCOC-mdm2 X2. Six hours after transfection cells were incubated at 32°C or 39°C for 24 h and then further cultured for 5 h without (−) or with (+) 200 μM dicoumarol (Dic.). (C) A31N-ts20 cells were transfected with 200 ng of pRc/CMV human wild-type p53 alone or cotransfected with 1.5 μg of pSUPER NQO1 (Su-NQO1) or pSUPER RFX (Su-RFX) with and without 600 ng (lanes 4 and 5) or 1 μg (lanes 6 and 7) of pEFIRES-HA-NQO1. Six hours after transfection, cells were incubated at 32°C or 39°C for 24 h. (D) A31N-ts20 cells were incubated at the permissive (32°C) or nonpermissive (39°C) temperature for 48 h without (−) or with (+) 50 μM dicoumarol (Dic.). Cell cycle analysis was performed by using fluorescence-activated cell sorter, and percent of sub-G1 (apoptotic) cells was determined.
Inhibition of NQO1 Activity Inhibits Apoptosis in A31N-ts20 Cells at the Temperature Without Ubiquitination.
Accumulation of p53 can lead to induction of apoptosis (5). We have previously shown that dicoumarol-induced p53 degradation in normal thymocytes and myeloid leukemic cells protects these cells against p53-mediated apoptosis (20). Culture of A31N-ts20 cells at 39°C for 48 h caused p53 accumulation and induced apoptosis (Fig. 5D and ref. 34). At this temperature, when ubiquitination is suppressed, dicoumarol induced p53 degradation (Fig. 5D, lanes 3 and 4) and completely inhibited apoptosis (Fig. 5D). These results indicate that the ability of dicoumarol to induce degradation of p53 can rescue cells from induction of apoptosis under conditions without ubiquitination.
Discussion
The tumor suppressor p53 is a short-lived protein that accumulates after different stress signals that decrease the ability of Mdm-2 to ubiquitinate p53. This accumulation can be achieved by posttranslational modifications of p53 and Mdm-2 or by their binding to other proteins (3, 4). By using an NQO1-specific inhibitor and overexpression of wild-type NQO1 or an inactive polymorphic NQO1, we have shown that NQO1 activity regulates p53 stability (15, 20). We now further show that reduction of endogenous NQO1 level by an NQO1-directed siRNA is sufficient to decrease p53 levels. p73 is also stabilized by NQO1 and our results can explain the recent demonstration of reduced p53 and p73 levels in NQO1 knockout mice (26).
DNA damage or oxidative stress can cause p53 stabilization because of inhibition of Mdm-2 activity. Overexpression of NQO1 showed even higher p53 stabilization under these conditions and did not inhibit p53 degradation induced by overexpression of Mdm-2 (15). These results suggested that p53 stabilization by NQO1 may not be mediated by inhibition of Mdm-2 activity. This possibility was now tested by using an Mdm-2-resistant mutant p53 (p53[22, 23]). Our results indicate that induction of p53 degradation by inhibition of NQO1 activity is independent of Mdm-2. By using in vitro degradation assays and cells defective in ubiquitination, we show that this NQO1-regulated and Mdm-2-independent p53 degradation is also ubiquitin-independent. Thus, the NQO1-regulated p53 degradation pathway requires neither the E3-ubiquitin ligase Mdm-2 nor any E3 or other proteins involved in the ubiquitination process. Our results show that p53 can undergo proteasomal degradation by two alternative pathways: one is ubiquitin-dependent and regulated by Mdm-2, whereas the other is ubiquitin-independent and regulated by NQO1. This finding implies that p53 stabilization is not solely dependent on inhibition of p53–Mdm-2 interaction but also requires NQO1 activity.
Both viral and cellular proteins can stabilize p53 by a protein-sequestration mechanism. The SV40 LT binds to p53 and prevents complex formation with Mdm-2 (11, 12), and the tumor suppressor p14ARF binds to Mdm-2 with the same outcome (16, 17). The adenovirus E1A oncogene stabilizes p53 by inducing p14ARF (29). Despite the lack of involvement of Mdm-2 in the NQO1-regulated p53 proteasomal degradation pathway, the ability of p14ARF to inhibit p53 degradation was maintained in this alternative pathway. Because p14ARF is known to function by interacting with Mdm-2, its role in the NQO1-regulated pathway was unexpected. However, at least under certain conditions, a direct interaction of wild-type p53 and p53[22,23] with p14ARF was observed (35), which may explain our data. Thus, p14ARF has a “double-lock” activity that inhibits p53 degradation by the Mdm-2-regulated and the NQO1-regulated pathways, which may ensure maximal p53 accumulation after different types of stress. The viral oncogenes SV40 LT and adenovirus E1A also seem to have a similar “double-lock” activity.
Cells accumulate p53 after exposure to oxidative stress, and NQO1 promotes p53 stabilization especially under oxidative stress (15). Accumulated p53 induces expression of the PIG3 (quinone oxidoreductase homolog) and FDXR genes and stabilizes the p66Shc protein, which increases intracellular production of reactive oxygen species and promotes apoptosis of damaged cells (36, 37). Reactive oxygen species increase NQO1 activity (38), which in turn further stabilizes p53. This sequence of events is in agreement with the proposed feed-forward loop for p53 stabilization by reactive oxygen species (36).
Our finding that NQO1 stabilizes p53 and p73 has clinical implications. NQO1-deficient mice have reduced p53 and p73 levels and develop myeloid hyperplasia (26) and are more susceptible to development of carcinogen-induced cancer (39). Humans carrying the polymorphic C609T NQO1 gene that encodes a biologically inactive enzyme that fails to stabilize p53 (15) are more susceptible to tumor development (40, 41). The absence of a NQO1-regulated pathway for stabilization of tumor suppressors such as p53 and p73 can thus lead to tumor formation.
Acknowledgments
We thank S. Budilovsky, Z. Bercovich, and R. Kama for their assistance, Dr. T. Unger for the pRc/CMV-human p53 plasmid, Dr. M. Oren for anti-p53 antibodies, the monoclonal anti-Mdm-2 antibody and the pCOC-mdm2 X2, pRc/CMV-E6, pCDNA3-HA-p14ARF, and pRc/CMV-human p53[22,23] plasmids, Dr. R. Agami for the p-Super plasmid, Dr. B. Vogelstein for the p53-null HCT116 cells, and Dr. H. Ozer for A31N and A31N-ts20 cells. This work was supported by the Israel Academy of Sciences and Humanities and by research grants from the Benoziyo Institute of Molecular Medicine, the Dolfi and Lola Ebner Center for Biomedical Research, the Otolaryngology Research Foundation, and Mrs. Bernice Gershenson.
Abbreviations
- HA
hemagglutinin
- NQO1
NAD(P)H:quinone oxidoreductase 1
- siRNA
short interfering RNA
- SV40
simian virus 40
- LT
large T antigen
References
- 1.Vogelstein B, Kinzler K W. Cell. 1992;70:523–526. doi: 10.1016/0092-8674(92)90421-8. [DOI] [PubMed] [Google Scholar]
- 2.Oren M. FASEB J. 1992;6:3169–3176. doi: 10.1096/fasebj.6.13.1397838. [DOI] [PubMed] [Google Scholar]
- 3.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 4.Vogelstein B, Lane D, Levine A J. Nature (London) 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 5.Yonish-Rouach E, Resnitzky D, Lotem J, Sachs L, Kimchi A, Oren M. Nature (London) 1991;352:345–347. doi: 10.1038/352345a0. [DOI] [PubMed] [Google Scholar]
- 6.Lotem J, Sachs L. Blood. 1993;82:1092–1096. [PubMed] [Google Scholar]
- 7.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. Nature (London) 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 8.Clarke A R, Purdie C A, Harrison D J, Morris R G, Bird C C, Hooper M L, Wyllie A H. Nature (London) 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 9.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 10.Kubbutat M H, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 11.Reihsaus E, Kohler M, Kraiss S, Oren M, Montenarh M. Oncogene. 1990;5:137–145. [PubMed] [Google Scholar]
- 12.Tiemann F, Zerrahn J, Deppert W. J Virol. 1995;69:6115–6121. doi: 10.1128/jvi.69.10.6115-6121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dasgupta G, Momand J. Exp Cell Res. 1997;237:29–37. doi: 10.1006/excr.1997.3766. [DOI] [PubMed] [Google Scholar]
- 14.Peng Y, Chen L, Li C, Lu W, Chen J. J Biol Chem. 2001;276:40583–40590. doi: 10.1074/jbc.M102817200. [DOI] [PubMed] [Google Scholar]
- 15.Asher G, Lotem J, Kama R, Sachs L, Shaul Y. Proc Natl Acad Sci USA. 2002;99:3099–3104. doi: 10.1073/pnas.052706799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Xiong Y, Yarbrough W G. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 17.Honda R, Yasuda H. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hershko A. Trends Biochem Sci. 1996;21:445–449. doi: 10.1016/s0968-0004(96)10054-2. [DOI] [PubMed] [Google Scholar]
- 19.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 20.Asher G, Lotem J, Cohen B, Sachs L, Shaul Y. Proc Natl Acad Sci USA. 2001;98:1188–1193. doi: 10.1073/pnas.021558898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chowdary D R, Dermody J J, Jha K K, Ozer H L. Mol Cell Biol. 1994;14:1997–2003. doi: 10.1128/mcb.14.3.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin J, Chen J, Elenbaas B, Levine A J. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 23.Brummelkamp T R, Bernards R, Agami R. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 24.Bercovich Z, Rosenberg-Hasson Y, Ciechanover A, Kahana C. J Biol Chem. 1989;264:15949–15952. [PubMed] [Google Scholar]
- 25.Pariat M, Carillo S, Molinari M, Salvat C, Debussche L, Bracco L, Milner J, Piechaczyk M. Mol Cell Biol. 1997;17:2806–2815. doi: 10.1128/mcb.17.5.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long D J, II, Gaikwad A, Multani A, Pathak S, Montgomery C A, Gonzalez F J, Jaiswal A K. Cancer Res. 2002;62:3030–3036. [PubMed] [Google Scholar]
- 27.Balint E, Bates S, Vousden K H. Oncogene. 1999;18:3923–3929. doi: 10.1038/sj.onc.1202781. [DOI] [PubMed] [Google Scholar]
- 28.Dobbelstein M, Roth J. J Gen Virol. 1998;79:3079–3083. doi: 10.1099/0022-1317-79-12-3079. [DOI] [PubMed] [Google Scholar]
- 29.de Stanchina E, McCurrach M E, Zindy F, Shieh S Y, Ferbeyre G, Samuelson A V, Prives C, Roussel M F, Sherr C J, Lowe S W. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huibregtse J M, Scheffner M, Howley P M. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuchs S Y, Adler V, Buschmann T, Yin Z, Wu X, Jones S N, Ronai Z. Genes Dev. 1998;12:2658–2663. doi: 10.1101/gad.12.17.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Querido E, Blanchette P, Yan Q, Kamura T, Morrison M, Boivin D, Kaelin W G, Conaway R C, Conaway J W, Branton P E. Genes Dev. 2001;15:3104–3117. doi: 10.1101/gad.926401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jennissen H P. Eur J Biochem. 1995;231:1–30. [PubMed] [Google Scholar]
- 34.Monney L, Otter I, Olivier R, Ozer H L, Haas A L, Omura S, Borner C. J Biol Chem. 1998;273:6121–6131. doi: 10.1074/jbc.273.11.6121. [DOI] [PubMed] [Google Scholar]
- 35.Kamijo T, Weber J D, Zambetti G, Zindy F, Roussel M F, Sherr C J. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang P M, Bunz F, Yu J, Rago C, Chan T A, Murphy M P, Kelso G F, Smith R A, Kinzler K W, Vogelstein B. Nat Med. 2001;7:1111–1117. doi: 10.1038/nm1001-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trinei M, Giorgio M, Cicalese A, Barozzi S, Ventura A, Migliaccio E, Milia E, Padura I M, Raker V A, Maccarana M, et al. Oncogene. 2002;21:3872–3878. doi: 10.1038/sj.onc.1205513. [DOI] [PubMed] [Google Scholar]
- 38.Bello R I, Gómez-Díaz C, Navarro F, Alcaín A J, Villalba J M. J Biol Chem. 2001;276:44379–44384. doi: 10.1074/jbc.M107168200. [DOI] [PubMed] [Google Scholar]
- 39.Long D J, II, Waikel R L, Wang X J, Roop D R, Jaiswal A K. J Natl Cancer Inst. 2001;93:1166–1170. doi: 10.1093/jnci/93.15.1166. [DOI] [PubMed] [Google Scholar]
- 40.Smith M T. Proc Natl Acad Sci USA. 1999;96:7624–7626. doi: 10.1073/pnas.96.14.7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith M T, Wang Y, Kane E, Rollinson S, Wiemels J L, Roman E, Roddam P, Cartwright R, Morgan G. Blood. 2001;97:1422–1426. doi: 10.1182/blood.v97.5.1422. [DOI] [PubMed] [Google Scholar]